Abstract
The transcription factor SREBP1c (sterol-regulatory-element-binding protein 1c) is highly expressed in adipose tissue and plays a central role in several aspects of adipocyte development including the induction of PPARγ (peroxisome-proliferator-activated receptor γ), the generation of an endogenous PPARγ ligand and the expression of several genes critical for lipid biosynthesis. Despite its significance, the regulation of SREBP1c expression during adipogenesis is not well characterized. We have noted that in several models of adipogenesis, SREBP1c expression closely mimics that of known C/EBPβ (CCAAT/enhancer-binding protein β) targets. Inhibition of C/EBP activity during adipogenesis by expressing either the dominant-negative C/EBPβ LIP (liver-enriched inhibitory protein) isoform, the co-repressor ETO (eight-twenty one/MTG8) or using siRNAs (small interfering RNAs) targeting either C/EBPβ or C/EBPδ significantly impaired early SREBP1c induction. Furthermore, ChIP (chromatin immunoprecipitation) assays identified specific sequences in the SREBP1c promoter to which C/EBPβ and C/EBPδ bind in intact cells, demonstrating that these factors may directly regulate SREBP1c expression. Using cells in which C/EBPα expression is inhibited using shRNA (short hairpin RNA) and ChIP assays we show that C/EBPα replaces C/EBPβ and C/EBPδ as a regulator of SREBP1c expression in maturing adipocytes. These results provide novel insight into the induction of SREBP1c expression during adipogenesis. Moreover, the findings of the present study identify an important additional mechanism via which the C/EBP transcription factors may control a network of gene expression regulating adipogenesis, lipogenesis and insulin sensitivity.
Keywords: adipocyte, adipogenesis, CCAAT/enhancer-binding protein (C/EBP), lipid, sterol-regulatory-element-binding protein 1c (SREBP1c), transcription
INTRODUCTION
Adipose tissue is a complex, highly active metabolic and endocrine organ [1]. While the adverse metabolic consequences of excessive adiposity are well known, pathologically decreased lipid accumulation or impaired adipogenesis in lipodystrophic subjects also has similar deleterious metabolic consequences including insulin resistance, dyslipidaemia and the associated risk of cardiovascular disease [2,3]. Thus optimal metabolic health probably requires the restraint of adipose tissue mass while still maintaining the capacity to respond accurately to substrate availability to increase adipose mass when required. A fuller understanding of the pathways regulating the formation and maintenance of adipocytes is therefore likely to inform therapeutic strategies for the treatment of syndromes involving either decreased or increased adiposity.
The development of new adipocytes from pluripotent precursors involves a complex and tightly orchestrated programme of gene expression [1]. Key early regulators of this process are C/EBPβ and C/EBPδ (CCAAT/enhancer-binding proteins β and δ). Acting alongside multiple co-activators, co-repressors and other transcription factors they play a central role in the subsequent induction of PPARγ (peroxisome-proliferator-activated receptor γ) and C/EBPα, transcription factors that have been dubbed the ‘master regulators of adipogenesis’ owing to their critical role in this process. The targets of these transcription factors include the promoters of many genes of the mature lipogenic and insulin-sensitive adipocyte such as aP2 (adipocyte protein 2), PEPCK (phosphoenolpyruvate carboxykinase), lipoprotein lipase, adiponectin and GLUT4 (glucose transporter 4) [4–6]. Thus the C/EBP family of transcription factors has a critical role in adipocyte development and lipid accumulation. Studies investigating the importance of C/EBPβ and C/EBPδ have demonstrated that loss of one or both of these factors can lead to decreased adipose mass in mice and decreased adipogenesis in cellular models [7,8]. In addition C/EBP factors may directly influence lipogenesis by controlling the early induction of the key lipogenic enzyme DGAT2 (diacylglycerol acyltransferase 2) during adipogenesis [9].
SREBP1c (sterol-regulatory-element-binding protein 1c) is another important pro-adipogenic transcription factor that can directly regulate the expression of several key genes of lipid metabolism [10]. Moreover, in adipocyte differentiation SREBP1c appears to contribute both to the expression of PPARγ and the production of an endogenous PPARγ ligand [11,12]. SREBP1c expression and activity, via cleavage and nuclear translocation, are acutely responsive to insulin [10,13]. In addition to controlling genes involved in lipid metabolism, the regulation of the expression of the adipokines leptin and adiponectin by insulin is also mediated by SREBP1c [14,15]. Thus in both developing and mature adipocytes, SREBP1c can potentially integrate information of nutritional and metabolic status to control new adipocyte formation, lipid metabolism, insulin sensitivity and, via adipokines, whole-body energy homoeostasis and appetite. Despite the importance of SREBP1c and its established role in adipocyte development, relatively little is known about the factors controlling its expression during adipogenesis, although LXRα (liver X receptor α) has been shown to be important for SREBP1c expression in these cells [16]. In the present study, we demonstrate that the C/EBP family of transcription factors also play a critical role in both the early induction of SREBP1c and the maintenance of its expression in maturing adipocytes.
EXPERIMENTAL
Preadipocyte isolation and cell culture
Human preadipocytes were grown from the stromovascular fraction of collagenase-digested abdominal subcutaneous adipose tissue as previously described [17]. At various times following induction of differentiation, cells were harvested, and RNA was extracted. 3T3-L1 preadipocytes were maintained and differentiated as described in [18]. 3T3-L1 preadipocyte cells stably expressing the LIP (liver-enriched inhibitory protein) isoform of C/EBPβ or ETO (eight-twenty one/MTG8) were as described previously [9]. Differentiating 3T3-L1 cells were assessed for lipid content by staining with Oil Red O as described in [18]. Murine E14 (where E is embryonic day) ES (embryonic stem) cells were cultured and differentiated as described in [19].
siRNA (small interference RNA) knockdown
Synthetic double-stranded siRNAs against C/EBPβ or C/EBPδ mRNAs were purchased from Ambion. 3T3-L1 preadipocytes were plated at a density 1 × 105 cells per well in 12-well plates the day before siRNA transfection. siRNA/liposome mixes containing 2 μg of Lipofectamine™ 2000 (Invitrogen) and 100 nM siRNA/well were incubated with cells for 6 h in the absence of serum. Medium was replaced with serum containing 3T3-L1 growth medium for 18 h prior to the induction of differentiation. 3T3-L1 preadipocytes stably expressing shRNA (short hairpin RNA) sequences targeting C/EBPα were generated using the pSiren retroQ kit (BD Biosciences) according to the manufacturer’s protocol. Retrovirus production, 3T3-L1 infection and selection were essentially as described in [18].
RNA isolation, cDNA synthesis and real-time PCR
Total RNA was extracted from cell cultures using an RNeasy kit (Qiagen). Adipose tissue was isolated from Cebpb-null mice or their wild-type littermates and RNA was isolated as previously described [20]. All procedures were approved by the UCHSC Animal Care and Use Committee. Primer Express, version 1.0 software (PerkinElmer Applied Biosystems), was used to design the probes and primers for real-time quantitative PCR to determine Srebp1c, Srebp1a, Dgat2, Pparg2, aP2, Glut4, Cebpb and Cebpd mRNA expression. A primer/probe mix to assay Cebpa was obtained from Applied Biosystems. RNA was reverse-transcribed using Moloney-murine-leukaemia virus reverse transcriptase and random hexamer primers (Promega). The resulting cDNA was used in 12 μl PCR mixtures, in which 300 nmol/l forward and reverse primers and, where applicable, 150 nmol/l fluorogenic probe were used in combination with ABI Taqman or SYBR® Green Master Mix (Applied Biosystems). Reactions were carried out in duplicate for each sample on an ABI 7900 sequence detection system (PerkinElmer Biosystems) according to the manufacturer’s instructions. The relative quantities of amplified cDNAs were analysed with the SDS software (Applied Biosystems) and target values were normalized to 18S rRNA (tissue samples) or cyclophilin A mRNA (cell culture samples).
ChIP (chromatin immunoprecipitation) assay
3T3-L1 preadipocytes in 35-mm wells were differentiated for various times as indicated. The DNA and protein were cross-linked in situ with 0.5 % formaldehyde at 37 °C for 5 min. Soluble chromatin was prepared using a ChIP assay kit (Upstate Biotechnology). The lysate was sonicated four times for 10 s at 4 °C. The lysates were precipitated with either 5 μl of antiC/EBPβ, anti-C/EBPδ or anti-C/EBPα antibody (Santa Cruz Biotechnology) overnight before Protein A–agarose beads were added. DNA was recovered by digesting with 10 μg/ml proteinase K at 45 °C for 30 min and purification using a QIAquick PCR purification kit (Qiagen). The presence of DNA sequences associated with the immunoprecipitated proteins was determined using specific primers amplifying DNA sequences, including the binding sites being assayed, and SYBR® Green Master Mix according to the manufacturer’s protocol. Values obtained from immunoprecipitated samples were normalized to those from input samples.
Western blot analysis
Protein samples were extracted by scraping in lysis buffer [50 mM Hepes (pH 7.4), 150 mM NaCl, 10 mM EDTA, 1 mM Na3VO4, 30 mM NaF, 10 mM Na4P2O7, 2.5 mM benzamidine, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 μg/ml antipain, 0.5 mM PMSF and 1 % Triton X-100] containing 1 % Nonidet P40, followed by sonication as described previously [18]. After centrifugation for 10 min at 13 000 g, samples of supernatant containing 30 μg of protein were denatured and analysed by Western blotting. All antibodies were from Santa Cruz Biotechnology.
Statistics
Statistical analyses were performed using Student’s t test or ANOVA for multiple comparisons.
RESULTS
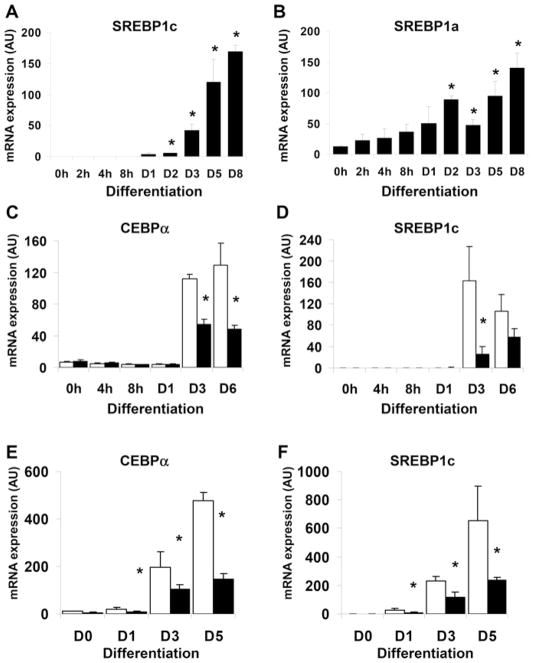
The gene Srebpf1 encodes the proteins SREBP1a and SREBP1c which have highly overlapping specificities for target sequences, but appear to differ in their transactivation capacities [21]. While both SREBP1a and SREBP1c are expressed in adipose tissue in vivo, SREBP1c is the more highly expressed in this tissue in both mice and humans [22]. However, initial studies suggested that only the SREBP1a isoform was expressed in differentiating 3T3-L1 cells [22], although other investigators have reported substantial expression of SREBP1c [12]. We therefore examined the expression of each isoform in differentiating 3T3-L1 preadipocytes. As shown in Figure 1(A), SREBP1c mRNA expression was almost undetectable in preadipocytes, but dramatically induced during adipogenesis and abundant in 3T3-L1 adipocytes. In contrast SREBP1a was clearly detectable in preadipocytes and, while also induced further during differentiation, did not show the dramatic increases observed for SREBP1c (Figure 1B). These assays do not permit quantitative comparison of SREBP1a versus SREBP1c levels, however, they do show that SREBP1c is highly induced and abundant during adpogenesis, making 3T3-L1 cells suitable to study the transcriptional regulation of SREBP1c. When searching for candidate pathways that might regulate SREBP1c we noted that the time course of its induction was similar to that observed for known C/EBPβ targets such as C/EBPα, 11βHSD1 (11-β-hydroxysteroid dehyrogenase type 1) and DGAT2 [9,23,24]. In addition we found that SREBP1c and the C/EBPβ target DGAT2 were induced with similar time courses in both murine ES cells undergoing adipogenic differentiation (Supplementary Figures S1A and S1B at http://www.BiochemJ.org/bj/425/bj4250215add.htm) and differentiating isolated human preadipocytes (Supplementary Figures S1C and S1D).
Figure 1. Inhibtion of C/EBPβ activity by constitutive LIP or ETO expression inhibits the induction of SREBP1c during adipogenesis.
3T3-L1 preadipocytes were induced to differentiate for various times up to 8 days (D8), RNA was isolated and the expression of SREBP1c (A) or SREBP1a (B) was determined by real-time PCR. Results shown are normalized to cyclophilin A, and are means ± S.E.M. (n = 4). (C) 3T3-L1 preadipocytes were stably transfected with LIP (solid bars) or empty vector (open bars), and confluent cells were induced to differentiate for the times shown. RNA was isolated and the expression of C/EBPα mRNA determined by real-time PCR. (D) Identical samples were also assayed for SREBP1c mRNA. (E) C/EBPα mRNA expression was determined in differentiating 3T3-L1 preadipocytes stably transfected with empty vector (open bars) or ETO (solid bars). (F) SREBP1c mRNA expression was also determined in the same samples. All results shown are normalized to cyclophilin, and are means ± S.E.M. (n = 4). *P < 0.05, compared with mock-transfected cells at the same time point. AU, arbitrary units.
To further investigate whether C/EBPβ might regulate SREBP1c expression we examined its expression in cells over-expressing the inhibitory LIP isoform of C/EBPβ and cells constitutively expressing the C/EBPβ inhibitor ETO [9]. As shown in Figures 1(C) and 1(D) respectively, LIP expression inhibited not only C/EBPα expression but also the induction of SREBP1c mRNA. Likewise, inhibition of C/EBP activity with ETO impaired the induction of C/EBPα (Figure 1E) and SREBP1c expression (Figure 1F) to a similar degree. Taken together these results support a role for C/EBPβ in the regulation of SREBP1c during adipogenesis.
LIP is known to inhibit multiple C/EBP isoforms, whereas the specificity of ETO is not fully defined. Therefore we next used specific siRNA oligonucleotides to knockdown individual C/EBP isoforms to examine their contribution to the regulation of SREBP1c. Cells were transfected with siRNAs targeting C/EBPβ or C/EBPδ or a mixture of both prior to induction of differentiation. As shown in Figure 2(A), the increase in C/EBPβ expression following the induction of differentiation was inhibited by transfection with C/EBPβ siRNA, but not C/EBPδ siRNA. A similar degree of inhibition was observed when C/EBPβ siRNA was co-transfected with C/EBPδ siRNA. Likewise C/EBPδ siRNA significantly impaired the induction of C/EBPδ expression, but did not prevent C/EBPβ induction (Figure 2B). C/EBPδ siRNA was similarly effective when co-transfected with C/EBPβ siRNA, substantially inhibiting C/EBPδ expression.
Figure 2. siRNA knockdown of C/EBP β and/or C/EBPδ activity inhibits SREBP1c induction during adipogenesis.
(A) 3T3-L1 preadipocytes were transfected with control siRNA (open bars) or siRNA targeting C/EBPβ (light grey bars), C/EBPδ (dark grey bars) or a mixture of siRNA targeting both C/EBPβ and C/EBPδ (solid bars), and were subsequently induced to differentiate for various times. C/EBPβ mRNA expression was assayed by real-time PCR. (B) C/EBPδ mRNA expression was assayed in the same samples. In both cases values are means ± S.E.M. (n = 4). *P < 0.05, compared with control siRNA transfected cells at the same time point. (C) Control, C/EBPβ (siCβ), C/EBPδ (siCδ) and C/EBPβ+C/EBPδ (siCβ+δ) siRNA-transfected cells were also assayed for C/EBPβ [LIP/LAP (liver-enriched activating protein)] and C/EBPδ protein expression by Western blot analysis. (D) Lipid accumulation was assessed by Oil Red O staining in control, C/EBPβ, C/EBPδ and C/EBPβ+C/EBPδ siRNA-transfected cells following differentiation for 8 days. (E–H) Control (open bars), C/EBPβ (light grey bars), C/EBPδ (dark grey bars) and C/EBPβ+C/EBPδ (solid bars) siRNA-transfected cells were also differentiated for various times, RNA was isolated and assayed for the expression of C/EBPα (E), DGAT2 (F), SREBP1c (G) and SREBP1a (H) by real-time PCR. Results shown are normalized to cyclophilin, and are means ± S.E.M. (n = 4). *P < 0.05, compared with control siRNA-transfected cells at the same time point.
Lysates from identically transfected cells differentiated for various times were analysed by Western blotting to determine either C/EBPβ or C/EBPδ protein expression (Figure 2C). Knockdown of C/EBPβ or C/EBPδ with siRNA resulted in an almost complete loss of the induction of C/EBPβ (top panel) or C/EBPδ (bottom panel) protein respectively, consistent with the effects observed at the RNA level. Again, co-transfection of cells with siRNA targeting both C/EBPβ and C/EBPδ led to almost complete inhibition of the expression of both of these proteins. As we have previously reported [9], knockdown of C/EBPβ led to an almost complete inhibition of lipid accumulation in differentiating 3T3-L1 cells as assessed by Oil Red O staining (Figure 2D). Knockdown of C/EBPδ expression had a less marked effect on lipid accumulation in these cells. The knockdown of both C/EBPβ and C/EBPδ together led to the most dramatic inhibition of lipid accumulation.
We next examined the effect of C/EBPβ and C/EBPδ knockdown on a well-characterized C/EBP target gene, C/EBPα. As predicted by previous cellular and in vivo studies, C/EBPα expression was most strongly affected by inhibition of C/EBPβ, although C/EBPδ knockdown also substantially inhibited its induction during adipogenesis (Figure 2E). Knockdown of both C/EBPβ and C/EBPδ in the same cells further impaired the induction of C/EBPα mRNA. Having shown that C/EBPβ regulates DGAT2 induction during adipogenesis in a previous study [9], we also examined whether C/EBPδ could regulate DGAT2 expression. Indeed, knockdown of C/EBPδ significantly inhibited DGAT2, which was reduced to 45 % and 50 % of the levels seen in control cells at day 3 and day 5 of differentiation respectively (Figure 2F). In addition, the combined knockdown of both C/EBPβ and C/EBPδ caused an even greater reduction in DGAT2 expression. These results suggest that, in addition to C/EBPβ, C/EBPδ makes an important contribution to the regulation of DGAT2 expression during early adipogenesis.
Analysis of SREBP1c expression in these samples revealed that the regulation of SREBP1c was very similar to that of C/EBPα and DGAT2. Knockdown of C/EBPβ alone led to inhibition of SREBP1c expression to approx. 30 % and 50 % of the levels seen in control cells at day 3 and day 5 respectively, following differentiation of these cells (Figure 2G). Inhibition of C/EBPδ using siRNA gave very similar results, suggesting that both C/EBPβ and C/EBPδ are important for the induction of SREBP1c during adipogenesis. Moreover, siRNA knockdown of both C/EBPβ and C/EBPδ in the same cells reduced expression of SREBP1c at day 3 and day 5 further to approx. 20 % and 35 %of those in control cells respectively.
To determine the specificity of these effects, the effect of C/EBPβ and C/EBPδ inhibition on SREBP1a expression was also determined. As shown in Figure 2(H), the inhibition of C/EBPβ, C/EBPδ or combined inhibition of both factors by siRNA did not significantly affect SREBP1a expression at any of the time points tested. These results show that, as SREBP1a is normally induced in these cells, C/EBPβ/δ knockdown does not inhibit all changes associated with adipogenesis, but rather is specific to genes downstream of these factors. These data also indicate that the two isoforms of SREBP are regulated by different factors during adipogenesis.
To determine whether our observations in 3T3-L1 cells may extend to in vivo adipocyte development, we examined gene expression in white adipose tissue isolated from Cebpb-knockout mice. Cebpb mRNA was undetectable in these samples (Figure 3A), while the expression of Cebpa was significantly reduced (Figure 3B), as previously reported [9]. Consistent with our results from cultured cells, Srebp1c expression was significantly decreased in the white adipose tissue of these mice (Figure 3C). This suggests that C/EBPβ is also involved in the expression of SREBP1c in adipocytes in vivo. In addition, the expression of Srebp1a was not affected in mice lacking Cebpb (Figure 3D), providing in vivo support for our previous findings in cultured preadipocytes that SREBP1c, but not SREBP1a, is selectively regulated by C/EBP factors. We next sought to determine whether C/EBPβ and C/EBPδ could directly regulate SREBP1c through binding to its promoter. Examination of the putative promoter of SREBP1c revealed several potential C/EBP consensus-binding sites within the 4 kb upstream of the transcriptional start site. To assess binding to these putative sites we performed ChIP analysis, immunoprecipitating C/EBPβ or C/EBPδ, and using real-time PCR to quantify binding to specific DNA sequences. Of nine putative binding sites identified, three showed significant binding of either C/EBPβ or C/EBPδ, which was responsive to the induction of adipogenesis, whereas the other six did not (results not shown). The three responsive sites were designated site 1, site 2 and site 3, and are schematically represented in Figure 4(A). As shown in Figures 4(B)–4(D), C/EBPβ binding to all three sites was increased during early differentiation, as C/EBPβ expression increases. Similarly these sites also bound C/EBPδ with similar time courses in identically treated cells (Figures 4E–4G). Taken together, these results strongly suggest that both C/EBPβ and C/EBPδ are direct upstream regulators of SREBP1c expression, directly binding to the SREBP1c promoter during early adipogenesis.
Figure 3. SREBP1c, but not SREBP1a, expression is reduced in adipose tissue of mice lacking C/EBPβ.
RNA was isolated from white adipose tissue of wild-type (WT) or Cebpb-knockout (KO) mice, reverse-transcribed and the expression of Cebpb (A), Cebpa (B), Srebp1c (C) and Srebp1a (D) was assayed by real-time PCR. Results are normalized to 18S rRNA, and are means ± S.E.M. (n = 3). *P < 0.05, compared with wild-type mice. AU, arbitrary units.
Figure 4. Identification of C/EBPβ- and C/EBPδ-binding sites in the SREBP1c promoter.
(A) Analysis of the 4 kb upstream of the first exon of SREBP1c by TESS (transcription element search software) revealed three potential C/EBPβ-binding sites, denoted site 1, site 2 and site 3. 3T3-L1 preadipocytes were induced to differentiate for various times and ChIP analysis performed to assess binding of C/EBPβ to the putative site 1 (B), site 2 (C) and site 3 (D). C/EBPδ binding to the same sites was also determined (E–G). C/EBPβ- and C/EBPδ-bound DNA in immunoprecipitates was quantified by real-time PCR. Values in each sample were normalized to total genomic DNA of the same sequence in the ‘input’ starting sample prior to immunoprecipitation. Values are means ± S.E.M. from four independent experiments. *P < 0.05, compared with binding at 0 h. AU, arbitrary units.
As has been previously described for the C/EBPα and DGAT2 promoters, among others, the maximal binding of C/EBPβ and C/EBPδ to the SREBP1c promoter lags behind the induction of these factors by several hours, probably due to the binding of inhibitory factors such as CHOP10 (C/EBP-homologous protein 10) and ETO. Similarly, the maximal binding of C/EBPβ and C/EBPδ precedes the peak of SREBP1c expression as has previously been observed for other well-characterized C/EBPβ/δ targets, probably due to the formation of inactive promoter-bound complexes that must be de-repressed for SREBP1c expression to occur.
We have previously observed that C/EBPα replaces C/EBPβ as a major regulator of DGAT2 expression when the expression of the latter diminishes in the later stages of adipogenesis [9]. Thus we investigated whether C/EBPα might similarly take over the regulation of SREBP1c expression from C/EBPβ and C/EBPδ as adipogenesis progresses. To test this we generated 3T3-L1 cells stably expressing shRNA targeting C/EBPα to inhibit its expression. This led to a significant inhibition of C/EBPα induction, which was reduced by 80 % or greater following induction of differentiation (Figure 5A). Consistent with the reciprocal regulation of PPARγ and C/EBPα during adipogenesis, C/EBPα inhibition also led to significant reduction in the expression of PPARγ in cells differentiated for 5 days (Figure 5B). The expression of key markers of the maturing adipocyte, including the fatty-acid-binding protein aP2 and the insulin-sensitive GLUT4, were also suppressed in cells in which C/EBPα had been knocked down. Similarly, the expression of DGAT2 was significantly inhibited in these cells, consistent with our previous study using preadipocytes transiently transfected with siRNA targeting C/EBPα [9]. In the same cells SREBP1c expression was found to be inhibited by approx. 70 %at day 5 of differentiation. In contrast, the loss of C/EBPα had no significant effect on the expression of the SREBP1a isoform.
Figure 5. C/EBPα regulates SREBP1c expression in the later stages of adipogenesis.
(A) 3T3-L1 preadipocytes stably expressing a control shRNA (open bars) or shRNA targeting C/EBPα (solid bars) were induced to differentiate for various times, RNA was extracted and the expression of C/EBPα mRNA was determined by real-time PCR. (B) Day 5 (D5) differentiated samples from cells expressing control (open bars) or C/EBPα-targeting shRNA (solid bars) were assayed for the expression of PPARγ2 (Pγ2), aP2, GLUT4 (GLT4), DGAT2 (DGT2), SREBP1c (SR1c) and SREBP1a (SR1a). Values are means ± S.E.M., normalized to cyclophilin (n = 3). *P < 0.05, compared with control shRNA-transfected cells at the same time point. (C–E) 3T3-L1 preadipocytes were induced to differentiate for various times and subjected to ChIP analysis to assess binding of C/EBPα to the putative site 1 (C), site 2 (D) and site 3 (E) indicated in Figure 4(A). C/EBPα-bound DNA in immunoprecipitates was quantified by real-time PCR. Values in each sample were normalized to total genomic DNA of the same sequence in the ‘input’ starting sample prior to immunoprecipitation. Values are means ± S.E.M. from four independent experiments. *P < 0.05, compared with binding at 0 h. AU, arbitrary units.
To determine whether the regulation of SREBP1c by C/EBPα could occur through direct binding of the same sites in the SREBP1c promoter occupied in early adipogenesis by C/EBPβ and C/EBPδ, we performed ChIP assays. C/EBPα was immunoprecipitated from 3T3-L1 preadipocytes differentiated for various times and bound DNA sequences corresponding to putative C/EBP-binding sites were assayed by real-time PCR. This revealed that site 1 inducibly binds C/EBPα as adipogenesis proceeds, replacing the binding by C/EBPβ and C/EBPδ 96 h after induction of differentiation (Figure 5C). In contrast, relatively weak binding was observed for C/EBPα to sites 2 and 3, and the marginal increases detected as adipogenesis progressed were not significant (Figures 5D and 5E).
Taken together, these results suggest that, during the later stages of adipogenesis, C/EBPα may substitute for C/EBPβ and C/EBPδ in the control of SREBP1c expression and that this involves binding to site 1 approx. 3.5 kb upstream of the transcriptional start site in the SREBP1c promoter.
DISCUSSION
SREBP1c plays a central role in lipid metabolism, particularly in the liver and adipose tissue. While the regulation of SREBP1c in the liver has been extensively studied, the factors regulating the induction of SREBP1c in developing adipocytes has received less attention, despite the important role acknowledged for this protein in adipocyte development. Our results are the first to show a key direct role for C/EBP factors in the regulation of SREBP1c with C/EBPβ and C/EBPδ initially binding to the SREBP1c promoter and subsequent regulation by C/EBPα. Selective siRNA- or shRNA-mediated knockdown demonstrated that the loss of any of these C/EBP factors significantly impairs the induction of SREBP1c. In addition, our results suggest that, at least for C/EBPβ, this regulatory mechanism is likely to operate in vivo, as mice lacking C/EBPβ also have reduced SREBP1c levels in adipose tissue.
While the present study places C/EBPβ upstream of SREBP1c during adipogenesis, the C/EBPβ promoter has conversely been described as a direct target of SREBP1c in mature adipocytes [25]. In this instance SREBP1c appears to be at least in part responsible for the induction of C/EBPβ in response to insulin. This illustrates that, rather than acting in a fixed canonical cascade, these genes function in a complex inter-regulatory network, the order of which will depend on factors including extracellular signals and the differentiation status of the cells involved.
From the present study it is difficult to discriminate the relative importance of C/EBPβ and C/EBPδ activity directly compared with the effects of consequent reduced expression of C/EBPα in the control of SREBP1c and lipogenesis. Forced expression of PPARγ in fibroblasts from Cebpa-null mice permits appropriate induction of adipocyte genes such as aP2, GLUT4 and adiponectin during adipogenesis, but with significantly reduced lipid accumulation [26,27]. We have previously shown that DGAT2, a key enzyme of lipogenesis, is a direct target of C/EBPα in adipocytes [9]. We now show that the effect of DGAT2 loss in cells with impaired C/EBP function may be exacerbated by the decreased expression of SREBP1c and the panoply of lipogenic genes it regulates. The precise relative importance of direct C/EBPα-mediated compared with indirect SREBP1c-mediated pathways in the control of lipogenesis will require the specific knockdown of SREBP1c.
Given the complex interacting network of transcription factors involved in adipocyte differentiation, it is also difficult to determine the contribution of direct compared with indirect actions of C/EBP factors in regulating the SREBP1c promoter. C/EBPs have important roles in inducing the expression of many proadipogenic transcription factors, notably PPARγ, and several of these are likely to regulate SREBP1c expression themselves. However, our ChIP assay data, clearly showing physical binding of C/EBPs to the SREBP1c promoter in intact differentiating adipocytes, strongly suggests that the C/EBPs make a significant contribution to SREBP1c expression as primary regulators. Moreover, the observation that knockdown of C/EBPβ expression does not affect C/EBPδ induction and vice versa strengthens the case that SREBP1c inhibition in each case does not result from an overall impairment of adipogenesis, but rather a selective inhibition of downstream genes.
It is interesting that C/EBP inhibition selectively inhibited SREBP1c, but not SREBP1a, expression. The relative importance of the SREBP1a and SREBP1c isoforms in different cultured models of adipogenesis has been controversial. Although specific inhibition of these two isoforms has not been compared in cultured preadipocytes, the results of the present study are consistent with a more important role for SREBP1c in adipogenesis. In the absence of C/EBPβ, C/EBPδ and/or C/EBPα induction, lipogenesis, adipogenesis and SREBP1c expression are coordinately impaired, while SREBP1a expression is unaffected. Although this does not demonstrate that loss of SREBP1c alone would replicate this phenotype, it does suggest that, of the two isoforms, SREBP1c is more tightly linked to adipocyte development and lipogenesis. Given that both SREBP1a and SREBP1c appear to have near identical target specificities and that SREBP1a is the more potent, at least in cultured hepatocytes [21], it is not clear why this is the case. Cellular studies selectively inhibiting the SREBP1 isoforms during adipogenesis using shRNA will be valuable in dissecting their relative importance, particularly as attempts to understand specific SREBP1 function in vivo using animal models has given confusing results. Loss of both SREBP1 isoforms caused significant embryonic lethality and up-regulation of SREBP2 in surviving mice [28], while selective ablation of SREBP1c led to reduced hepatic expression of lipogenic gene expression but no overt adipose phenotype, although epididymal fat mass was reduced [29]. Paradoxically, adipose-specific overexpression of constitutively active nuclear SREBP1 in mice led to a complex syndrome of lipodystrophy [30]. Other important regulators of adipogenesis, such as PPARγ and C/EBPα, have required more complex in vivo models, including chimaeric and hypomorphic mice and inducible postnatal knockouts to circumvent mortality in utero [31–33]. Similar models may be required to clarify the true importance of SREBP1c in adipose tissue development in vivo.
From a pathophysiological perspective, SREBP1c levels may be decreased in obese and Type 2 diabetic subjects [34,35], while altered SREBP1c levels have also been reported in lipodystrophic HIV patients undergoing antiretroviral therapy [36]. Furthermore, altered SREBP1c function may underlie or exacerbate lipodystrophy in patients with mutations in lamin A/C [37,38]. Thus a fuller understanding of how SREBP1c expression and/or activity may be modulated could be of significant therapeutic benefit in these conditions.
In summary, in the present study we have demonstrated for the first time that SREBP1c is regulated directly by C/EBP factors during adipocyte differentiation. This provides novel insight into the poorly defined transcriptional regulation of SREBP1c in developing adipose tissue, a key site of its action. Given the pleiotropic effects of SREBP1c, it identifies an additional mechanism via which C/EBP factors can indirectly influence lipid homoeostasis and insulin action. Such further delineation of the complex network controlling adipocyte development is important for the development of therapeutic strategies to treat diseases featuring altered adipose tissue mass or function, including obesity and lipodystrophies.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the Wellcome Trust [grant number 078986/Z/06/Z (to V. A. P. and S. O. R.)]; the Dorothy Hodgkin Postgraduate Award Scheme [grant number EP/P500796 (to W.-S. A.)]; the British Heart Foundation [grant number FS/05/092 (to J. J. R.)]; the Medical Research Council [grant number GO800203 (to J. J. R.)]; the Cambridge National Institutes of Health Research Comprehensive Biomedical Research Centre [grant number CG50826 METABOLISM (to C. E. L.); the Medical Research Council Centre for Obesity and Related Medical Diseases [grant number GO600717]; and the National Institutes of Health [grant number DK059767 (to J. E. F. and S. M. R.). J. J. R. and S. O. R. are members of the EUGENE2 Consortium (European Network on Functional Genomics of Type 2 Diabetes).
Abbreviations used
- aP2
adipocyte protein 2
- C/EBP
CCAAT/enhancer-binding protein
- ChIP
chromatin immunoprecipitation
- DGAT2
diacylglycerol acyltransferase 2
- ES cell
embryonic stem cell
- ETO
eight-twenty one/MTG8
- GLUT4
glucose transporter 4
- LIP
liver-enriched inhibitory protein
- PPAR
peroxisome-proliferator-activated receptor
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SREBP
sterol-regulatory-element-binding protein
Footnotes
AUTHOR CONTRIBUTION
Victoria Payne and Wo-Shing Au performed the majority of the experimental work, and Christopher Lowe performed some minor additional experiments. Shaikh Rahman and Jacob Friedman provided samples from Cebpb-knockout mice. Stephen O’Rahilly provided additional advice, while Justin Rochford initiated the study, designed and supervised the experiments and drafted the manuscript, All authors contributed to the discussion of data, proposed improvements to the experimental work and revision of the manuscript.
References
- 1.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet. 2006;7:175–199. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- 3.Hegele RA, Joy TR, Al-Attar SA, Rutt BK. Thematic review series: Adipocyte Biology. Lipodystrophies: windows on adipose biology and metabolism. J Lipid Res. 2007;48:1433–1444. doi: 10.1194/jlr.R700004-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Rangwala SM, Lazar MA. Transcriptional control of adipogenesis. Annu Rev Nutr. 2000;20:535–559. doi: 10.1146/annurev.nutr.20.1.535. [DOI] [PubMed] [Google Scholar]
- 5.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 6.Semple RK, Chatterjee VK, O’Rahilly S. PPARγ and human metabolic disease. J Clin Invest. 2006;116:581–589. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto H, Kurebayashi S, Hirose T, Kouhara H, Kasayama S. Reduced IRS-2 and GLUT4 expression in PPARγ 2-induced adipocytes derived from C/EBPβ and C/EBPδ-deficient mouse embryonic fibroblasts. J Cell Sci. 2002;115:3601–3607. doi: 10.1242/jcs.00044. [DOI] [PubMed] [Google Scholar]
- 9.Payne VA, Au WS, Gray SL, Nora ED, Rahman SM, Sanders R, Hadaschik D, Friedman JE, O’Rahilly S, Rochford JJ. Sequential regulation of diacylglycerol acyltransferase 2 expression by CAAT/enhancer-binding protein beta (C/EBPβ) and C/EBPα during adipogenesis. J Biol Chem. 2007;282:21005–21014. doi: 10.1074/jbc.M702871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 12.Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARγ through the production of endogenous ligand. Proc Natl Acad Sci USA. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 14.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo JB, Moon HM, Noh MJ, Lee YS, Jeong HW, Yoo EJ, Kim WS, Park J, Youn BS, Kim JW, et al. Adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element-binding protein 1c regulates mouse adiponectin expression. J Biol Chem. 2004;279:22108–22117. doi: 10.1074/jbc.M400238200. [DOI] [PubMed] [Google Scholar]
- 16.Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor expression. Mol Cell Biol. 2004;24:3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laudes M, Christodoulides C, Sewter C, Rochford JJ, Considine RV, Sethi JK, Vidal-Puig A, O’Rahilly S. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J Biol Chem. 2004;279:11711–11718. doi: 10.1074/jbc.M310240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochford JJ, Semple RK, Laudes M, Boyle KB, Christodoulides C, Mulligan C, Lelliott CJ, Schinner S, Hadaschik D, Mahadevan M, et al. ETO/MTG8 is an inhibitor of C/EBPβ activity and a regulator of early adipogenesis. Mol Cell Biol. 2004;24:9863–9872. doi: 10.1128/MCB.24.22.9863-9872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne VA, Grimsey N, Tuthill A, Virtue S, Gray SL, Dalla Nora E, Semple RK, O’Rahilly S, Rochford JJ. The human lipodystrophy gene BSCL2/seipin may be essential for normal adipocyte differentiation. Diabetes. 2008;57:2055–2060. doi: 10.2337/db08-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, Croniger C, Arizmendi C, Harada-Shiba M, Ren J, Poli V, Hanson RW, Friedman JE. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPβ gene. J Clin Invest. 1999;103:207–213. doi: 10.1172/JCI4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amemiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res. 2002;43:1220–1235. [PubMed] [Google Scholar]
- 22.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang QQ, Jiang MS, Lane MD. Repressive effect of Sp1 on the C/EBPα gene promoter: role in adipocyte differentiation. Mol Cell Biol. 1999;19:4855–4865. doi: 10.1128/mcb.19.7.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams LJ, Lyons V, MacLeod I, Rajan V, Darlington GJ, Poli V, Seckl JR, Chapman KE. C/EBP regulates hepatic transcription of 11β-hydroxysteroid dehydrogenase type 1. A novel mechanism for cross-talk between the C/EBP and glucocorticoid signaling pathways. J Biol Chem. 2000;275:30232–30239. doi: 10.1074/jbc.M001286200. [DOI] [PubMed] [Google Scholar]
- 25.Le Lay S, Lefrere I, Trautwein C, Dugail I, Krief S. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1C) regulation of gene expression in 3T3-L1 adipocytes. Identification of CCAAT/enhancer-binding protein β as an SREBP-1C target. J Biol Chem. 2002;277:35625–35634. doi: 10.1074/jbc.M203913200. [DOI] [PubMed] [Google Scholar]
- 26.Gustafson B, Jack MM, Cushman SW, Smith U. Adiponectin gene activation by thiazolidinediones requires PPARγ 2, but not C/EBPα: evidence for differential regulation of the aP2 and adiponectin genes. Biochem Biophys Res Commun. 2003;308:933–939. doi: 10.1016/s0006-291x(03)01518-3. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 28.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 30.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutnikova H, Cock TA, Watanabe M, Houten SM, Champy MF, Dierich A, Auwerx J. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPARγ hypomorphic mice. Proc Natl Acad Sci USA. 2003;100:14457–14462. doi: 10.1073/pnas.2336090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR7γ is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Croniger CM, Lekstrom-Himes J, Zhang P, Fenyus M, Tenen DG, Darlington GJ, Hanson RW. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein α. J Biol Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- 34.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14:1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sewter C, Berger D, Considine RV, Medina G, Rochford J, Ciaraldi T, Henry R, Dohm L, Flier JS, O’Rahilly S, Vidal-Puig AJ. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-α. Diabetes. 2002;51:1035–1041. doi: 10.2337/diabetes.51.4.1035. [DOI] [PubMed] [Google Scholar]
- 36.Bastard JP, Caron M, Vidal H, Jan V, Auclair M, Vigouroux C, Luboinski J, Laville M, Maachi M, Girard PM, et al. Association between altered expression of adipogenic factor SREBP1 in lipoatrophic adipose tissue from HIV-1-infected patients and abnormal adipocyte differentiation and insulin resistance. Lancet. 2002;359:1026–1031. doi: 10.1016/S0140-6736(02)08094-7. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum Mol Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- 38.Maraldi NM, Capanni C, Mattioli E, Columbaro M, Squarzoni S, Parnaik WK, Wehnert M, Lattanzi G. A pathogenic mechanism leading to partial lipodistrophy and prospects for pharmacological treatment of insulin resistance syndrome. Acta Biomed. 2007;78:207–215. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.