Abstract
Recognition of microbial danger signals by toll-like receptors (TLR) causes re-programming of macrophages. To investigate kinase cascades triggered by the TLR4 ligand lipopolysaccharide (LPS) on systems level, we performed a global, quantitative and kinetic analysis of the phosphoproteome of primary macrophages using stable isotope labelling with amino acids in cell culture, phosphopeptide enrichment and high-resolution mass spectrometry. In parallel, nascent RNA was profiled to link transcription factor (TF) phosphorylation to TLR4-induced transcriptional activation. We reproducibly identified 1850 phosphoproteins with 6956 phosphorylation sites, two thirds of which were not reported earlier. LPS caused major dynamic changes in the phosphoproteome (24% up-regulation and 9% down-regulation). Functional bioinformatic analyses confirmed canonical players of the TLR pathway and highlighted other signalling modules (e.g. mTOR, ATM/ATR kinases) and the cytoskeleton as hotspots of LPS-regulated phosphorylation. Finally, weaving together phosphoproteome and nascent transcriptome data by in silico promoter analysis, we implicated several phosphorylated TFs in primary LPS-controlled gene expression.
Keywords: macrophage, nascent RNA, phosphoproteome, SILAC, toll-like receptors
Introduction
Macrophages reside in all tissues and continuously sample their environment by phagocytosis and endocytosis. They sense invading pathogens through pattern recognition receptors (PRRs) that bind common microbial structures. The best characterised group of PRRs is the evolutionary conserved family of toll-like receptors (TLRs), transmembrane proteins expressed on the cell surface (e.g. TLR2, TLR4, TLR5) or in the endosome (e.g. TLR7–9) (Takeda and Akira, 2004). Stimulation of macrophages in vitro with the TLR4 agonist lipopolysaccharide (LPS) of Gram-negative bacteria causes within a few hours substantial re-programming of gene expression (Huang et al, 2001; Lang et al, 2002; Nau et al, 2002; Foster et al, 2007; Mages et al, 2007). This rapid response is pivotal for control of pathogen replication, and includes production of chemokines, which recruit leukocytes to the site of infection, anti-microbial effector molecules and cytokines that initiate and control the adaptive immune response.
After recognition of microbial ligands, TLR signalling is initiated by binding of the adapter molecule MyD88 to the cytoplasmic Toll/IL-1R domain present in all TLRs. Recruitment of IL-1R-associated kinases (IRAK4, IRAK1) and the adapter protein TNF-receptor-associated factor 6 triggers kinase cascades that result in activation of the MAPK and NFkB pathways (Takeda and Akira, 2004). This core pathway has been shown by many pieces of pharmacological and genetic evidence, and controls gene expression by activation of latent transcription factors (TFs) (e.g. NFkB proteins and CREB family members) and by effects on mRNA stability (Hao and Baltimore, 2009). Phosphorylation has an essential role in TF activation: in the case of NFkB, phosphorylation of the NFkB-bound inhibitor IkB by the IKK complex is the first step in the process leading to IkB degradation, release of active NFkB and translocation to the nucleus (Vallabhapurapu and Karin, 2009); CREB and IRF family TFs form dimers after phosphorylation, enabling them to enter the nucleus and transactivate promoters of cytokine and chemokine genes (Honda and Taniguchi, 2006).
To avoid excessive inflammation, macrophage activation is controlled by endogenous regulators, such as the immunosuppressive cytokine IL-10 (Lang, 2005; Liew et al, 2005). Down-regulation is reflected on the level of signal transduction by the transient activation of key signalling modules. NFkB signalling, for example, is down-regulated by re-synthesis of IkB protein and export of NFkB from the nucleus (Vallabhapurapu and Karin, 2009). De-phosphorylation and inactivation of MAPKs is brought about by members of the MAPK phosphatase family, for example, by Dual specificity phosphatase 1 (DUSP1), that is induced by LPS in macrophages and prevents excessive cytokine production by deactivating p38 MAPK (Chi et al, 2006; Hammer et al, 2006; Salojin et al, 2006; Zhao et al, 2006).
TLR signalling has been extensively studied. A recent review of TLR signalling compiled a network of 340 proteins and 444 reactions involved in TLR signalling (Oda and Kitano, 2006). However, a comprehensive analysis of phosphorylation events in macrophages in response to TLR stimulation is missing. A former study restricted to tyrosine phosphorylation was further limited by its non-quantitative nature and did not use primary cells (Aki et al, 2005). Therefore, it is unknown (1) whether the canonical pathways described above comprise the main phosphorylation events, kinases and TFs for gene expression re-programming, and (2) which other molecular functions and biological processes are regulated by phosphorylation in LPS-activated macrophages.
Recent progress in mass spectrometry-based proteomics driven by leaps in instrument performance and advances in computational proteomics has opened the possibility to quantitatively investigate global changes in protein abundance and post-translational modifications (Cox and Mann, 2007). Stable isotope labelling with amino acids in cell culture (SILAC) allows mixing of samples before enrichment and fractionation steps, and has proved especially useful for direct comparison of phosphopeptide abundance in time course or treatment analyses (Olsen et al, 2006; Kruger et al, 2008; Pan et al, 2008).
Here, we combined SILAC, phosphopeptide enrichment and high-accuracy mass spectrometry to analyse the phosphoproteome changes in resting versus LPS-activated primary bone marrow-derived macrophages. We report the identification of nearly 7000 phosphorylation sites on more than 1800 phosphoproteins in macrophages, with a large fraction of up-regulated and down-regulated phosphorylation sites in response to LPS activation. Bioinformatic analyses found enrichment of pathways associated with TLR signalling, in addition revealed the cytoskeleton as a hotspot for phosphorylation in macrophages, and highlighted other biological processes and functions. In parallel, we analysed LPS-induced de novo transcription by Affymetrix microarrays of purified 4-thiouridine (4sU)-tagged RNA (‘nascent RNA’) (Dolken et al, 2008). By integrating TF phosphorylation with nascent transcriptome data using in silico promoter analysis we identified transcriptional regulators previously not implicated in TLR-induced gene expression.
Results
Quantitative phosphoproteome analysis of primary macrophages
Our global and quantitative analysis of phosphorylation sites in macrophages builds on a previously described strategy combining SILAC for quantification, strong cation exchange chromatography (SCX) and titanium dioxide (TiO2) chromatography for phosphopeptide enrichment and high-accuracy mass spectrometric characterisation (Olsen et al, 2006), which we optimised for use with primary bone marrow-derived macrophages (Figure 1A).
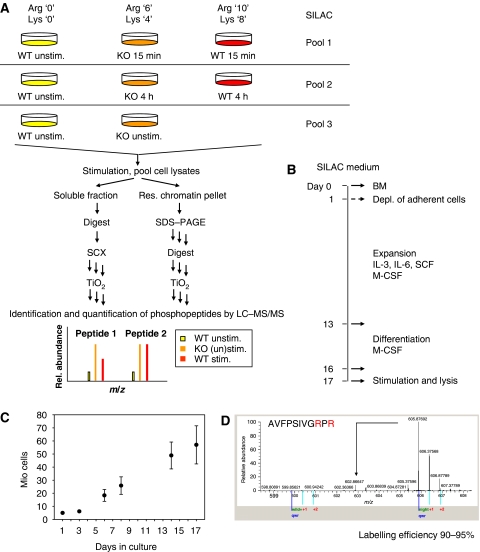
Figure 1.
Experimental system and design. (A) Strategy for global and quantitative analysis of LPS-induced phosphorylation. Bone marrow cells from wild type (WT) and Dusp1-deficient (KO) mice were SILAC encoded with normal and stable isotope-substituted arginine and lysine amino acids, creating three states distinguishable by mass ((m/z) mass/charge). Each population was stimulated with LPS for 15 min or 4 h or left un-treated. Unstimulated wild-type cells were included in all three pools as a common reference point. Cell lysates to be directly compared were pooled, fractionated and enzymatically digested into peptides, and phosphopeptides were enriched on TiO2 beads and analysed by online LC-MS/MS. Owing to the mass shifts introduced by the SILAC amino acids mass spectra of labelled peptides revealed SILAC triplets (same peptide from the three cell populations), with the intensities of the peaks reflecting the relative amounts of a peptide in the three conditions. This SILAC-based approach allowed high-accuracy quantification of phosphopeptides and, in most cases, localisation of the phosphate group with single amino acid accuracy. Two independent experiments were performed. (B) Optimised protocol for SILAC of bone marrow-derived macrophages. (C) Cell proliferation under the SILAC protocol. Total number of cells at different time points during SILAC labelling (mean±standard deviation from two independent experiments). (D) Labelling efficiency. Representative peptide containing two arginine residues. The arrow indicates the position of partially labelled peptide.
SILAC requires sufficient time of cell culture for a full labelling of all proteins with heavy isotope versions of essential amino acids. We therefore adapted the standard protocol for generation of bone marrow-derived macrophages by inducing expansion of progenitor cells with the cytokines IL-3, IL-6 and SCF in the presence of macrophage colony stimulating factor (M-CSF). After expansion, cells were differentiated into macrophages with M-CSF only (Figure 1B). This 17-day protocol yielded large numbers of cells (Figure 1C) and resulted in a high-labelling efficiency (Figure 1D). Macrophages obtained by the standard or SILAC-adapted protocol were comparable in terms of surface marker expression (F4/80, CD11b), and responded equally to LPS stimulation with activation of p38 MAPK and production of inflammatory cytokines (Supplementary Figure S1).
Macrophages were SILAC encoded with both arginine and lysine using three distinct isotopic forms. Pooling samples from three different labelling conditions for further preparation ensures equal sample treatment and highly accurate quantification. Comparison of more than three conditions can be achieved by including a common reference lysate in several pools, which is used for calculation of phosphopeptide ratios. Here, we analysed the phosphoproteome of macrophages in response to LPS. Pools of lysates were prepared from WT and Dusp1-deficient macrophages stimulated with LPS for 15 min or 4 h (Figure 1A). After fractionation, tryptic digest and phosphopeptide enrichment, online liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed as described in ‘Materials and methods’.
We reproducibly identified 6956 phosphorylation sites on 1850 proteins with single amino acid accuracy (according to the PTM score; Olsen et al, 2006), more than 60% of which were novel with respect to the phosphorylation site database Expasy (containing all Swiss-Prot/TrEMBL entries; http://www.expasy.ch) and a recent phosphoproteome study in the mouse liver cell line Hepa1-6 (Pan et al, 2008) (Supplementary Table S1). The overlap between our two completely independent experiments was 63–89%, depending on the experiment referred to (Figure 2A). For bioinformatic analyses, we focused on reproducibly identified phosphorylation sites, if not indicated otherwise. Validation of phosphosites identified by mass spectrometry can be done by immunoblotting in cases where phosphorylation site-specific antibodies are available. We confirmed the regulated phosphorylation of GSK3β at S9 and ribosomal protein S6 at S235/236 (Supplementary Figure S2), the phosphorylation of p38 MAPK (Mapk14) at T180 and Y182 (Supplementary Figure S1) and of ERK1 MAPK (Mapk3) at T203 and Y205 (4D). In addition, the strong phosphorylation of ATF2 and TTP (Zfp36) at various residues was reflected by the higher molecular weight bands observed for LPS-treated samples in western blot analysis (Supplementary Figure S2). We also confirmed that macrophages grown using the SILAC protocol were very similar to macrophages obtained with the standard protocol under M-CSF, in extent and kinetics of phosphorylation of ribosomal protein S6 and Stat1 (Supplementary Figure S3). In accordance with previous reports from other cellular systems (Olsen et al, 2006; Villen et al, 2007; Pan et al, 2008) most phosphorylation sites were on serine (84%) and threonine residues (14%), whereas tyrosine phosphorylation occurred only in 2% of the cases (Figure 2B).
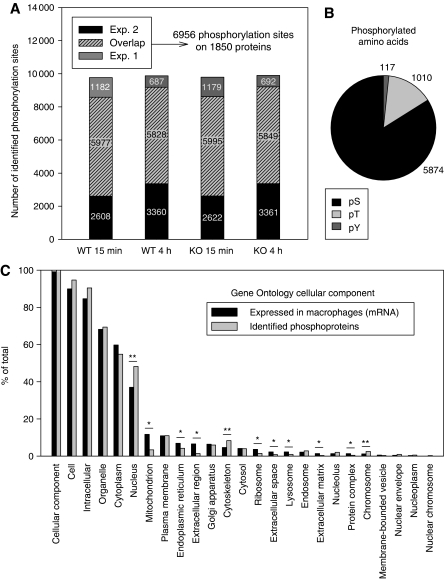
Figure 2.
Macrophage phosphorylation sites and proteins. (A) Overlap of phosphorylation sites identified in two independent experiments. Depicted are phosphorylation sites that could be quantified in the indicated conditions relative to unstimulated wild type. All following bioinformatic analyses focus on reproducibly identified phosphorylation sites, if not indicated otherwise. (B) Distribution of phosphorylated amino acids. Total numbers of quantified serine- (pS), threonine- (pT), tyrosine- (pY) phosphorylation sites. (C) Distribution of phosphorylated proteins in cellular compartments. Genes expressed in macrophages (see Supplementary information) and identified phosphoproteins were assigned to GOSlim GO terms for cellular component using the GO browser of Spotfire Decision Site. The number of proteins associated with each GO term is referred to the total number of proteins in the respective list (% of total). Significantly over- and under-represented GO terms are marked (odds ratio ⩾1.3 (**) or ⩽0.76 (*) and corrected P-value ⩽0.05).
We detected phosphoproteins from all cellular compartments. A comparison of the Gene Ontology (GO) annotation for cellular component between the identified phosphoproteins and genes expressed in macrophages showed an overall similar distribution (Figure 2C). As expected, extracellular proteins were under-represented among phosphoproteins. We also observed a relative paucity of proteins from the mitochondria, ribosomes, endoplasmatic reticulum and lysosomes among phosphorylated proteins. This is in line with previous observations of low phosphorylation (Olsen et al, 2006) and protein kinases expression (Pagliarini et al, 2008) in the mitochondria. Surprisingly, the plasma membrane was well represented among phosphoproteins. Of note, we observed an over-representation of the terms ‘nucleus’, ‘chromosome’ and ‘cytoskeleton’ among phosphoproteins.
Dynamics of the phosphoproteome after TLR4 activation
We investigated changes in phosphorylation dependent on LPS, time and the MAPK phosphatase DUSP1. Deletion of DUSP1 had only a small impact on the phosphoproteome, with <1.3% of all phosphopeptides hyper-phosphorylated in resting and 2% in LPS-activated Dusp1-deficient macrophages (data not shown). However, in contrast to the strong and reproducible effects of LPS in wild-type cells (see below), we observed a high degree of variability between experiments in Dusp1-deficient macrophages. We therefore decided to focus here on the effect of LPS on the phosphoproteome in wild-type cells.
Stimulation with LPS strongly affected the phosphoproteome at both time points. Overall, phosphorylation of 24% of all sites was up-regulated and of 9% was down-regulated in response to LPS in wild-type cells (Figure 3A and B). The extent of regulation by LPS was very similar between short and prolonged stimulation (Supplementary Figure S4). Analysis of the kinetic profiles (Figure 3C) revealed that the phosphorylation status was sustained over time for only 36% of up-regulated and 25% of down-regulated phosphorylation sites (Figure 3D, black bars; Figure 3C). All other sites changed their phosphorylation status over time (Figure 3D, light grey bars; Figure 3C) or were detected at one of the time points only (Figure 3D), arguing against presence at the other time point in high amounts. Taken together, the changes in phosphorylation were of a highly dynamic nature for a large fraction of regulated phosphorylation sites.
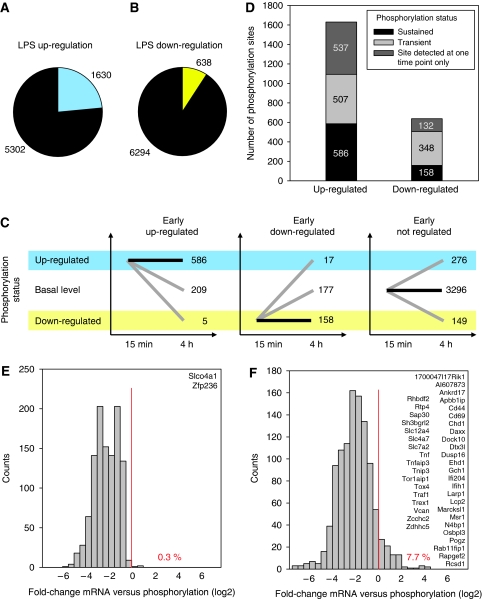
Figure 3.
Regulation of phosphorylation. (A, B) Extent of regulation by LPS. A fold-change of at least 1.5 in both experiments was used as threshold to define (A) up-regulated and (B) down-regulated phosphorylation sites in WT cells. These criteria can be considered very stringent, as <0.5% of the identified non-phosphorylated peptides were found to change more than 1.5-fold within 15 min after stimulation (data not shown). The distribution of ratios for phosphopeptides and non-phosphorylated peptides is shown in Supplementary Figure S8. (C, D) Kinetics of regulation. (C) Kinetic profiles of phosphorylation sites identified in wild-type cells at both time points. (D) Phosphorylation sites were detected at one of the time points only (dark grey) or at both time points, with either sustained (black) or transient phosphorylation status (light grey). (C) Kinetic profiles of phosphorylation sites identified in wild-type cells at both time points. (D) Kinetic status of phosphorylation for LPS-regulated sites. (E, F) Contribution of gene expression changes to regulation of the phosphoproteome. Transcriptome analyses using Affymetrix Mouse Gene ST 1.0 microarrays were performed on LPS-treated macrophages cultured under identical conditions as for the phosphoproteome experiments (except SILAC). Changes in gene expression (total RNA; 45 min and 4.5 h LPS treatment) relative to changes in phosphorylation (15 min, 4 h) are shown for LPS up-regulated phosphorylation sites (E) early and (F) late after stimulation (log2 ratio of mean fold-changes from two independent experiments, 0.5-bins). The percentage of phosphorylation sites with stronger change in gene expression than in phosphorylation and gene symbols of affected proteins are indicated.
To estimate the contribution of LPS-induced changes in gene expression to changes in phosphorylation, transcriptome analyses were performed on macrophages cultured under identical conditions and stimulated with LPS for 45 min or 4.5 h. Early after stimulation, only two genes with up-regulated protein phosphorylation showed a stronger increase in total mRNA levels (Figure 3E). At the late time point, regulation of gene expression was more common, but more than 90% of all up-regulated phosphorylation sites had a stronger change in phosphorylation than in gene expression (Figure 3F), indicating that changes in gene expression in most cases do not account for the increased phosphorylation.
Kinase activity induced by LPS
Each of the phosphorylation sites identified here is the substrate of one or more kinases. To obtain footprints of kinase activation in response to TLR ligation, we searched the phosphopeptide sequences for the known substrate specificities of 33 human kinases (http://www.phosida.com), which have been shown to match well with murine phosphorylation sites (Pan et al, 2008), and determined motifs enriched among LPS up-regulated compared to non-regulated phosphorylation sites (Table I). Fifteen minutes after stimulation the strongest over-representation was observed for the ERK/MAPK motif, which is in accordance with the known role of the MAPK module in TLR signalling. Other over-represented motifs were associated with kinases recently described in the context of TLR signalling: PKD has a role in TLR9 and TLR5 signalling (Ivison et al, 2007; Park et al, 2009); activation of AKT and its targets GSK3 and the mammalian target of rapamycin (mTOR) have recently been shown in response to TLR stimulation (Cao et al, 2008; Schmitz et al, 2008); Ca2+-dependent activation of CAMK2 is required for the expression of many LPS-target genes (Liu et al, 2008). Other kinases, among them the DNA damage-activated kinases ATM/ATR and the cell cycle-associated kinases AURORA and CHK1/2, have not been linked to the response to LPS. Many of the associated kinases were expressed in macrophages and some had LPS-regulated phosphorylation sites themselves potentially modulating kinase activity (Table I). Interestingly, over-representation of several motifs, including the ERK/MAPK motif, was also observed for down-regulated phosphorylation sites (data not shown), suggesting that TLR signalling also triggers down-regulation of this type of phosphorylation by activation of phosphatases or degradation of the phosphorylated proteins.
Table 1. Kinases activated during TLR4 signalling.
| Time | Motif | Enrichment (odds ratio) | Kinase expression | Kinase phosphorylation |
|---|---|---|---|---|
| Kinase motifs (http://www.phosida.com) over-represented in LPS up-regulated compared to non-regulated phosphorylation sites (odds ratio ⩾1.3 and corrected P-value ⩽0.05), suggesting kinase activation in response to LPS. Expression of associated kinases determined in the corresponding microarray experiments and identification of phosphopeptides from the kinases themselves are indicated by superscripts ‘a’ and ‘b’. | ||||
| aPhosphorylation regulated (⩾1.5-fold) at the respective time point. | ||||
| bExpression regulated (⩾2-fold). | ||||
| 15 min | ERK/MAPK | 3.7 | Mapk1, 3, 4, 6-9, 11-15 | Mapk3a, 6, 9, 10a, 14a |
| ATM/ATR | 3.5 | Atm, Atr | — | |
| PKD | 3.4 | Pkd1, Pkdl1-3, Pkd2 | — | |
| CHK1 | 3.2 | Chkab, Chkb-cpt1b | — | |
| AURORA | 2.5 | Aurora-c, Aurkaip1 | — | |
| CAMK2 | 2.1 | Camk2a, b, d, g, n1, n2 | Camk2d, Camkk2 | |
| PLK | 1.9 | Plk1, 2b, 3, 4 | — | |
| NEK6 | 1.7 | Nek1-9 | Nek3, 9 | |
| PKA | 1.6 | Prkaa1-2, -b1-2, -ca-b, -g1-3, -r1a-b, -r2a-b | Prkaa1a, -b1a, -g2a, r1a, -r2a, -r2b | |
| CK1 | 1.6 | Ckb, Ckm, Ckmt1-2 | — | |
| GSK3 | 1.6 | Gsk3a, Gsk3b | Gsk3a, Gsk3ba | |
| AKT (PKB) | 1.4 | Akt1, Akt2, Akt3, Akts1, Aktip | Akt, Akt1s1, Aktip | |
| 4 h | PKD | 2.0 | Pkd1, Pkdl1-3, Pkd2 | — |
| CHK1 | 1.9 | Chka, Chkb-cpt1b | — | |
| ERK/MAPK | 1.8 | Mapk1, 3, 4, -6-9, 11-15 | Mapk3a, 6, 9, 10, 14 | |
Association of LPS-regulated phosphoproteins with signalling pathways and functional annotation
To test whether TLR4-induced phosphorylation preferentially targets specific signalling pathways and cellular processes, we made use of two annotation systems: InnateDB, a database integrating pathway information from several other sources (http://www.innateDB.ca) and the GOSlim GO annotation (http://www.geneontology.org/GO) for molecular functions and biological processes.
We identified 48 phosphoproteins annotated as members of the murine or human TLR, MAPK or NFKB signalling pathways in InnateDB or on the innate immunity signalling poster compiled by Latz and Fitzgerald (2008), 31 of which showed LPS-regulated phosphorylation (Supplementary Table S2). The pathway annotation ‘TLR signalling’ showed a trend for enrichment among LPS-regulated phosphoproteins compared to non-regulated phosphoproteins (odds ratio 2.4; P-value 0.15). Significant over-representation was found for MAPK signalling members and pathways recently described as activated downstream of TLRs, for example, the AKT and mTOR pathways and the Rho GTPase cycle (Ruse and Knaus, 2006) (Table IIA; Supplementary Figure S5). GO analysis showed enrichment of the terms ‘signal transduction’, ‘cell communication’ and ‘kinase activity’ (Table IIB; Supplementary Table S3). Interestingly, functional annotation terms associated with the cytoskeleton were also significantly enriched among LPS-regulated phosphoproteins. ‘Cell proliferation’ showed a trend for over-representation (odds ratio 4.6; corrected P-value 0.12), consistent with over-representation of motifs for cell cycle-associated kinases observed above. In summary, unbiased statistical analyses of kinase motifs, signalling pathways and functional GO annotation highlighted known and novel players of TLR signalling and linked TLR activation to the cytoskeleton and cell proliferation (see overview in Figure 4).
Table 2. Signalling pathways, molecular functions and biological processes targeted by LPS-regulated phosphorylation.
| Enrichment (odds ratio) |
|||
|---|---|---|---|
| Overall | 15 min | 4 h | |
| (A) Over-represented signalling pathways. Signalling pathways (http://www.innateDB.ca; Lynn et al, 2008) with at least five identified phosphoproteins were analysed for over-representation among LPS-regulated phosphoproteins compared to non-regulated phosphoproteins (odds ratio ⩾1.3 and P-value ⩽0.05; below cut-off: P-value criterion not met, details see text). Depicted are over-represented pathways that did not show more than 75% overlap with MAPK, AKT and mTOR signalling. | |||
| (B) Over-represented Gene Ontology terms. Phosphoproteins were assigned to GOSlim Gene Ontology terms for molecular functions and biological processes (http://www.geneontology.org/GO), and over-representation among LPS-regulated phosphoproteins compared to non-regulated phosphoproteins was determined (odds ratio ⩾1.3 and corrected P-value ⩽0.05; below cut-off: P-value criterion not met, details see text) for terms with at least three identified phosphoproteins. | |||
| (A) Pathway name | |||
| MTOR signalling pathway | >16.0 | >17.9 | 6.5 |
| Adipocytokine signalling pathway | >11.0 | > 8.0 | |
| AKT phosphorylates targets in the cytosol | >8.0 | > 6.0 | |
| AKT (PKB)-Bad signalling | 2.5 | ||
| EGFR1 | >8.0 | ||
| Caspase-mediated cleavage of cytoskeletal proteins | >8.0 | 8.5 | 7.8 |
| TGF-β signalling pathway | >7.0 | ||
| TNF-α | >7.0 | ||
| Insulin signalling pathway | 3.0 | 2.6 | |
| MAPK signalling pathway | 2.0 | ||
| Rho GTPase cycle | 2.8 | ||
| Below cut-off | |||
| Toll-like receptor signalling pathway | 2.4 | 1.4 | 3.3 |
| (B) Gene ontology term | |||
| Signal transduction | 3.1 | 2.6 | 1.9 |
| Cell communication | 2.8 | 2.6 | 2.1 |
| Actin binding | 2.3 | 2.9 | |
| Cytoskeletal protein binding | 2.3 | 2.4 | |
| Kinase activity | 1.7 | ||
| Below cut-off | |||
| Cell proliferation | 4.6 | 2.1 | 3.0 |
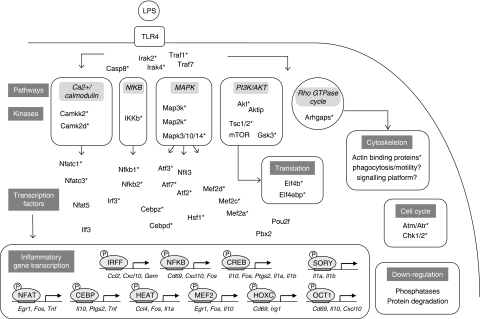
Figure 4.
The phosphoproteome of LPS-activated macrophages—overview. Hotspots of TLR4-induced phosphorylation at the level of pathways, kinases and biological processes are summarised. Various signalling modules, cytoskeletal re-arrangement, cell cycle proteins and the translation machinery use the reversible protein modification for controlled activation that is both rapid and transient, as accomplished by phosphatase activity and phosphoprotein degradation. Most importantly, changes in phosphorylation activate TFs followed by inflammatory gene transcription indispensable for host defence. Selected examples of phosphoproteins are shown for each process. Depicted TFs have enriched evolutionary conserved binding sites in the promoters of highly induced LPS-target genes or were experimentally validated. Asterisks indicate LPS-regulated phosphorylation on a protein or kinase target.
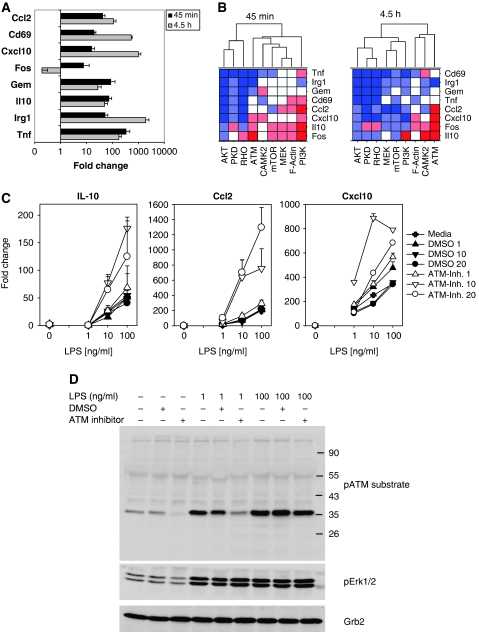
To investigate the functional relevance of kinases (PI3K, AKT, CAMK2, ATM, PKD, MEK1, mTOR) and pathways (cytoskeletal rearrangement, Rho GTPases) enriched among LPS-regulated phosphoproteins, we used a panel of pharmacological inhibitors and determined the expression of a set of eight LPS-inducible genes (Figure 5A and B). Inhibition of AKT and Rho had the strongest inhibitory effect. On the other hand, inhibition of PI3K increased expression of IL-10, CCL-2 and Fos, consistent with a regulatory role of PI3K in LPS-induced gene expression (Fukao and Koyasu, 2003). Surprisingly, pharmacological inhibition of the ATM kinase boosted the mRNA levels of Fos, CCL-2, CXCL-10 and IL-10, especially after 4.5 h. The role of ATM in innate immune activation has not been investigated before. We therefore confirmed the results of the inhibitor screen for ATM on the expression of IL-10, CCL-2 and CXCL-10 using additional concentrations of LPS and inhibitor (Figure 5C). ATM kinase motifs were enriched among LPS-regulated phosphorylation sites (Table I). By immunoblotting with an antibody recognising phosphorylated ATM substrate proteins, we validated that LPS induced ATM kinase activity (Figure 5D). Pharmacological ATM inhibition reduced the intensity of ATM substrate phosphorylation, especially when lower concentrations of LPS were used (Figure 5D). Together, these data indicate a functional role of LPS-induced ATM kinase activity in the negative regulation of a subset of LPS-target genes.
Figure 5.
Pharmacological inhibition of LPS-activated pathways differentially impacts gene expression. (A) Induction of selected LPS-target genes. Strongly LPS-induced genes were identified by microarray analysis of nascent RNA (see Figure 6). The changes in expression of 10 selected target genes were analysed by qRT–PCR with Roche Universal Probe Library reagents in independent experiments using total RNA from macrophages generated by the standard protocol after 45 min and 4.5 h. Shown are mean and s.d. of quadruplicate PCR results from a representative experiment of two performed. (B) Effects of pharmacological inhibitors on LPS-induced gene expression. Macrophages were treated and analysed as in (A); the pharmacological inhibitors were added 2 h before stimulation with LPS. Fold-changes induced by LPS were calculated relative to the untreated samples using the ΔΔCT method and subsequently normalised to the effect of LPS in the absence of inhibitor. The experiment was performed two times. For visualisation of the inhibitor effects, the data were colour coded to indicate inhibition (blue) or increased induction (red) by a factor of >2 in both (dark colour) or one of the experiments (light colour). (C) ATM-inhibition increases expression of IL-10, CCL2 and CXCL10. Macrophages were pretreated with different concentrations of ATM inhibitor (μM) or solvent (DMSO) for 2 h, followed by stimulation with titrated amounts of LPS for 4.5 h. qRT–PCR data of quadruplicate samples from one representative experiment of two to three performed are shown. (D) Increased ATM substrate phosphorylation after LPS is blocked by ATM inhibitor. After pretreatment with DMSO or ATM inhibitor (10 μM) for 2 h, macrophages were stimulated for 1 h with LPS as indicated. Phosphorylated ATM substrate proteins were detected with Cell Signaling antibody Cat. #2851.
Connecting TF phosphorylation with LPS-induced transcriptional activation
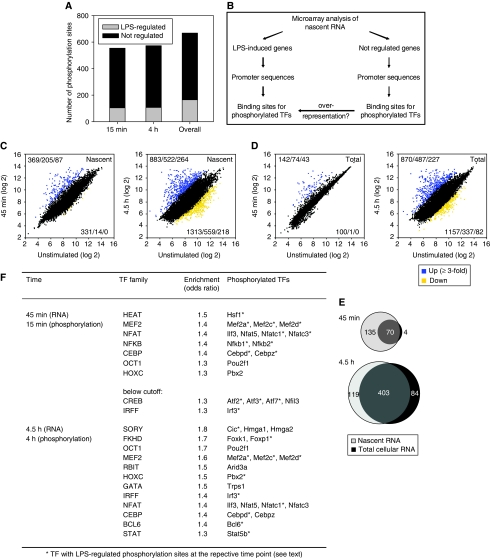
One major function of signal transduction is regulation of gene expression. Phosphorylation controls TF translocation, association with binding partners, binding to DNA or transcriptional activation capacity (Karin, 1991). We detected 187 phosphoproteins annotated as transcriptional regulators (Genomatix Matrix Library 7.1) with 668 phosphorylation sites, 25% of which were regulated by LPS (Figure 6A). We hypothesised that the frequencies of binding sites for phosphorylated TFs may be increased in promoters of LPS-regulated genes (Figure 6B). To identify transcriptionally regulated genes with high sensitivity, we isolated nascent RNA after metabolic labelling with 4-thiouridine during the last 35 min before cell harvest, as described recently (Dolken et al, 2008). Microarray analyses of nascent RNA identified substantially more probe sets as up-regulated after 45 min of LPS stimulation than parallel analyses of total cellular RNA (Figure 6C–E). In contrast, 4.5 h after stimulation, up-regulated genes in total and nascent RNA largely overlapped (Figure 6E). This approach therefore allowed a much more sensitive detection of early changes in transcription, and the respective genes are likely to be direct targets of LPS-regulated TFs.
Figure 6.
LPS-induced TF phosphorylation and changes in the nascent transcriptome—in silico promoter analysis. (A) Phosphorylation sites on TFs. Phosphorylation sites up-regulated or down-regulated more than 1.5-fold in both experiments are indicated in grey. (B) Workflow for integration of phosphoproteome and transcriptome data. Microarray analyses of metabolically labelled nascent and of total cellular RNA were performed on macrophages. Promoter sequences of LPS-regulated genes (induction ⩾3-fold) and of genes that were not transcriptionally altered in response to LPS (2000 least regulated probe sets) were retrieved with Genomatix Gene2Promoter. Promoters were analysed for the presence of binding sites for all identified phosphorylated TF families with Genomatix RegionMiner, and significant over-representation in LPS-regulated promoters was determined (odds ratio ⩾1.3; corrected P-value ⩽0.05). (C–E) Increase in microarray sensitivity by analysis of nascent RNA. Microarray analysis of (C) nascent and (D) total cellular RNA from two independent experiments. Macrophages were left un-treated or stimulated with LPS for 45 min or 4.5 h. Nascent RNA was labelled by addition of 4sU during the last 35 min of stimulation and purified after extraction of total cellular RNA. For each comparison, the number of probe sets induced at least two-/three-/five-fold is represented in the upper left corner, the number of probe sets repressed at least two-/three-/five-fold in the lower right corner. (E) Venn diagrams illustrate the increased sensitivity for early changes in transcription for at least three-fold-induced probe sets of nascent compared to total RNA analyses. (F) TF families with over-represented binding sites in the promoters of transcriptionally LPS-regulated genes.
In silico promoter scanning for binding sites for all 50 TF families with phosphorylated members was used to test for enrichment in transcriptionally induced genes. Forty-five minutes after LPS, we found significant over-representation of binding sites for NFkB, an established mediator of LPS-induced transcription; two other canonical LPS-activated TF families, CREB and IRFF, showed a trend for enrichment (odds ratio 1.3; corrected P-value 0.10 and 0.08, respectively). Significant enrichment for CEBP, MEF2, NFAT and HEAT binding sites suggested a more genuine role for the associated TFs, which have been described as activators of individual LPS-target genes (Tanaka et al, 1995; Han et al, 1997; Matsumoto et al, 1999; Zhu et al, 2003; Inouye et al, 2004, 2007). In addition, OCT1 and HOXC family members, which to date have not been assigned a role in LPS-induced transcription, were significantly enriched (Figure 6F). In contrast, analysis of promoters of genes induced in total cellular RNA after 45 min did not reveal any significant over-representation (data not shown). For IRFF, CEBP, MEF2, NFAT, OCT1, HOXC over-representation was still observed at 4.5 h in genes regulated on nascent and total RNA levels (Figure 6F and data not shown), suggesting an enduring role for these factors, whereas NFkB binding sites were not enriched any more.
Phosphorylation sites on many TF family members with binding site enrichment were LPS-regulated and more than half of them have not been reported earlier (Figure 6F; Supplementary Table S4). Although the technically complex extraction of proteins from chromatin pellet fractions allowed detection of several TF phosphorylation sites in one of the experiments only, potential functional relevance was suggested by the fact that most regulated sites are evolutionary conserved (89 and 96% at 15 min and 4.5 h, respectively; Supplementary Table S4), as are associated TF binding sites in several of the top 20 LPS-induced target genes (selected examples in Supplementary Figure S6).
Our in silico integration of phosphoproteome and nascent transcriptome data confirmed canonical and identified a number of novel candidate TFs driving TLR-induced gene expression. To obtain initial information about the involvement of TFs with binding site enrichment in the expression of LPS-target genes, we silenced expression of Cebpz (CEBP matrix family), Hsf1 (HEAT), Atf7 (CREB) and Cic (SORY) in primary macrophages using siRNA knockdown (Wiese et al, 2010) (Supplementary Figure S7A). Analysis of direct changes in 128 critical TLR-regulated genes with the nCounter system (Geiss et al, 2008; Amit et al, 2009) suggested Il1a and Il1b as potential target genes (Supplementary Figure S7B). Indeed, qRT–PCR confirmed that the LPS-induced expression of Il1a and Il1b was significantly reduced when the novel TF CIC or the CREB family member ATF7 was knocked down (Supplementary Figure S7C). None of the TF knockdowns had a significant effect on Tnf expression (Supplementary Figure S7C). These results validate our in silico identification of CIC and ATF7 as transcriptional regulators of the TLR pathway. Future knockout investigations of these factors will address the role of these and other enriched phosphorylated TFs in innate immune function.
Discussion
This study provides the first unbiased and quantitative investigation of the macrophage phosphoproteome and its dynamic changes in response to TLR activation. We adapted an SILAC labelling approach to primary macrophages that, coupled with TiO2-based phosphopeptide enrichment and high-accuracy mass spectrometry, enabled us to reproducibly identify and quantify a large number of serine, threonine and tyrosine phosphorylation sites with high confidence. The substantial phosphoproteome regulation on LPS stimulation is comparable in extent to the transcriptional re-programming of macrophages (Foster et al, 2007; Mages et al, 2007; data from this study), and reflects the important role of phosphorylation cascades in TLR signalling. Our parallel phosphoproteome and transcriptome analyses underline the notion that widespread phosphorylation precedes massive transcriptional changes; integration of these two sets of systems level data by in silico promoter mapping and TF binding site enrichment analysis identified phosphorylated TFs as candidate regulators of TLR-induced gene expression.
The dynamic nature of phosphorylation cascades in TLR-stimulated macrophages is evident from the kinetic differences. Decreased phosphorylation in response to LPS, or return of increased phosphorylation after an early peak, may be caused by protein degradation or through phosphatase activity. The M-CSF receptor is an example for the first mechanism, with decreases in a phosphorylated peptide as well as in three non-phosphorylated peptides, consistent with earlier reports on LPS- and IFNγ-induced M-CSF receptor degradation (Baccarini et al, 1992; Sester et al, 1999; Trost et al, 2009). Evidence for phosphatase activity is provided by our observation that for many phosphoproteins with a down-regulated site other phosphopeptides were unchanged or increased. Progress in mass spectrometry should allow to generate quantitative proteome data in the near future to definitively determine which changes in phosphorylation are influenced by differences in protein levels (Cox and Mann, 2007). However, our parallel transcriptome analysis already suggests that only a minor fraction of induced phosphorylation results from increased expression of the protein.
Hotspots of TLR-induced phosphorylation
Bioinformatic analyses of the regulated phosphorylation sites and proteins for over-representation of kinase motifs and functional annotation found the major canonical TLR-activated molecular players, including the kinases IRAK2, IRAK4, MAPKs and upstream kinases, and the NFkB-activating kinase IKKβ, and revealed a number of less well appreciated and novel signalling components. Although we observed regulated phosphorylation of many known TLR signalling components, it should be noted that some established players (e.g. IRAK1; IKBKE) were not detected in this screen. This observation indicates that despite the reproducible identification of nearly 7000 phosphorylation sites the screen is not yet saturated. In addition, the time points chosen here may not fit to the phosphorylation kinetics of some pathway molecules such as IRAK1 that is known to be phosphorylated early and then degraded rapidly (Li et al, 2001; Kollewe et al, 2004); similarly, the expected IFNβ-induced Stat1 tyrosine phosphorylation (Thomas et al, 2006) was observed by immunoblotting in standard and SILAC-protocol macrophages with a strong peak between 2 and 3 h but nearly gone after 4 h (Supplementary Figure S3). The large fraction of new phosphorylation sites on known pathway components (65%, Supplementary Table S2) points to new regulatory aspects of TLR signalling. The identification of these trademark TLR pathway modules by unbiased statistical testing strengthens the validity of our experimental data, which are summarised in the form of a model in Figure 6.
The PI3K/AKT pathway, together with its diverging downstream kinases GSK3 and mTOR, was prominently enriched among LPS-regulated phosphoproteins. Ser9-phosphorylation of GSK3 leads to increased production of IL-10 (Hu et al, 2006) and may thereby mediate the described negative regulatory role of PI3K/AKT activation (Fukao and Koyasu, 2003). GSK3 kinase motif-bearing phosphoproteins identified here may contribute to down-regulation of macrophage activity. On the other hand, the strong enrichment of mTOR pathway proteins highlights the importance of this pathway in innate immune signalling, consistent with recent reports showing its important role in IFN type I production (Cao et al, 2008), IL-10 expression (Ohtani et al, 2008; Weichhart et al, 2008) and Stat3-dependent control of Caspase-1 (Schmitz et al, 2008). How exactly mTOR controls these effects is unclear at present, but an mTOR-dependent increase in translational efficiency is involved in the regulation of IRF7 expression (Colina et al, 2008). Our observed phosphorylation of translation initiation factors and of multiple ribosomal proteins after LPS may be linked to mTOR activation.
Already 20 years ago, it was described that microbial stimuli block macrophage proliferation (Hume et al, 1987). The enrichment of the GO term ‘cell proliferation’ among LPS-regulated phosphoproteins and evidence for activation of the cell cycle relevant kinases ATM/ATR and CHK1/2 among the LPS-regulated phosphorylation sites suggest a potential phosphorylation-dependent mechanism for inhibition of proliferation. ATM and ATR, usually activated by genotoxic stress, phosphorylate the cell cycle checkpoint kinases Chk2 and Chk1, respectively (Abraham, 2001). Interestingly, the p38-activated kinase mapkap kinase-2 (MK2) has a very similar kinase motif and is a functional analogue of Chk1/2 (Manke et al, 2005); therefore, it is possible that the enrichment for the CHK1/2 kinase motif observed here is the footprint of LPS-induced, p38-dependent MK2-activation. A functional role for ATM kinase in the negative regulation of some LPS-induced cytokines is suggested by the effects of a pharmacological ATM inhibitor on expression of IL-10, CCL2 and CXCL10. How exactly ATM kinase influences inflammatory gene expression and which ATM substrate proteins (Matsuoka et al, 2007) are phosphorylated in response to TLR4 stimulation will be the subject of future studies.
That cytoskeletal and actin binding proteins are targeted by TLR4-induced phosphorylation was unexpected, as the cytoskeleton is usually not part of TLR pathway models (Oda and Kitano, 2006). However, two key features of macrophages, motility and phagocytosis, depend on cytoskeletal remodelling and are enhanced by TLR stimulation (Blander and Medzhitov, 2004; West et al, 2004) through MAPK-dependent pathways. Rho family GTPases has a major role in actin remodelling (Aderem and Underhill, 1999; Greenberg and Grinstein, 2002), and we find enrichment of the InnateDB pathway term ‘Rho GTPase cycle’. Our identification of multiple phosphorylation sites on cytoskeletal proteins should be useful in the investigation of cytoskeletal remodelling and phagocytosis. The prominence of actin binding protein phosphorylation could also indicate a genuine function of the cytoskeleton in providing a platform for recruitment and spatial targeting of signalling molecules; reversible phosphorylation could be a control switch for this process.
Integration of TF phosphorylation and transcriptional activation data
Here, we present the first study integrating TF phosphorylation and nascent transcriptome data through in silico promoter analysis of binding site enrichment. At the early 45 min time point the majority of transcriptional changes probably represents direct target genes of LPS-activated TFs. Sensitive and unbiased detection of these changes required the analysis of nascent RNA (Dolken et al, 2008). This approach confirmed the known role of NFkB and CREB TFs in early LPS-induced gene expression and of the Trif dependence, later acting IRFF TFs, but in addition identified a number of less established (HEAT, MEF2, CEBP, NFAT) and in the context of TLR-signalling new transcriptional regulators, such as OCT, HOXC and SORY family proteins. NFAT is a key TF in T cells; only recently, a requirement for NFAT activation in DC and macrophages was shown for Dectin-1-dependent gene expression (Goodridge et al, 2007). Of note, binding of NFATc1 to a site in the IL-12p40 promoter has been demonstrated after TLR stimulation (Zhu et al, 2003). Our identification of NFAT family TFs with LPS-regulated phosphorylation together with binding site enrichment in promoters of TLR4-activated genes suggests a broader role for the calcineurin/NFAT pathway. In this context, our finding of pronounced enrichment of the CAMK2 motif among LPS-regulated phosphoproteins is supported by recent reports showing LPS triggered increase in Ca2+ levels and activation of Camk2 (Liu et al, 2008) and Ca2+/calmodulin-dependent expression of many LPS-target genes (Lai et al, 2009).
Computational approaches for the inference of transcriptional networks from microarray gene expression have used a combination of hierarchical clustering of time-course transcriptome data and promoter motif scanning to associate TFs with groups of co-expressed genes (Nilsson et al, 2006; Ramsey et al, 2008). However, the fact that TF binding site motifs usually are recognised by more than one TF protein and the tendency of TF binding sites to co-occur impede the unambiguous identification of the TF from enrichment analysis. Furthermore, many TFs are regulated not on the level of expression but post translationally, and are therefore missed by these approaches. Our global phosphorylation data on TF activation in response to LPS help to fill these gaps and allowed us to implicate novel phosphorylated regulators of macrophage transcriptional responses. This approach recognised the best characterised LPS-activated TFs in macrophages (NFkB, CREB) and identified the recently reported regulatory TF CEBPD (Litvak et al, 2009) as enriched. Importantly, siRNA-mediated knockdown of the CREB family TF ATF7 and of the SORY binding protein CIC demonstrated a non-redundant contribution of these phosphorylated TFs in the LPS-induced expression of Il1a and Il1b (Supplementary Figure S7). This experimental validation of a functional role for ATF7 and CIC makes us confident that also other enriched phosphorylated TFs identified here will be verified as true regulators of LPS-induced transcription in ongoing studies.
Conclusion
Taken together, this study provides a new, global perspective on innate immune activation by TLR signalling. We quantitatively detected a large number of site-specific phosphorylation events, which are now publicly available through the Phosida database (http://www.phosida.com). By combining different data mining approaches, we consistently identified canonical and novel TLR-activated signalling modules. In particular, the PI3K/AKT and the related mTOR pathway were highlighted; furthermore, DNA damage–response-associated ATM/ATR kinases and the cytoskeleton emerged as unexpected hotspots for phosphorylation. Finally, weaving together corresponding phosphoproteome and nascent transcriptome datasets through the loom of in silico promoter analysis we identified several TFs acting at the intersection of TLR-induced kinase activation and gene transcription.
Materials and methods
Mice, SILAC of bone marrow-derived macrophages
Wild-type and Dusp1-deficient mice on a C3H/HeN background were bred under pathogen-free conditions at the animal facility of the Institute of Medical Microbiology, Immunology and Hygiene at Technische Universität München, Germany. Bone marrow cells were isolated and cultured in SILAC medium for 17 days: After overnight depletion of adherent cells non-adherent cells were expanded by addition of recombinant murine IL-3 (10 mg/l), IL-6 (10 mg/l) and SCF (50 mg/l) (Tebu-Bio) in the presence of 10% L-cell conditioned medium (LCCM) as a source of M-CSF on 10 cm bacteriological plates, starting with 1 × 107 cells per plate. These cytokines have a role in macrophage development in vivo (Metcalf, 1997) and have been used to stimulate proliferation of bone marrow cells for retroviral infections (Holst et al, 2006). M-CSF was included in the cultures from the beginning to favour the differentiation of macrophages. Cultures were split every 2–3 days. After 13 days, cells were plated in medium with 10% LCCM without cytokines to complete differentiation into macrophages for 3 days. On day 16, non-adherent cells were discarded and 25 × 106 adherent cells were plated on 15 cm cell culture plates (Falcon) without LCCM for stimulation the next day. Details on the splitting procedure are given in Supplementary information.
SILAC medium
Dulbecco's modified Eagle's medium with stable glutamine deficient in L-arginine and L-lysine (custom made, Biochrom AG), supplemented with 1% penicillin/streptomycin (Biochrom AG), 0.1% 2-mercaptoethanol (Gibco), 10% dialysed fetal bovine serum (Gibco), 84 g/l L-arginine HCl labelled with 13C6 (Arg ‘6’) or 13C615N4 (Arg ‘10’), 146 g/l L-lysine HCl labelled with 2D4 (Lys ‘4’) or 13C615N2 (Lys ‘8’) (EurisoTop GmbH) or their non-labelled counterparts (Arg ‘0’ and Lys ‘0’) (Sigma Aldrich). Thirty g/l non-labelled L-proline (Sigma Aldrich) was added to reduce conversion of labelled arginine to proline (<5%, data not shown).
Stimulation, cell lysis, fractionation and phosphopeptide preparation
Per condition, 50 × 106 SILAC-encoded macrophages were left un-treated or stimulated with 100 ng/ml Escherichia coli LPS (Sigma Aldrich) for 15 min or 4 h. Cells were washed with PBS, lysed in ice-cold modified RIPA buffer for 15 min and scraped. Lysates were pooled, vortexed for 2 min and centrifuged to separate soluble and chromatin pellet fractions (17 000 g, 15 min).
The soluble fraction was precipitated overnight at −20°C by adding 4 volumes of ice-cold acetone. The acetone precipitate was re-solubilised in 8 M urea (6 M urea/2 M thiourea, Sigma Aldrich). Reduction, alkylation, enzymatic digest in-solution and SCX of soluble protein mixtures were performed essentially as described (Olsen et al, 2006). Proteins from the insoluble chromatin pellet were extracted by DNA digest with benzonase (Merck) and re-solubilisation in 8 M urea followed by incubation with loading dye under rotation at 95°C for 5 min. Proteins were reduced and alkylated, resolved by SDS–PAGE on a gradient gel (4–15% Tris–HCl Ready Gel Precast Gel, Bio-Rad), stained with Coomassie and digested in situ essentially as described (Shevchenko et al, 2006). From all fractions, phosphopeptides were enriched on TiO2 beads (GL Sciences, Japan) in the presence of 2,5-dihydrobenzoic acid essentially as described in Olsen et al (2006). Details are given in Supplementary information.
Modified RIPA buffer
One Percent Igepal CA-630, 0.1% sodium deoxycholate, 150 mM sodium chloride, 1 mM EDTA, 50 mM Tris (pH 7.5), supplemented with 1 mM sodium ortho-vanadate, 5 mM sodium fluoride and 5 mM β-glycerophosphate for inhibition of phosphatases and complete protease inhibitors (Roche Applied Science) directly before use.
Mass spectrometric analysis
Phosphopeptide mixtures were analysed by online nanoflow LC-MS/MS as described earlier (Olsen et al, 2006) with a few modifications. All LC-MS analysis were performed with 2 h gradients on an EASY-nLC system (Proxeon Biosystems) directly coupled to an LTQ-Orbitrap XL instrument (Thermo Electron) that was operated in the data-dependent acquisition mode to automatically switch between orbitrap full scan MS and LTQ MS/MS using a top10 method. Raw files were analysed and quantified using the MaxQuant software suite (Cox and Mann, 2008), peptides were identified by Mascot and filtered for <1% false discovery rate (FDR) in MaxQuant. Phosphorylation sites were localised inside the identified peptide sequences using the PTM score algorithm (Olsen et al, 2006). Phosphopeptide ratios were calculated, referring to unstimulated wild types were calculated for each genotype and time point, and were normalised such that the median of log-transformed ratios of all identified peptides was zero, to correct for unequal sample mixing. Specific details on the MS acquisition and the downstream analysis are given in Supplementary information. The phosphoproteome dataset is also accessible in the Phosida database (http://141.61.102.18/phosida/specificprojects/login.aspx?project=219&).
Metabolic labelling, purification and analysis of nascent RNA
Metabolic labelling and purification of nascent RNA were performed essentially as described (Dolken et al, 2008), with minor modifications for use with primary macrophages that are described in Supplementary information. The microarray dataset has been deposited as series GSE20674 in the Gene Expression Omnibus database and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20674.
Bioinformatic analyses
Contaminating FCS and human keratin proteins were excluded as described in Supplementary information. Analyses on the phosphoprotein level were performed on all phosphorylated proteins, regardless of the probability for right localisation of the phosphate group within a peptide according to the PTM score developed by Olsen et al (2006) (note that the likelihood that such a peptide is phosphorylated is still ⩾99%). Analyses on the phosphorylation site level included only sites for which the phosphate group could be located within the peptide with single amino acid accuracy (class I sites).
Contribution of gene expression changes to regulation of the phosphoproteome
Changes in gene expression at the level of total cellular RNA were correlated with changes in phosphorylation for all proteins with LPS up-regulated phosphorylation (log2 ratio of mean fold-changes from two independent experiments). If several probe sets existed for one gene, the probe set with the highest expression value was selected. For 35 proteins with LPS up regulated phosphorylation no corresponding probe set was found or RNA expression was not above background level.
GO analysis
Although most GO analysis tools calculate an over-representation of GO terms over the genomic background we developed a strategy for direct comparison of different lists against each other: Numbers of phosphoproteins associated with each GO term were determined using the GO Browser in Spotfire Decision Site (Tibco), the generic GOSlim ontology file (OBO-Edit version 1.101) and the GO annotation file for mouse (version 11/7/2008), downloaded from http://www.geneontology.org/GO. To determine statistically significant over-representation of terms, odds ratio ((number of matches list A/number of non-matches list A)/(number of matches list B/number of non-matches list B)) and Fisher's exact probability using the R Statistics package (http://www.r-project.org) were calculated for each GO term, comparing proteins with LPS-regulated and non-regulated phosphorylation sites. P-values were corrected for multiple testing using the Benjamini–Hochberg method for controlling FDR (Benjamini and Hochberg, 1995). Only GO terms with at least three identified phosphoproteins were analysed. GO terms with an odds ratio ⩾1.3 or ⩽0.67 and a corrected P-value ⩽0.05 were considered significant.
Kinase motifs
Phosphorylation sites were matched to the known substrate specificities (linear sequence motifs) for 33 human kinases (http://www.phosida.com). To determine statistically significant over-representation of a motif among LPS-induced phosphorylation sites the number of sites that matched the pattern was determined among LPS-induced phosphorylation sites and among phosphorylation sites that were not up-regulated in response to LPS. Odds ratios and Fisher's exact probabilities, which were corrected for multiple testing, were calculated as described for the GO analysis. Motifs with an odds ratio ⩾1.3 and a corrected P-value ⩽0.05 were considered significant. All enriched kinase motifs matched at least 10 phosphorylation sites.
Signalling pathways
Phosphoproteins were assigned to signalling pathways through ENSEMBL identifiers using InnateDB (Lynn et al, 2008) (http://www.innateDB.ca, version 29/1/2009), which provides pathway annotation from many different databases and calculates over-representation over the genomic background. For a direct comparison of LPS-regulated and not LPS-regulated phosphoproteins, the number of phosphoproteins associated with each pathway was determined with InnateDB, and odds ratio and Fisher's exact probability were calculated as described for the GO analysis. Only pathways for which we identified at least five phosphoproteins were included in the analysis. Signalling pathways with an odds ratio ⩾1.3 and a P-value ⩽0.05 were considered significant.
Gene symbols of over-represented signalling pathways were extracted from InnateDB and loaded into the database STRING 8.0 (Jensen et al, 2009) (http://string.embl.de) for extraction of functional interaction networks. Reported interactions include direct (physical) and indirect (functional) interactions based on experimental evidence from high-throughput studies, co-regulation of gene expression, same genomic context or co-citation in the literature. Pathway networks were visualised with Cytoscape v.2.6.2 (http://www.cytoscape.org). Only interactions with a minimum STRING combined score of 0.400, which represents the default medium confidence level in STRING, were kept.
TF binding sites
To determine over-represented TF binding sites in LPS-regulated promoters, promoter sequences of LPS-induced genes (⩾3-fold) and of genes not regulated by LPS (2000 probe sets with the least regulation; to minimise background noise, only probe sets with GeneID and a maximal expression value of at least 50) were retrieved with Genomatix Gene2Promoter through GeneIDs (http://www.genomatix.de, large-scale option, database version ElDorado 07-2008) and searched for the presence of binding sites for 50 TF families with phosphorylated members, with Genomatix RegionMiner (http://www.genomatix.de, matrix library version 7.1). RegionMiner determines the number of hits (we considered the number of promoters with a binding site, not the number of binding sites within a promoter) and calculates over-representation over the genomic background. To determine binding site over-representation in promoters of LPS-regulated genes compared to promoters of genes not regulated by LPS, we calculated odds ratios and Fisher's exact P-values, which were corrected for multiple testing, as described for the GO analysis. TF families with an odds ratio ⩾1.3 and a corrected P-value ⩽0.05 were considered significant.
To determine evolutionary conservation of TF binding sites, the promoters of the 20 most strongly induced genes in nascent RNA (45 min, ranked mean fold-change from two independent experiments) were compared to orthologous vertebrate promoters (retrieved with Genomatix Gene2Promoter, database version ElDorado 12-2009) with Genomatix MatInspector (Cartharius et al, 2005), and similar positions of TF binding sites relative to the transcriptional start sites were determined by eye in Genomatix-aligned promoters.
Supplementary Material
Table S1. Reproducibly identified macrophage phosphorylation sites.
Table S2. Phosphorylation sites on known TLR signalling molecules.
Table S3. LPS-regulated phosphoproteins associated with enriched Gene Ontology terms
Table. S4. Phosphorylation sites on transcription factors with binding site enrichment in LPS-regulated promoters.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB576-A10 and SFB643-A10 to RL), the ELAN fonds of the Medical Faculty at FAU Erlangen-Nürnberg (to RL), the German Federal Ministry of Education and Research (NGFN plus grant 01GS0801 to LD), the Max-Planck Society and by the European Union (Interaction Proteome LSHG-CT-2003-505520 to MM). The Center for Protein Research is funded by a generous grant from the Novo Nordisk Foundation. We thank C Bogdan (Erlangen) and H Wagner (Munich) for helpful comments on the paper, and F Gnad (Munich) for upload of the dataset to Phosida.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Aderem A, Underhill DM (1999) Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 17: 593–623 [DOI] [PubMed] [Google Scholar]
- Aki D, Mashima R, Saeki K, Minoda Y, Yamauchi M, Yoshimura A (2005) Modulation of TLR signalling by the C-terminal Src kinase (Csk) in macrophages. Genes Cells 10: 357–368 [DOI] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE et al. (2009) Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarini M, Dello Sbarba P, Buscher D, Bartocci A, Stanley ER (1992) IFN-gamma/lipopolysaccharide activation of macrophages is associated with protein kinase C-dependent down-modulation of the colony-stimulating factor-1 receptor. J Immunol 149: 2656–2661 [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Blander JM, Medzhitov R (2004) Regulation of phagosome maturation by signals from toll-like receptors. Science 304: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B (2008) Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 9: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, Flavell RA (2006) Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci USA 103: 2274–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, Makrigiannis AP, Bell JC, Sonenberg N (2008) Translational control of the innate immune response through IRF-7. Nature 452: 323–328 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2007) Is proteomics the new genomics? Cell 130: 395–398 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH (2008) High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978 [DOI] [PubMed] [Google Scholar]
- Fukao T, Koyasu S (2003) PI3K and negative regulation of TLR signaling. Trends Immunol 24: 358–363 [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Goodridge HS, Simmons RM, Underhill DM (2007) Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol 178: 3107–3115 [DOI] [PubMed] [Google Scholar]
- Greenberg S, Grinstein S (2002) Phagocytosis and innate immunity. Curr Opin Immunol 14: 136–145 [DOI] [PubMed] [Google Scholar]
- Hammer M, Mages J, Dietrich H, Servatius A, Howells N, Cato AC, Lang R (2006) Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J Exp Med 203: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386: 296–299 [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D (2009) The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DA (2006) Generation of T-cell receptor retrogenic mice. Nat Protoc 1: 406–417 [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6: 644–658 [DOI] [PubMed] [Google Scholar]
- Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, Ivashkiv LB (2006) IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24: 563–574 [DOI] [PubMed] [Google Scholar]
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N (2001) The plasticity of dendritic cell responses to pathogens and their components. Science 294: 870–875 [DOI] [PubMed] [Google Scholar]
- Hume DA, Allan W, Fabrus B, Weidemann MJ, Hapel AJ, Bartelmez S (1987) Regulation of proliferation of bone marrow-derived macrophages. Lymphokine Res 6: 127–139 [PubMed] [Google Scholar]
- Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A (2007) Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem 282: 33210–33217 [DOI] [PubMed] [Google Scholar]
- Inouye S, Izu H, Takaki E, Suzuki H, Shirai M, Yokota Y, Ichikawa H, Fujimoto M, Nakai A (2004) Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem 279: 38701–38709 [DOI] [PubMed] [Google Scholar]
- Ivison SM, Graham NR, Bernales CQ, Kifayet A, Ng N, Shobab LA, Steiner TS (2007) Protein kinase D interaction with TLR5 is required for inflammatory signaling in response to bacterial flagellin. J Immunol 178: 5735–5743 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (2009) STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M (1991) Signal transduction and gene control. Curr Opin Cell Biol 3: 467–473 [DOI] [PubMed] [Google Scholar]
- Kollewe C, Mackensen AC, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin MU (2004) Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem 279: 5227–5236 [DOI] [PubMed] [Google Scholar]
- Kruger M, Kratchmarova I, Blagoev B, Tseng YH, Kahn CR, Mann M (2008) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc Natl Acad Sci USA 105: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Wan M, Wu J, Preston-Hurlburt P, Kushwah R, Grundstrom T, Imbalzano AN, Chi T (2009) Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc Natl Acad Sci USA 106: 1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ (2002) Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol 169: 2253–2263 [DOI] [PubMed] [Google Scholar]
- Lang R (2005) Tuning of macrophage responses by Stat3-inducing cytokines: molecular mechanisms and consequences in infection. Immunobiology 210: 63–76 [DOI] [PubMed] [Google Scholar]
- Latz E, Fitzgerald KA (2008) Innate immunity: sensing and signalling http://www.nature.com/nri/poster/innate.
- Li X, Commane M, Jiang Z, Stark GR (2001) IL-1-induced NFkappa B and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK). Proc Natl Acad Sci USA 98: 4461–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O’Neill LA (2005) Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol 5: 446–458 [DOI] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A (2009) Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 10: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao M, Li N, Wang C, Zheng Y, Cao X (2008) CaMKII promotes TLR-triggered proinflammatory cytokine and type I interferon production by directly binding and activating TAK1 and IRF3 in macrophages. Blood 112: 4961–4970 [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, Lo R, Naseer M, Que J, Yau M, Acab M, Tulpan D, Whiteside MD, Chikatamarla A, Mah B, Munzner T et al. (2008) InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol 4: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mages J, Dietrich H, Lang R (2007) A genome-wide analysis of LPS-tolerance in macrophages. Immunobiology 212: 723–737 [DOI] [PubMed] [Google Scholar]
- Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB (2005) MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell 17: 37–48 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Tanaka T, Kaisho T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Akira S (1999) A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J Immunol 163: 5039–5048 [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Metcalf D (1997) The molecular control of granulocytes and macrophages. Ciba Found Symp 204: 40–50; discussion 50-46 [DOI] [PubMed] [Google Scholar]
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA (2002) Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA 99: 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Bajic VB, Suzuki H, di Bernardo D, Bjorkegren J, Katayama S, Reid JF, Sweet MJ, Gariboldi M, Carninci P, Hayashizaki Y, Hume DA, Tegner J, Ravasi T (2006) Transcriptional network dynamics in macrophage activation. Genomics 88: 133–142 [DOI] [PubMed] [Google Scholar]
- Oda K, Kitano H (2006) A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2: 2006 0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M, Nagai S, Kondo S, Mizuno S, Nakamura K, Tanabe M, Takeuchi T, Matsuda S, Koyasu S (2008) Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood 112: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Gnad F, Olsen JV, Mann M (2008) Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics 8: 4534–4546 [DOI] [PubMed] [Google Scholar]
- Park JE, Kim YI, Yi AK (2009) Protein kinase D1 is essential for MyD88-dependent TLR signaling pathway. J Immunol 182: 6316–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, Navarro G, Roach JC, Rosenberger CM, Rust AG, Yudkovsky N, Aderem A, Shmulevich I (2008) Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol 4: e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruse M, Knaus UG (2006) New players in TLR-mediated innate immunity: PI3K and small Rho GTPases. Immunol Res 34: 33–48 [DOI] [PubMed] [Google Scholar]
- Salojin KV, Owusu IB, Millerchip KA, Potter M, Platt KA, Oravecz T (2006) Essential role of MAPK phosphatase-1 in the negative control of innate immune responses. J Immunol 176: 1899–1907 [DOI] [PubMed] [Google Scholar]
- Schmitz F, Heit A, Dreher S, Eisenacher K, Mages J, Haas T, Krug A, Janssen KP, Kirschning CJ, Wagner H (2008) Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur J Immunol 38: 2981–2992 [DOI] [PubMed] [Google Scholar]
- Sester DP, Beasley SJ, Sweet MJ, Fowles LF, Cronau SL, Stacey KJ, Hume DA (1999) Bacterial/CpG DNA down-modulates colony stimulating factor-1 receptor surface expression on murine bone marrow-derived macrophages with concomitant growth arrest and factor-independent survival. J Immunol 163: 6541–6550 [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Takeda K, Akira S (2004) TLR signaling pathways. Semin Immunol 16: 3–9 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T (1995) Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell 80: 353–361 [DOI] [PubMed] [Google Scholar]
- Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN (2006) Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem 281: 31119–31130 [DOI] [PubMed] [Google Scholar]
- Trost M, English L, Lemieux S, Courcelles M, Desjardins M, Thibault P (2009) The phagosomal proteome in interferon-gamma-activated macrophages. Immunity 30: 143–154 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M (2009) Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27: 693–733 [DOI] [PubMed] [Google Scholar]
- Villen J, Beausoleil SA, Gerber SA, Gygi SP (2007) Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA 104: 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, Müller M, Säemann MD (2008) The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29: 565–577 [DOI] [PubMed] [Google Scholar]
- West MA, Wallin RP, Matthews SP, Svensson HG, Zaru R, Ljunggren HG, Prescott AR, Watts C (2004) Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodeling. Science 305: 1153–1157 [DOI] [PubMed] [Google Scholar]
- Wiese M, Castiglione K, Hensel M, Schleicher U, Bogdan C, Jantsch J (2010) Small interfering RNA (siRNA) delivery into murine bone marrow- derived macrophages cells by electroporation. J Immunol Methods 353: 102–110 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang X, Nelin LD, Yao Y, Matta R, Manson ME, Baliga RS, Meng X, Smith CV, Bauer JA, Chang CH, Liu Y (2006) MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. J Exp Med 203: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Rao K, Xiong H, Gagnidze K, Li F, Horvath C, Plevy S (2003) Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J Biol Chem 278: 39372–39382 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Reproducibly identified macrophage phosphorylation sites.
Table S2. Phosphorylation sites on known TLR signalling molecules.
Table S3. LPS-regulated phosphoproteins associated with enriched Gene Ontology terms
Table. S4. Phosphorylation sites on transcription factors with binding site enrichment in LPS-regulated promoters.