Abstract
Genetic screens for phenotypic similarity have made key contributions to associating genes with biological processes. With RNA interference (RNAi), highly parallel phenotyping of loss-of-function effects in cells has become feasible. One of the current challenges however is the computational categorization of visual phenotypes and the prediction of biological function and processes. In this study, we describe a combined computational and experimental approach to discover novel gene functions and explore functional relationships. We performed a genome-wide RNAi screen in human cells and used quantitative descriptors derived from high-throughput imaging to generate multiparametric phenotypic profiles. We show that profiles predicted functions of genes by phenotypic similarity. Specifically, we examined several candidates including the largely uncharacterized gene DONSON, which shared phenotype similarity with known factors of DNA damage response (DDR) and genomic integrity. Experimental evidence supports that DONSON is a novel centrosomal protein required for DDR signalling and genomic integrity. Multiparametric phenotyping by automated imaging and computational annotation is a powerful method for functional discovery and mapping the landscape of phenotypic responses to cellular perturbations.
Keywords: DNA damage response signalling, massively parallel phenotyping, phenotype networks, RNAi screening
Introduction
Forward genetic screens for visual phenotypes in model organisms have proven a powerful method for associating phenotypes with genes and for the prediction of functional relationships (Jorgensen and Mango, 2002; St Johnston, 2002). Aggregating genes by similarity of their loss-of-function phenotype has provided key insights into signalling pathways that have a conserved function from Drosophila to human (Nusslein-Volhard and Wieschaus, 1980; Bier, 2005). Complex visual phenotypes, such as defects in pattern formation during development, greatly facilitated the classification of genes into pathways, and phenotypic similarities in many cases predicted molecular relationships.
The possibility of depleting specific transcripts by RNA interference (RNAi) in cells has resulted in new screening techniques for associating genes with biological processes (Dorsett and Tuschl, 2004; Echeverri et al, 2006; Moffat et al, 2006; Boutros and Ahringer, 2008). Simple, univariate phenotypes, such as measurements of cell viability or specific reporter gene activity, have been applied to screen for modifiers on a genome-wide scale; however, the information content in finding different genes sharing the same univariate phenotype is limited (Boutros et al, 2004; DasGupta et al, 2005; Whitehurst et al, 2007). More complex multiparametric readouts from microscopy and automated image analysis promise broad coverage of distinct phenotypes. Such visual phenotypes could predict specific functional relationships between genes, while not being biased by the particular choice of phenotypic assay.
As the technology for carrying out large-scale screens by imaging of cells has been established in recent years, a main challenge is the automated analysis of images and subsequent interpretation of cellular phenotypes. Extracting functional relationships on a genome-wide scale is an unresolved problem. Cells respond to many extrinsic and intrinsic stimuli and change their shape, cell-cycle status and proliferation rate. Similar to patterning decisions in whole organisms, such changes are broad reflectors of cellular functions and have the potential to simultaneously provide a multitude of views on biological processes. Annotation of phenotypes by visual inspection has been effective (Kiger et al, 2003; Eggert et al, 2004; Sonnichsen et al, 2005), but creates a bottleneck for larger experiments. Several studies used automated extraction of multiparametric computational descriptors of cellular morphology to analyse the effects of small molecules or siRNAs on cultured cells (Perlman et al, 2004; Neumann et al, 2006; Bakal et al, 2007; Loo et al, 2007; Jones et al, 2009). Owing to the complexity of the multivariate data analysis, these studies focused on small sets of reagents. Genome-wide studies have been performed for simple, univariate phenotypes (Mukherji et al, 2006; Sims et al, 2009). Genome-wide analysis of high-dimensional, multiparametric descriptors, and hence the inference of more specific, multidimensional phenotypes, has remained a challenge.
In this study, we describe an experimental and computational approach to predict gene function genome-wide based on changes in the morphology of individual cells within cell populations. We establish a computational method to separate multivariate variation of interest from noise induced by the environment or intrinsic stochastic behaviour of the cells, and to generate multivariate phenotypic profiles that summarize similarity and dissimilarity of phenotypes at the level of treated cell populations. We applied this methodology to a genome-wide RNAi screen and produced a phenotypic map of 1820 siRNA-mediated perturbations that led to significant phenotypic variation. We investigated the genes DONSON, SON, CADM1 and CD3EAP that were found in phenotypic proximity to known components of cell-cycle regulation and DNA damage response (DDR). Further experiments characterized their roles in the maintenance of genomic integrity.
Results
Automated analysis for high-throughput imaging
We established a method for generating phenotypic profiles by automated microscopy, measuring the effects of RNAi-mediated knockdowns on the morphology of HeLa cells (Figure 1). For each perturbation, 1000 cells were reverse transfected with siRNAs spotted in 384-well plates. Each plate also contained negative controls and positive control siRNAs with known phenotypes. Forty-eight hours after siRNA transfection, cells were fixed, stained for DNA and the cytoskeletal proteins actin and tubulin, and imaged using an automated microscope (Figure 1A and B).
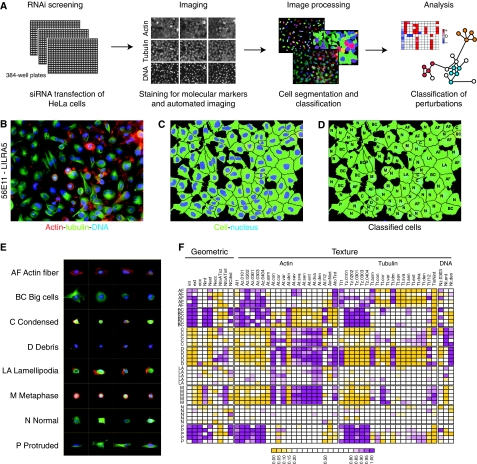
Figure 1.
Automated phenotyping of an RNAi screen. (A) Schematic representation of screening and analysis protocol. (B) Example image of cells transfected with siRNA targeting LILRA5 and stained after 48 h for DNA (blue) and the cytoskeletal proteins actin (red) and tubulin (green). (C) Cytoplasm (green) and nuclei (blue) boundaries were determined by automatic segmentation. (D) Cells after classification, using the labelling scheme of panel E. (E) Examples of cells from eight cell classes used for the training of the classifier. (F) Each cell is described by a vector of 51 numerical descriptors (cell and nucleus geometry, textures of actin, tubulin and DNA stains). Box colours correspond to quantiles of the cell descriptor distributions in the full dataset.
To identify distinct phenotypes, we used automated image analysis to classify all individual cells based on nuclear and cytoskeletal fluorescent markers. Nuclei and cytoplasm boundaries of cells (Figure 1C) were determined by a segmentation algorithm. We then computed 51 cell descriptors that quantified intensities, shape characteristics and texture (Figure 1F; Supplementary information). The descriptors were rotationally invariant, robust to pixel noise and served as input for classification of cells by a support vector machine (SVM) (Boser et al, 1992). Cells were classified into 1 of 10 classes, which included cells showing protrusion/elongation, cells in metaphase, large cells, condensed cells, cells with lamellipodia and cellular debris (Figure 1D and E; Supplementary Table II). Feature selection and sensitivity analysis showed that the classification performance was not driven by single descriptors, but rather depended on the joint behaviour of multiple descriptors (Supplementary Figure 2).
Under wild type and each perturbed condition, imaged cell populations always consisted of multiple phenotypic classes. We reasoned that phenotypic information could be inferred from changes in frequencies of the different classes and from descriptor summaries over the cell populations. For each siRNA experiment, we computed a phenotypic profile, which is a vector of 13 numbers representing cell count, median cellular descriptors computed on the population and proportions of each cell class (Supplementary Table VII).
To assess the performance of the approach, we first carried out a small siRNA screen by targeting approximately 800 transcripts in HeLa cells, including most kinases and controls with known phenotypes (Figure 2A). Quantitative descriptors of cells after perturbations (Figure 2B) were evaluated to assess reproducibility. The results showed that median values of the cellular descriptors, computed over cell populations, were highly reproducible across replicate experiments, with a median correlation coefficient of 0.80 (Figure 2C and D). Cell classification accuracy, based on cellular descriptors, was estimated by cross-validation to be 96–100%, depending on cell class (Table I). Median descriptors for the defined siRNA controls UBC, CLSPN and TRAPPC3 were well separated from those of the Renilla luciferase (Rluc) controls, showing multiparametric Z′ factors of 0.79, 0.62 and 0.60 (Figure 2D and E; Supplementary information).
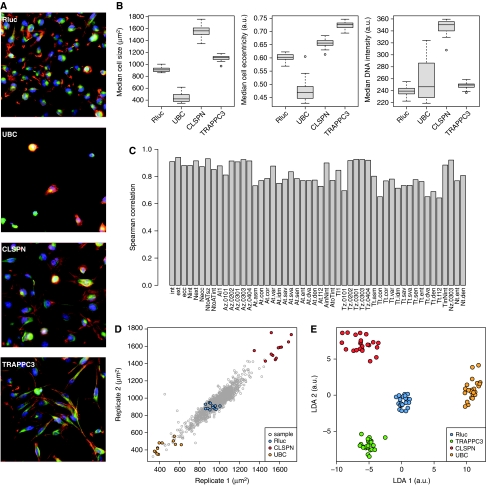
Figure 2.
Quantitative analysis of the cell descriptors. (A) Example images of cells transfected with negative (Rluc) and positive (UBC, CLSPN and TRAPPC3) control siRNAs. (B) Box plots of three median cell descriptors measured in control wells. Bars represent medians and rectangles show the interquartile ranges, n=24 wells per control. (C) Spearman correlation coefficients of the median cell descriptors of Figure 1F between replicated wells. One cellular descriptor from Figure 1, representing the distance between cell and nucleus centre of mass, had a Spearman correlation coefficient of −0.02 and was removed from the analysis. (D) Scatter plot of the descriptor ‘median cell size’ between replicate wells, showing a Spearman correlation coefficient of 0.94. (E) Linear discriminant analysis (LDA) projection of three median cell descriptors (cell size, nucleus eccentricity and nucleus intensity), computed on control wells. Corresponding multiparametric Z’ factors, computed between controls UBC, CLSPN and TRAPPC3 and negative control Rluc were 0.79, 0.62 and 0.60. Source data is available for this figure at www.nature.com/msb.
Table 1. Confusion table for the cell classification step, computed by five-fold cross-validation on the training set.
| True classes |
||||||
|---|---|---|---|---|---|---|
| BC | D | M | N | P | Z | |
| Cells were classified according to theirs descriptors, using an SVM with a training set of 1740 cells distributed in six classes: big cells (BC), debris (D), metaphase (M), normal (N), cells with protrusion (P) and telophase (Z). Average cell classification accuracy (Acc. %) per class is reported in the last row. | ||||||
| Predicted classes | ||||||
| BC | 110 | 0 | 0 | 0 | 0 | 0 |
| D | 1 | 253 | 0 | 2 | 0 | 1 |
| M | 0 | 0 | 240 | 0 | 0 | 0 |
| N | 1 | 3 | 0 | 640 | 4 | 4 |
| P | 0 | 0 | 0 | 0 | 252 | 0 |
| Z | 0 | 5 | 0 | 1 | 0 | 223 |
| Acc. % | 98.2 | 96.9 | 100 | 99.5 | 98.5 | 97.8 |
The experiments also demonstrated that changes in individual descriptors contained phenotypically relevant information. For example, though Rluc siRNA-treated cells showed a median size of 910 μm2, targeting the mitotic inhibitor kinase WEE1 led to significant smaller cells, with a median size of 453 μm2 (Wilcoxon rank sum test, P<10−15). Another example is PPP4C, whose depletion led to significantly elongated cells compared to Rluc treatment (median cell eccentricity of 0.66 compared to 0.59, Wilcoxon rank sum test, P<10−15). These results indicate high reproducibility of the experimental and computational approach, and its ability to quantitatively discern biologically relevant phenotypic variation.
Genome-wide RNAi screen to cluster genes based on cellular descriptors
To cluster genes and predict function on a genome-wide scale, we measured the effects of 22 839 siRNA-mediated knockdowns on HeLa cells (see Materials and methods). We acquired four images per perturbation, from which ∼6.5 million cells were automatically segmented, quantified and classified (a comprehensive track of the analysis is provided at http://www.cellmorph.org). Each siRNA effect was summarized by a phenotypic profile as described above. On average, 324 cells were recorded in Rluc siRNA-negative control treatments. siRNAs against PLK1, an essential cell-cycle regulator, reduced the number of cells eight-fold. siRNA-mediated depletions resulted in a cell population comprising a variety of cell classes. For example, negative control Rluc wells contained on average 57.3% cells classified as normal, 15.0% condensed cells and 2.0% metaphase cells, with the remainder distributed among other classes.
To quantify the differences between perturbation effects, we measured the similarity between phenotypic profiles. As profiles are multiparametric descriptors whose components have different dynamic ranges and signal-to-noise ratios, because of cell intrinsic stochastic behaviour and experimental factors, we transformed the profile values of each siRNA perturbation into a set of scores ranging between 0 and 1, using for this a parametric family of sigmoidal functions (see Supplementary information). We termed the vector of scores a phenoprint (Figure 3C) and defined the phenotypic distance between a pair of perturbations as the distance between their corresponding phenoprints. We reasoned that pairs of related proteins should be enriched for smaller phenotypic distances, when compared with random pairs, and used distance metric learning (Xing et al, 2003) to determine the sigmoid transformation parameters that led to maximal enrichment. We used the STRING 7.1 dataset (von Mering et al, 2007) as a set of related protein pairs, considering all interactions with a confidence score higher than 0.4, including pairs predicted with the STRING neighbourhood, gene fusion, co-occurence, co-expression, experiments, databases and text mining methods. Out of the genome-wide RNAi screen, 1820 siRNA perturbations produced non-zero phenoprints. To test whether individual genes in STRING influenced the computation of the phenoprints, we removed all information about the genes RRM1, TMEM61, CLSPN, CADM1, CD3EAP, CEP164, NUF2, DONSON and SON (as shown in Figure 3C), recomputed their phenoprints and found that they were unaltered (Supplementary Figure 8). This indicates that the distance learning method is based on agglomerative properties of the training data, not on presence of individual gene pairs. We estimate that the false-positive rate of the computational analysis is low because negative control siRNAs Rluc showed in 97.1% (132/136) of all cases a zero phenoprint; 0.8% (1/136) of positive controls (PLK1) scored as false negatives.
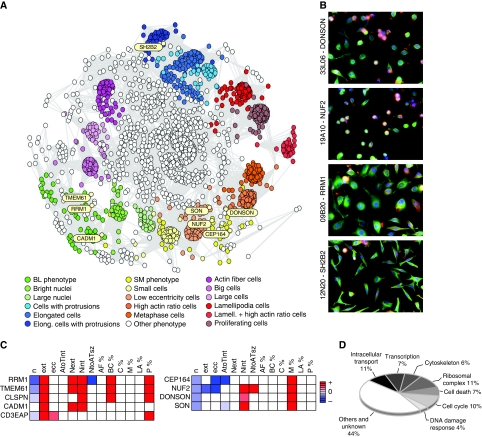
Figure 3.
Genome-wide two-dimensional map of the perturbation phenotypes. (A) Each of the 1820 nodes represents a perturbation and is characterized by a phenoprint. Nodes are linked by a grey edge when their phenotypic distance is below 0.16. Groups of nodes with small distances form clusters and are coloured according to their most prominent cell subpopulations. The phenotypic cluster BL is characterized by a decrease in the number of cells and increase of metaphase cells. The cluster SM shows an abundance of large cells with protrusions and bright nuclei. The map provides an overview over the variety of phenoprints and shows groups of phenotypically similar perturbations. An interactive version is provided on the companion website (see http://www.cellmorph.org). (B) Representative images of cell populations from different regions of the map show the similarity of phenotypes of neighbouring genes, and the gradual variation of phenotypes on paths through the map. (C) Heat maps of phenoprints of selected genes from the clusters BL and SM. The colour code represents the strength of the increase or decrease in phenotypic properties. (D) Functional categories of the 280 confirmed phenotypes; 44% (122 of 280) were functionally uncharacterized. Source data is available for this figure at www.nature.com/msb.
Visualization of phenotypic relationships and independent confirmation
To visualize the distribution of the 13-dimensional phenoprints, we projected them into a 2-dimensional map that is representative of the phenotypic similarity relationships (Figure 3A). The map shows the phenotypic landscape spanned by the genome-wide perturbations. Seventeen clusters of similar phenoprints were defined and added to the map, to highlight different phenotypic regions (see Supplementary information). These clusters included phenotypes characterized by an over-representation of cells with prominent lamellipodia (WNK3, ANXA4), cells with prominent actin fibres (ODF2, SOD3), abundance of large cells (CA14), many elongated cells (SH2B2, ELMO2), decrease in cell number (TPX2, COPB1, COPA), increase in number of cells in metaphase (BLR1, CIB2) and combinations of phenotypes such as presence of large cells with protrusions and bright nuclei (PTPRZ1, RRM1). Consistent with its phenoprint, ELMO2 has been described as an upstream regulator of the Rac1 GTPase (Gumienny et al, 2001), which controls the actin cytoskeleton organization. Other examples include COPB1 and COPA, which are essential for the maintenance of the Golgi architecture and are required for cell viability (Pepperkok et al, 1993; Guo et al, 1994; Neumann et al, 2006).
To test for biological reproducibility with independent siRNAs, we selected 604 out of 1820 siRNAs for retesting and confirmed that the phenotypes were reproduced for 310 (51.3%) genes in HeLa cells (Supplementary Table X), which is a rate similar to previously published reports (Tang et al, 2008). Furthermore, we assessed their phenotypes in a different cell type, osteosarcoma U2OS cells, in which 280 of the 310 siRNAs reproduced their phenotypes. Overall, 44% (122 of 280) were functionally uncharacterized; 31% (86 of 280) were implicated in cytoskeletal organization, intracellular transport, cell-cycle regulation or DDR (Figure 3D). We also monitored knockdown efficiency in a representative set of genes by real-time quantitative PCR (qPCR) and found that mRNA levels were reduced to at least 65% (Supplementary Figure 10). These experiments demonstrated that the screen identified functionally uncharacterized genes and reproducibly associated them with phenotypic clusters based on their cell population phenotypes.
Phenotypic proximity to predict gene function
Annotation of the 17 clusters, comprising 943 siRNA perturbations, indicated that genes having similar phenoprints tended to have similar biological functions (Supplementary Table XIII). For example, the cluster containing phenoprints characterized by bright nuclei includes several genes that are implicated in cell-cycle regulation or DNA replication such as CDCA8, CENPI, DTL and MCM10. Interestingly, the three subunits RPA1, RPA2 and RPA3 of the replication factor A, required for DNA replication and DDR, are also present in this cluster. Furthermore, the subunits of the Golgi coatomer complex COPB2, COPZ and COPA, and the essential cell-cycle kinase WEE1, all required for cell viability, had similar phenoprints. Comparisons of the phenotypic graph with functional networks including MouseNet (Kim et al, 2008) show significant enrichment of phenotypic similarities within network gene pairs (see Supplementary information).
To associate uncharacterized genes with biological processes, we selected candidate genes in proximity to known pathway components, and focused on phenotypic clusters that contained genes associated with DDR signalling and genomic integrity. We first examined a phenotypic cluster in proximity to CEP164 (Figure 3A and C). CEP164 is localized to the distal appendages of mature centrioles (Graser et al, 2007) and has been described as an important component in the DNA damage-activated signalling cascade (Sivasubramaniam et al, 2008). The phenotypic cluster included other proteins implicated in kinetochore-associated functions, such as NUF2 (DeLuca et al, 2002) (Figure 3B) and SGOL1 (Pouwels et al, 2007). In addition, we selected two largely uncharacterized factors: SON, a DNA binding protein localized at the mitotic spindles (Nousiainen et al, 2006), and DONSON (Figure 3B), which has not been studied earlier. The two genes are co-located in the human genome at 21q22. We observed that, among other quantitative descriptors, cell populations of this cluster were characterized by a relative decrease in cell number and an increase in metaphase cells.
A second, phenotypically distinct cluster included RRM1 (Figure 3A–C), which is part of the ribonucleoside–diphosphate reductase complex required for replication and DNA repair (Xue et al, 2003), and CLSPN (Figure 3B), a protein required for the activation of the checkpoint kinase CHEK1, a key mediator of DDR (Kumagai and Dunphy, 2000; Liu et al, 2006). This cluster also contained other DDR genes such as PRIM2 (Weiner et al, 2007) and SETD8 (Jorgensen et al, 2007). From this cluster, we selected the poorly characterized genes CADM1 and CD3EAP and hypothesized that they might be components of the DDR pathway.
To confirm target specificity, we compared the effects of four single siRNAs targeting a gene to that of the siRNA pool used in the screen and assessed their knockdown efficiency by qPCR (Supplementary Figure 11). These experiments showed that for each candidate, at least two out of the four independent siRNAs were effective in target knockdown and reproduced the phenotypes observed in the screen (Supplementary Table XI).
DONSON is required for cell-cycle progression and localizes to the centrosome
To further investigate the role of DONSON in cell-cycle progression, we performed time-course experiments with HeLa cells depleted for DONSON and measured the DNA content. As shown in Figure 4A, we observed a slower S-phase progression compared to controls, suggesting a DNA replication defect, followed by accumulation of cells in G2/M at 48 h. We then assessed whether DONSON is required for DNA replication during S-phase progression. U2OS cells were arrested with hydroxyurea (HU) in G1/S, and incorporation of bromodeoxyuridine (BrdU) was measured after release. As shown in Figure 4B, depletion of DONSON resulted in significant decrease in BrdU incorporation, indicating that DONSON is required for S-phase transition. We next asked whether the level of DONSON protein is regulated during the cell cycle. U2OS cells were arrested either in G1/S and G2/M by HU or nocodazole, respectively. After release from HU, we assessed DONSON protein levels for up to 24 h by immunoblot analysis. As shown in Figure 4C, DONSON levels peaked at 6 h after release and preceded the cyclin A expression as a marker for G2. Furthermore, when cells were released from G2/M-phase arrest, peak DONSON levels were observed later than phospho-H3 as a marker for M phase (Figure 4C). Taken together, these experiments indicate that DONSON levels are regulated during cell division, peaking during S phase and that DONSON is required for cell-cycle progression.
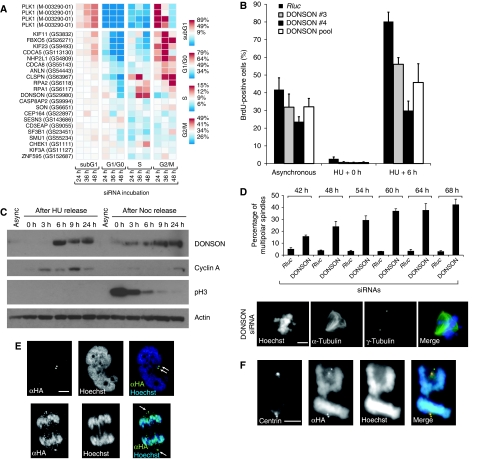
Figure 4.
Functional analysis of candidate genes for roles in cell-cycle progression and spindle organization. (A) Time-resolved cell-cycle analysis in HeLa cells transfected with indicated siRNAs at different time points. Box colours represent fractions of cells with DNA content corresponding to sub-G1, G1/G0, S and G2/M. HeLa cells transfected with siRNAs against DONSON showed a delay in S-phase progression. (B) Assessment of S-phase progression by BrdU incorporation; 48 h after siRNA transfection, U2OS cells were synchronized for 16 h with 1 mM hydroxyurea (HU) and released for 6 h in BrdU-containing medium. BrdU-positive cells were stained with anti-BrdU primary antibody (Calbiochem) and Alexa 488 secondary antibody. DNA was counterstained with propidium iodide. Imaging and quantification were performed with Acumen Explorer microplate reader. Values are shown as mean±s.d. of three biological replicates. (C) Cell-cycle-dependent protein expression of DONSON. U2OS cells were synchronized either in G1/S with HU or in G2/M with nocodazole (Noc). Cells were collected at different time points after release into cell cycle for western blot analysis. (D) DONSON depletion is associated with a 10-fold increase in multipolar spindles compared to control treatments. U2OS cells were transfected with a DONSON siRNA pool and immunostained for α- and γ-tubulin at indicated time points. U2OS cells transfected with Rluc siRNAs serving as negative control. Data represent mean±s.d. of three biological replicates. At least 200 metaphase spindles were counted in each experiment. Scale bar indicates 2.5 μm. (E) DONSON protein co-localizes with centrosomes. HeLa cells were transfected with HA-tagged DONSON for 48 h and immunostained with primary HA antibody and Alexa 488-conjugated secondary antibody. Arrows indicate the centrosomal staining of DONSON (green). DNA was counterstained with DAPI (blue). Scale bar indicates 2.5 μm. (F) DONSON co-localizes with centrin. U2OS cells were transfected with HA-tagged DONSON; 48 h after transfection, cells were immunostained with anti-centrin and anti-HA-tagged primary antibodies and Alexa 488-, Alexa 594-conjugated secondary antibodies, respectively. Scale bar indicates 2.5 μm. Source data is available for this figure at www.nature.com/msb.
To examine the subcellular localization of DONSON protein during the cell cycle, we expressed a DONSON-HA transgene, as we could not detect endogenous levels of DONSON in immunofluorescence experiments. We observed that DONSON localized in two perinuclear foci, which co-localized with the centrosomal markers γ-tubulin and centrin (Figure 4E and F; Supplementary Figure 15). In DONSON-depleted U2OS and HeLa cells, we observed a 10-fold increase of multipolar spindles in metaphase cells compared to control treatments (Figure 4D; Supplementary Figure 14). These results showed that DONSON protein is localized at the centrosome and is required for proper formation of bipolar spindles during cell-cycle progression.
DONSON, SON, CADM1 and CD3EAP are required for genomic integrity
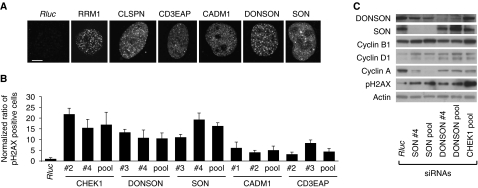
The bi-orientation attachment of each chromosome to both poles of the mitotic spindle is essential for genomic integrity (Loncarek et al, 2007). To test the involvement of DONSON in genomic integrity, we used quantitative immunofluorescence to measure the formation of γH2AX foci as a marker of DDR signalling (Figure 5A and B; Supplementary Figures 12 and 13). We observed that DONSON depletion led to the formation of γH2AX foci similar to the depletion of the known DDR effectors RRM1 and CHEK1. In addition, we checked other candidate genes regarding their involvement in genomic integrity. Similar to DONSON knockdown, we observed increased γH2AX formation in cells depleted of SON, CADM1 and CD3EAP (Figure 5A and B). One possible explanation of elevated γH2AX foci formation is the accumulation of cells in S phase (Tanaka et al, 2006). To test this hypothesis, we monitored cyclin levels in DONSON-depleted U2OS cells. As shown in Figure 5C, knock down of DONSON in U2OS cells led to increased cyclin D1 and decreased cyclin A and cyclin B1 levels, indicating that cells were arrested in G1 and that the elevated γH2AX foci formation observed in DONSON-depleted cells is not caused by an accumulation of cells in S phase. In contrast to DONSON, SON-depleted cells showed an enrichment of cyclin B1 protein, whereas cyclin A was decreased compared to the control treatment; this indicated that cells were arrested in G2/M (Figure 5C). In summary, these results suggest a role of DONSON, SON, CADM1 and CD3EAP in the maintenance of genomic integrity.
Figure 5.
Functional assays for candidate genes in maintenance of genomic integrity. (A) Depletion of RRM1, CLSPN, CD3EAP, CADM1, DONSON and SON induced γH2AX foci formation. U2OS cells were transfected with siRNA pools and immunostained with γH2AX antibody 72 h after transfection. Representative images of γH2AX foci formation are shown. Scale bar indicates 2.5 μm. (B) Quantification of γH2AX foci formation on depletion of candidate genes; 72 h after siRNA transfection, U2OS cells were fixed, immunostained for γH2AX and γH2AX-positive cells were quantified. Ratios of γH2AX-positive cells were normalized to the negative control Rluc siRNA treatment. Data are mean±s.d. of three biological replicates. (C) γH2AX accumulation in DONSON and SON-depleted cells is not caused by an accumulation of S-phase cells. U2OS cells were collected 72 h after siRNA transfection and cell lysates were analysed by western blotting using indicated antibodies. Source data is available for this figure at www.nature.com/msb.
DONSON, SON, CADM1 and CD3EAP are mediators of the DDR
The maintenance of genomic integrity is dependent on a functional DDR, a process comprising multiple signal transduction pathways that coordinate cell-cycle transitions, DNA replication, DNA repair and apoptosis (Cimprich and Cortez, 2008). The phosphorylation of CHEK1 is an early event in the DDR. To analyse the function of DONSON in DDR, we used immunoblot analysis to assess the phosphorylation of CHEK1 on γ irradiation in U2OS cells 48 h after transfection with DONSON. We observed that the phosphorylation of CHEK1 was strongly reduced in DONSON-depleted cells compared to negative control (Figure 6A). Moreover, DONSON-depleted cells showed an impaired phosphorylation of CHEK1 (Figure 6B and C) after UV exposure, similar to the effect of γ irradiation. Although ATM phosphorylation was not affected in DONSON-depleted cells, we observed an attenuated phosphorylation of RPA2, H2AX and NBS1 (Figure 6C). These results indicate that DONSON acts downstream of ATM and upstream of CHEK1 in the DDR signalling cascade.
Figure 6.
SON, DONSON and CD3EAP are required for the DNA damage response. (A) SON, DONSON and CD3EAP depletion leads to a decreased phosphorylation of CHEK1 on γ irradiation, similar to ATR; 48 h after siRNA transfection, cells were γ irradiated and collected 1 or 2 h later for immunoblot analysis, probing with indicated antibodies. (B) SON and DONSON depletion leads to attenuated phosphorylation of CHEK1 on UV exposure, similar to ATR depletion. U2OS cells were UVC irradiated (20 J/m2) 48 h after siRNA transfection. Cell lysates were collected for immunoblotting 2 h later and probed with indicated antibodies. (C) Knock down of DONSON and SON impairs RPA2 recruitment onto chromatin and the phosphorylation of ATR substrates. U2OS cells were transfected with siRNAs and UVC irradiated (20 J/m2). Subsequently, the chromatin-associated insoluble fraction was extracted 2 h after UV exposure, and the fractions were analysed for the indicated proteins by immunoblot.
To test whether DONSON is required for the recruitment of DDR-associated proteins onto chromatin, we extracted chromatin from UV-irradiated cells and analysed RPA2 levels by immunoblot (Liu et al, 2007). As shown in Figure 6C, we observed that the amount of RPA2 on the chromatin was significantly decreased on DONSON knockdown. As DONSON depletion leads to G1 arrest, we cannot fully exclude the possibility that cell-cycle arrest may in part contribute to the loss of RPA2, as shown previously for other factors (Szuts et al, 2003). However, these results suggest that DONSON is required for the recruitment of RPA2 to DNA lesions and for the activation of the DDR on γ and UV irradiation.
In addition to DONSON, we also tested several other phenotypically similar factors for a role in DDR signalling. We observed that knock down of SON, CADM1 and CD3EAP resulted in decreased phosphorylation of CHEK1 on γ irradiation (Figure 6A). Furthermore, depletion of SON impaired phosphorylation of RPA2, H2AX and NBS1 on UV irradiation. We also observed that RPA2 hyperphosphorylation is strongly attenuated after SON depletion in the chromatin fraction 2 h after UV exposure (Figure 6C).
In summary, our results suggest that SON, CADM1 and CD3EAP are components required for functional DDR response. Moreover, as ATM/ATR phosphorylation were not affected by the knock down of DONSON or SON, we integrate DONSON and SON in DDR signalling downstream the activation of ATM/ATR but upstream of RPA2 and NBS1.
Discussion
We present an experimental and computational approach to create a phenotypic map of a genome-wide set of RNAi-mediated perturbations, using automated phenotyping of cell populations by high-throughput imaging and multiparametric computational analysis. For ∼10% of targeted transcripts, we detected phenotypic changes on depletion of transcripts by RNAi. Similarity or dissimilarity of phenotypes was quantified from multiparametric descriptors by a computational method termed distance metric learning, which is able to learn a measure of (dis)similarity from a set of instances and to generalize this to unseen relationships. The derived measures for phenotypic similarity for each RNAi perturbation were visualized in a map and used to generate hypotheses about the function of genes. As the approach is unbiased, unexpected relationships can be uncovered. In this study, we investigated four previously poorly characterized genes, and described their roles in the DDR.
Cell morphology provides a broad reflection of many cellular processes, including cytoskeleton rearrangement, signalling, cell division and cell survival. Similar to forward genetic screens, the approach gives us the power to detect components in a wide range of processes. We believe that this approach ensures a coverage of many phenotypes and allows the analysis of multiple functions of the same gene, although it might miss modulators that could be detected with focused but more sensitive assays. However, it facilitates analysis of multiple functions of the same gene, and discovery of unanticipated functional relationships.
Quantifying similarity or dissimilarity between multiparametric phenotypic profiles is challenging, because of the different weights that can be given to the different parameters. For analysis on a genome-wide scale, an automated and objective approach is needed. We used distance metric learning to transform raw phenotypic profiles, which are a combination of genetic, environmental and stochastic variation, into biologically relevant phenoprints that capture the significant effects of the genetic perturbations. In this machine learning approach, the parameters used for the transformation are learned by presenting the algorithm with training sets of phenotypic profile pairs that are considered similar, as well as with pairs that are unrelated. The learned metric is then applied to unseen data. An advantage of this approach is that it does not rely on arbitrary cut-offs or ad hoc scoring functions. We note that distance metric learning has already been useful in other fields of biology. For instance, to compute the alignment score matrices used in protein sequence comparisons, such as PAM matrices (Dayhoff et al, 1978), their parameters are learned from a training set of naturally diverged sequences and the matrices are then used for inferring the degree of homology of protein sequences in general.
To test whether the prediction of gene function is accurate, we examined several candidates that were in phenotypic proximity to known components of the DDR and genomic integrity. We characterized the role of DONSON in DNA replication, proper spindle formation and the DDR. In agreement with our observation that knock down of DONSON led to the formation of multipolar spindles and the induction of G1 arrest, a loss of centrosomal integrity has been described earlier to result in checkpoint activation and inhibition of G1/S progression (Mikule et al, 2007). Moreover, several studies linked the loss of DDR components such as CHEK1 (Kramer et al, 2004) to impaired genomic integrity. We suggest that DONSON is part of a centrosomal network that integrates cell-cycle arrest and repairs in response to genotoxic stress (Tang et al, 2006; Loffler et al, 2007). This model is supported by the finding that the centrosomal protein CEP164 mediates the DDR by interacting with ATM and ATR, thus forming a link between the DDR and the centrosome (Sivasubramaniam et al, 2008). Furthermore, we showed that SON, CADM1 and CD3EAP are required for the DDR. Recently, the loss of SON has also been implicated in leukaemogenesis (Ahn et al, 2008). The reduced pCHEK1 response caused by CD3EAP knockdown is consistent with CD3EAP being a downstream target of ATR on induced DNA damage (Matsuoka et al, 2007). Interestingly, CD3EAP is encoded by a gene locus that overlaps with ERCC1, a DNA excision repair gene that is often mutated in colorectal cancer (Skjelbred et al, 2006). Loss of DDR components results in increased basal DNA damage and ultimately in a loss of genomic integrity. Consistently, knock down of the candidate genes DONSON, SON, CADM1 and CD3EAP led to increased γH2AX formation, supporting a function of these genes not only in the DDR but also in the maintenance of genomic integrity.
The results show that the calculated phenoprints generate useful hypotheses. The complete data set and computational methods are available at http://www.cellmorph.org and can serve as a resource for further functional discovery. Cell population-based phenotypic readouts are a sensitive method for deriving functional relationships and predict molecular functions.
Materials and methods
Cell culture
HeLa and U2OS cell lines were maintained in DMEM (Invitrogen) with 10% foetal bovine serum (FBS) (Invitrogen) and supplemented with penicillin (100 U/ml)/streptomycin (100 μg/ml) (Invitrogen). Cell lines were cultured at 37°C, 5% CO2 in a humidied incubator according to standard procedures.
Plasmids
An HA-tagged version of human DONSON was generated by modification of a DONSON cDNA clone (SC111799, OriGene). In brief, Don-HA forward and reverse primers (Supplementary Table I), and Phusion polymerase (Finnzymes) were used to amplify the DONSON coding sequence, cloned in frame with a C-terminal HA tag. The resulting PCR product was inserted into the pcDNA3.1(+) (Invitrogen) using NheI and EcoRI restriction sites. Cloning was confirmed by sequencing.
Generation of DONSON antibody
A rabbit anti-DONSON antibody was raised against a PGFRKPPEVVRLRRKRAR peptide (Charles River Laboratories). In western blots, the antibody specifically recognized a band corresponding to a size of 67 kDa. Specificity of the antibody was confirmed by siRNA knock down of DONSON.
Human siRNA library
The genome-wide siRNA library siGENOME (Dharmacon) was re-annotated against the NCBI RefSeq database (release 27). siRNA sequences were mapped to RefSeq transcripts and assigned to the corresponding gene. Out of the 21 061 siRNA pools, 17 145 were predicted to specifically target a single gene by perfect sequence identity.
High-throughput RNAi screening
For cell-based screening, the siRNA library was arrayed in black 384-well, clear bottom plates (Corning #3712 or BD Flacon #353962) using a Biomek FX200 liquid handling system (Beckman Coulter). Each well contained 5 μl of a 500 nM pool of four synthetic siRNA duplexes (dissolved in 1 × siRNA solution buffer, Dharmacon). Library siRNAs were spotted in columns 5–24; the remaining columns were used for controls. Positions A04 and B04 contained an siRNA pool targeting Rluc as negative control. Positions I04 and J04 contained an siRNA pool targeting PLK1. Reverse transfection of cells with siRNA pools was performed by delivering 15 μl of RPMI (Invitrogen) containing 0.05 μl of Dharmafect1 (Dharmacon). After 30 min of incubation at room temperature, 1000 HeLa cells in 30 μl of DMEM medium (Invitrogen) supplemented with 10% FBS (Invitrogen) were added to the siRNA transfection mix. All dispensing steps were performed with a Multidrop Combi dispensing system (Thermo). Plates were incubated for 48 h at 37°C/5% CO2.
For fluorescence microscopy, cells were fixed, permeabilized and immunostained for DNA, tubulin and actin using a Beckman FX200 liquid handling robot. First, cells were fixed with 5% paraformaldehyde (PFA) in PBS for 20 min at room temperature and then permeabilized with 0.2% Triton X-100 in PBS. Subsequently, cells were washed with 0.05% TX-100 in PBS and incubated with blocking buffer (0.05% TX-100/3% BSA in PBS). Cells were incubated overnight at 4°C with primary antibody anti-α-tubulin (Sigma, DM1A/T9026) diluted in PBS/0.05% TX-100/3% BSA. Then, cells were washed three times with PBS and incubated with Alexa 488 secondary antibody (1:500 in PBS, Invitrogen) for 45 min at room temperature. Actin was stained with TRITC-phalloidin (Sigma) in the secondary antibody solution, and DNA with Hoechst 33242 (Invitrogen). Cells were washed with PBS and stored therein. Batches of 10 384-well plates were processed per batch.
Automated imaging
Images were acquired on an automated BD Pathway 855 Bioimaging System (Becton Dickinson) with a 20 × objective (Olympus, NA=0.75) and a Hamamatsu monochrome digital black and white camera (Orca-ER). For high-throughput screening, plates were loaded onto the microscope with a Twister II Microplate robotic arm. Images of four different positions in each well were acquired, each containing channels for Hoechst (DNA), Alexa 488 (tubulin) and TRITC (actin). Each image had a resolution of 670 × 510 pixels with each channel at 12-bit intensity resolution. The total number of cells measured in a well was typically around 300. For the primary screen, a total of 22 839 wells were imaged in four spots each, using three channels, resulting in 274 068 grey scale images. The combination of the three channels leads to 91 356 three-colour images.
Automated phenotyping of cells
We used the EBImage package to perform the image processing operations (Pau et al, 2010). Images consisted of three channels (actin, tubulin and DNA). First, nuclei were segmented by adaptive thresholding of the DNA channel, distance map computation and watershed segmentation. Next, nuclei were regularized by morphological opening, and internal holes were removed by inverse flood fill (see Supplementary information). Cell boundaries were identified by Voronoi segmentation (Jones et al, 2009). Each cell was characterized by a set of 51 morphological descriptors, computed on different channels, which were grouped into 4 categories: 9 geometric features, 26 Haralick textural features, 11 Zernike moments and 5 miscellaneous features (see Supplementary information). All descriptors are translation and rotation invariant and are commonly used in cell imaging for classification purposes (Loo et al, 2007). Cells were classified into 1 of 10 classes (Supplementary Table II), using the ‘one-against-one’ multi-class strategy of the SVM algorithm, with a training set of 2545 manually annotated cells (Supplementary Tables III, IV, V and VI). Each well subject to siRNA perturbation was summarized by a vector of 13 numerical descriptors (Supplementary Table VII). Each value of a phenotypic profile was transformed to a [0,1] score using a set of parameterized monotonic functions, leading to a vector of 19 scores, which we termed a phenoprint. A phenoprint was considered a hit if at least one of its elements was larger than 0.5. The phenotypic distance between two perturbations was defined as the distance between two phenoprints, according to a modified L1 metric (see Supplementary information).
Assessment of the siRNA target specificity
To confirm siRNA-mediated phenotypic changes observed in the genome-wide screen, we used independent siRNA pools (Qiagen) for 604 gene transcripts (Supplementary Table X). The phenotype of the siRNA perturbation was monitored by microscopy as described above in HeLa and U2OS cells. Phenotypic changes were visually confirmed for 51% of target genes. For 60 of the 604 target genes, we additionally used individual siRNA duplexes (Dharmacon). In this study, 73% of phenotypic changes were visually confirmed with at least two out of four individual siRNAs (Supplementary Table XI). siRNA sequences are listed in Supplementary Table XII and can also be accessed at http://rnai.dkfz.de.
DNA content profiling using high-throughput immunocytometry
After transfection with siRNAs, cells were fixed by adding ice-cold 80% EtOH for 30 min. Cells were rehydrated by washing two times with PBS. After RNaseA digest (100 μg/ml) for 1 h at 37°C, cells were stained for 15 min in PBS containing 10 μg/ml propidium iodide (Molecular Probes). The stained cells were scanned with an Acumen Explorer eX3 microplate cytometer (TTP LabTech). The resulting DNA content histograms were manually gated.
Assessment of γH2AX foci formation
For qualitative analysis of γH2AX foci formation, cells were reverse transfected on glass coverslips. Cells were fixed, permeabilized and incubated with the primary antibody to phospho-H2AX (Upstate Biotechnology, 1:300) and subsequently with Alexa 488-conjugated secondary antibody (Invitrogen, 1:500). DNA was counterstained with Hoechst 33342 (Sigma, 1:1000 in PBS). Coverslips were mounted onto glass slides with Fluoromount G (Southern Biotech) mounting media. An Axioimager Z1 Microscope (Zeiss) equipped with a × 63 oil objective (Zeiss, NA=1.4) and an apotome were used for image acquisition.
Quantitative assessment of γH2AX foci formation was performed by reverse transfection of cells with siRNAs in 384-well plates. After incubation for 72 h, γH2AX foci formation was monitored by immunofluorescence staining of phospho-H2AX as described above and acquired using an Acumen Explorer microplate reader.
Western blot
Forty-eight hours after siRNA transfection, cells were either γ irradiated (10 Gy) or UVC irradiated (20 J/m2) and collected 1 and 2 h later. Whole-cell lysates were prepared in urea buffer (8 M urea, 0.1 M NaH2PO4, 10 mM TRIS–HCl, pH 7.5–8, protease inhibitors). Samples were subjected to SDS–PAGE (NuPage, 4–12% Bis–Tris; Invitrogen), transferred to PVDF membrane (Immobilon, Millipore) and subsequently incubated with antibodies. The following primary antibodies were used: CHEK1 (#2345), pCHEK1 (#2344), pNBS1 (#3001), NBS1 (#3000), cyclin A (#4656), pATM (#4526), ATM (#2873) and MCM2 (#4007) from Cell Signaling Technology, cyclin D1 (SC-8396) from Santa Cruz, RPA2 (NA19L) and PARP (AM30) from Calbiochem, phospho-H2AX (#07-164) and cyclin B1 (#05-373) from Millipore, actin (ab6276) from Abcam and SON (HPA023535) from Sigma. Secondary HRP-conjugated anti-mouse or rabbit antibodies were obtained from Amersham.
For chromatin isolation, cells were washed with PBS, resuspended in CSK buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 50 mM NaF, 0.1 mM Na-orthovanadate (Sigma-Aldrich), 0.1% Triton X-100 (Sigma-Aldrich) and protease inhibitors (Roche)), and incubated on ice for 10 min. Cytoplasmic proteins were separated from nucleic proteins by low-speed centrifugation at 1300 g for 5 min. Isolated nuclei proteins were washed once in CSK buffer and lysed in solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitors). After centrifugation (1700 g for 5 min), pellets were resuspended in urea buffer, 2 × SDS loading buffer was added and samples were boiled for 10 min.
Subcellular localization of DONSON
Cells were transfected with hDonson-HA plasmid using Trans-IT LT (Mirus) transfection reagent; 24 h after plasmid transfection, cells were either fixed in −20°C methanol/acetone (1:1) for 7 min or PFA after permeabilization with PBS supplemented with 0.05% Triton X-100. Immunofluorescence microscopy was performed with rabbit anti-HA polyclonal antibodies (Sigma #H6908) and mouse anti-α-tubulin (Sigma #T6557) or rabbit anti-centrin (Sigma #C7736) and mouse anti-HA antibodies (CST #2367). Alexa Fluor 488 goat anti-rabbit (Invitrogen #A-11008), Alexa Fluor 488 goat anti-mouse (Invitrogen #A-11001), Alexa Fluor 594 goat anti-mouse (Invitrogen #A-11005) and Alexa Fluor 594 goat anti-rabbit (Invitrogen #A-11012) were used as secondary antibodies. Images were acquired using an Axio CellObserver (Zeiss) equipped with a Colibri LED light source, standard fluorescence filters and a 63 × oil objective (Zeiss, NA=1.4).
Quantitative real-time PCR
Forty-eight hours after siRNA transfection, RNA was isolated with the RNeasy Mini kit (Qiagen) and 1 μg RNA was used as a template for cDNA synthesis using oligo-dT primer and the RevertAid H Minus First Stand cDNA Synthesis kit (Fermentas #K1632). Expression levels were normalized against GAPDH expression. The level of target gene knockdown was quantified in three independent experiments. Quantifications were performed using the Universal Probe Library (Roche) on a Light Cycler 480 (Roche). Primers for real-time PCR were designed using ProbeFinder (Roche) and are provided in Supplementary Table I.
Supplementary Material
Supplementary text, Supplementary figures S1–15, Supplementary tables SI–XII
Acknowledgments
We are grateful to Moritz Gilsdorf and Zeynep Arziman for bioinformatic support, Aennas Abbas for technical support and Richard Bourgon for helpful comments on the paper. TH is supported by a fellowship of the Studienstiftung. The research was supported in part by a Marie Curie Excellence Grant from the European Commission, the Helmholtz Association Alliance for Systems Biology, the BMBF and a Research Grant from the Human Frontiers Sciences Program Organization.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE (2008) Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA 105: 17103–17108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C, Aach J, Church GM, Perrimon N (2007) Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316: 1753–1756 [DOI] [PubMed] [Google Scholar]
- Bier E (2005) Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet 6: 9–23 [DOI] [PubMed] [Google Scholar]
- Boser BE, Guyon IM, Vapnik VN (1992) A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA [Google Scholar]
- Boutros M, Ahringer J (2008) The art and design of genetic screens: RNA interference. Nat Rev Genet 9: 554–566 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon RT, Perrimon N (2005) Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308: 826–833 [DOI] [PubMed] [Google Scholar]
- Dayhoff M, Schwartz B, Orcutt B (1978) A model of evolutionary change in protein. Atlas Protein Seq Struct 5: 345–352 [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T (2004) siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 3: 318–329 [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J et al. (2006) Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3: 777–779 [DOI] [PubMed] [Google Scholar]
- Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM (2004) Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol 2: e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA (2007) Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41 [DOI] [PubMed] [Google Scholar]
- Guo Q, Vasile E, Krieger M (1994) Disruptions in Golgi structure and membrane traffic in a conditional lethal mammalian cell mutant are corrected by epsilon-COP. J Cell Biol 125: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM (2009) Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci USA 106: 1826–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen EM, Mango SE (2002) The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet 3: 356–369 [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N (2003) A functional genomic analysis of cell morphology using RNA interference. J Biol 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Krumpelman C, Marcotte EW (2008) Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol 9: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J (2004) Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol 6: 884–891 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (2000) Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell 6: 839–849 [DOI] [PubMed] [Google Scholar]
- Liu E, Lee AY, Chiba T, Olson E, Sun P, Wu X (2007) The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J Cell Biol 179: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bekker-Jenen S, Mailand N, Lukas C, Bartek J, Lukas J (2006) Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26: 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H, Bochtler T, Fritz B, Tews B, Ho AD, Lukas J, Bartek J, Kramer A (2007) DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle 6: 2541–2548 [DOI] [PubMed] [Google Scholar]
- Loncarek J, Sluder G, Khodjakov A (2007) Centriole biogenesis: a tale of two pathways. Nat Cell Biol 9: 736–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LH, Wu LF, Altschuler SJ (2007) Image-based multivariate profiling of drug responses from single cells. Nat Methods 4: 445–453 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S (2007) Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol 9: 160–170 [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, Sahasrabudhe S, Reinhardt M, Natt F, Hall J, Mickanin C, Labow M, Chanda SK, Cho CY, Schultz PG (2006) Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA 103: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J (2006) High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods 3: 385–390 [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R (2006) Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA 103: 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Pau G, Fuchs F, Sklyar O, Boutros M, Huber W (2010) EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics 26: 979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Scheel J, Horstmann H, Hauri HP, Griffiths G, Kreis TE (1993) Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell 74: 71–82 [DOI] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ (2004) Multidimensional drug profiling by automated microscopy. Science 306: 1194–1198 [DOI] [PubMed] [Google Scholar]
- Pouwels J, Kukkonen AM, Lan W, Daum JR, Gorbsky GJ, Stukenberg T, Kallio MJ (2007) Shugoshin 1 plays a central role in kinetochore assembly and is required for kinetochore targeting of Plk1. Cell Cycle 6: 672–678 [DOI] [PubMed] [Google Scholar]
- Sims D, Duchek P, Baum B (2009) PDGF/VEGF signaling controls cell size in Drosophila. Genome Biol 10: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY (2008) Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev 22: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjelbred CF, Saebo M, Nexo BA, Wallin H, Hansteen IL, Vogel U, Kure EH (2006) Effects of polymorphisms in ERCC1, ASE-1 and RAI on the risk of colorectal carcinomas and adenomas: a case control study. BMC Cancer 6: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- St Johnston D (2002) The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet 3: 176–188 [DOI] [PubMed] [Google Scholar]
- Szuts D, Kitching L, Christov C, Budd A, Peak-Chew S, Krude T (2003) RPA is an initiation factor for human chromosomal DNA replication. Nucleic Acids Res 31: 1725–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kurose A, Huang X, Traganos F, Dai W, Darzynkiewicz Z (2006) Extent of constitutive histone H2AX phosphorylation on Ser-139 varies in cells with different TP53 status. Cell Prolif 39: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Erikson RL, Liu X (2006) Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1). Proc Natl Acad Sci USA 103: 11964–11969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L (2008) A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci USA 105: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P (2007) STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35: D358–D362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem 282: 33444–33451 [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA (2007) Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446: 815–819 [DOI] [PubMed] [Google Scholar]
- Xing EP, Ng AY, Jordan I, Russel S (2003) Distance metric learning, with application to clustering with side-information. Adv Neural Inf Process Syst 15: 505–512 [Google Scholar]
- Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y (2003) Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res 63: 980–986 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary figures S1–15, Supplementary tables SI–XII