Abstract
Aberrant organ development is associated with a wide spectrum of disorders, from schizophrenia to congenital heart disease, but systems-level insight into the underlying processes is very limited. Using heart morphogenesis as general model for dissecting the functional architecture of organ development, we combined detailed phenotype information from deleterious mutations in 255 genes with high-confidence experimental interactome data, and coupled the results to thorough experimental validation. Hereby, we made the first systematic analysis of spatio-temporal protein networks driving many stages of a developing organ identifying several novel signaling modules. Our results show that organ development relies on surprisingly few, extensively recycled, protein modules that integrate into complex higher-order networks. This design allows the formation of a complicated organ using simple building blocks, and suggests how mutations in the same genes can lead to diverse phenotypes. We observe a striking temporal correlation between organ complexity and the number of discrete functional modules coordinating morphogenesis. Our analysis elucidates the organization and composition of spatio-temporal protein networks that drive the formation of organs, which in the future may lay the foundation of novel approaches in treatments, diagnostics, and regenerative medicine.
Keywords: genetics, organ developmental networks, polygenic common disorders, proteomics, systems biology
Introduction
Insight into the biology of molecular networks driving organ development is an important and emerging field as aberrations in these systems underlie a wide spectrum of highly polygenic human disorders, ranging from schizophrenia (Walsh et al, 2008) to congenital heart disease (CHD) (Bruneau, 2008). Understanding the functional architecture of networks that orchestrate the development of organs may also lay the foundation of novel approaches in regenerative medicine, because manipulation of these systems will be necessary for the success of tissue-engineering technologies and stem cell therapy (Chien et al, 2008).
We used heart development and CHD as a general model for dissecting the functional protein networks underlying a developing organ and its related, genetically complex, human phenotypes. The heart is particularly suitable for such an analysis, because it is among the most studied of all organs, it is the organ most susceptible to disease, and its developmental processes and genes are extraordinarily conserved enabling straight forward integration of data between humans and model organisms (Olson, 2004; Srivastava, 2006).
Genetic studies in humans and model organisms have identified hundreds of genes involved in heart development. In mice, phenotypes caused by targeted mutations can be organized into hierarchical morphological subgroups, which point at the spatio-temporal function of the disrupted genes. These results have led to a hypothesis suggesting that during organ development, autonomous anatomical substructures are coordinated by discrete protein complexes or pathways (i.e. functional modules) integrating into higher-order functional networks, and that evolutionary newer anatomical structures might recycle parts of the networks used in more ancient structures (Fishman and Olson, 1997). Although transcription factors have been identified as central players in these processes (Kornberg and Tabata, 1993; Basson et al, 1997; Fishman and Olson, 1997; Schott et al, 1998; Weatherbee et al, 1998; Benson et al, 1999; Gaudet and Mango, 2002; Garg et al, 2003; Pourquie, 2003; Gaudet et al, 2004; Srivastava, 2006), we currently lack overviews of how most genes integrate into functional modules and networks during the different developmental stages. Our lack of understanding of this biological architecture is exemplified by the knowledge that genetic factors contribute significantly to CHD (Bruneau, 2008), but <5% of CHD patients have mutations within the few identified causal human genes, suggesting that many genetic principles of the molecular networks driving heart development remain to be understood.
Results and discussion
First, we manually curated a set of 255 cardiac developmental (CD) genes, in which targeted mutation leads to heart phenotypes in mouse models, from the Mouse Genome Database ver. 3.44 (Bult et al, 2008). We used the Inparanoid orthology database (O'Brien et al, 2005) to find the orthologous 255 human genes, and then identified their corresponding human proteins. We used InParanoid as this method, several times, has been shown to be superior to other methods for mapping functional orthologs (Hulsen et al, 2006; Chen et al, 2007). We refer to this set of human proteins as CD proteins. The 255 proteins are stratified into a total of 19 morphological subgroups reflecting the specific phenotype associated with their mutation (Supplementary Table S1), which can be used as an indicator for the spatio-temporal function of the individual genes. For each of the 19 sets of proteins, we constructed functional networks (Supplementary Figures S1, S2, S3 and S4) using their interaction patterns in refined experimental proteomics data (see Materials and methods), and indeed several novel modules not previously associated with heart development emerged from our analysis (see below). Randomization tests of the resulting networks show that 18 of the 19 gene sets significantly interact at the protein level after adjustment for multiple testing using a Bonferroni correction (see Materials and methods; Supplementary Figures S1, S2, S3 and S4), indicating that the genes involved in each developmental stage (and its corresponding specific phenotype) have a strong tendency to directly interact at the protein level, or are part of connected pathways. In total, the resulting interaction networks consist of 629 unique proteins, have both time and tissue resolution, and describe a wide variety of developmental stages and anatomical structures in the developing heart.
These data represent a new framework for the study of organ development at the systems level, and they extend considerably our understanding of the highly polygenic nature of organ developmental processes, which has been shown previously at the level of gene expression (Maduro and Rothman, 2002).
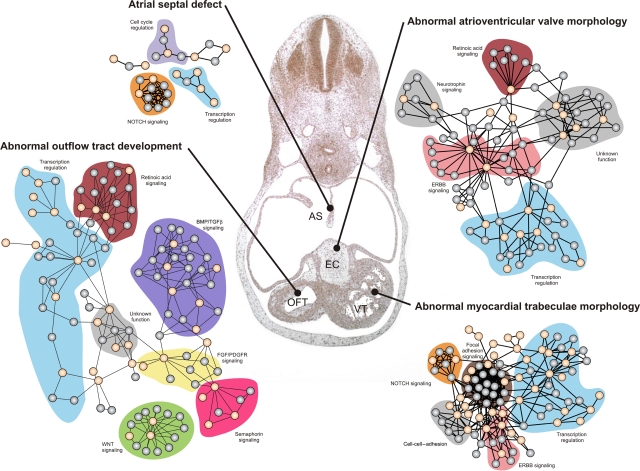
We manually annotated the functional clusters in the networks by literature curation. We chose manual literature curation over automated gene ontology analyses, to exploit the considerable experience and expertise in our group on developmental programs. This analysis revealed several functional modules, which are novel in relation to heart development, including focal adhesion signaling modules and a module of unknown function, which include Sorting nexin 9 (SNX9; Supplementary Information; Supplementary Figures S2C and S3A). The quality of the data was confirmed by the existence of many known functional modules in the networks (e.g. NOTCH signaling in the development of the ventricular trabeculae; Grego-Bessa et al, 2007). Examples of four networks are shown in Figure 1; the proteins involved in four phenotypes and their interaction partners fall into distinct modules, represented as highly interconnected subclusters in the networks. For example, the data show that WNT, semaphorin, FGF/PDGFR, BMP/TGFbeta, and retinoic acid signaling are involved in the development of the outflow tract and suggest that extensive communication takes place within and between modules.
Figure 1.
Examples of four functional networks driving the development of different anatomical structures in the human heart. These four networks constructed by analyzing the interaction patterns of four different sets of cardiac development (CD) proteins corresponding to the morphological groups ‘atrial septal defects,' ‘abnormal atrioventricular valve morphology,' ‘abnormal myocardial trabeculae morphology,' and ‘abnormal outflow tract development' (Supplementary Table S1). CD proteins from the relevant groups are shown in orange and their interaction partners are shown in gray. Functional modules annotated by literature curation are indicated with a colored background. High-resolution figures (including protein names) can be seen in Supplementary Figures S2A, S2C, S2D, and S3B, respectively. Centrally in the figure is a haematoxylin-eosin stained frontal section of the heart from a 37-day human embryo, where tissues affected by the four networks are marked; AS (developing atrial septum), EC (endocardial cushions, which are anatomical precursors to the atrioventricular valves), VT (developing ventricular trabeculae), and OFT (developing outflow tract). The entire set of 19 networks is shown in detail in Supplementary Figures S1, S2, S3 and S4, and can be downloaded from http://www.cbs.dtu.dk/suppl/dgf/.
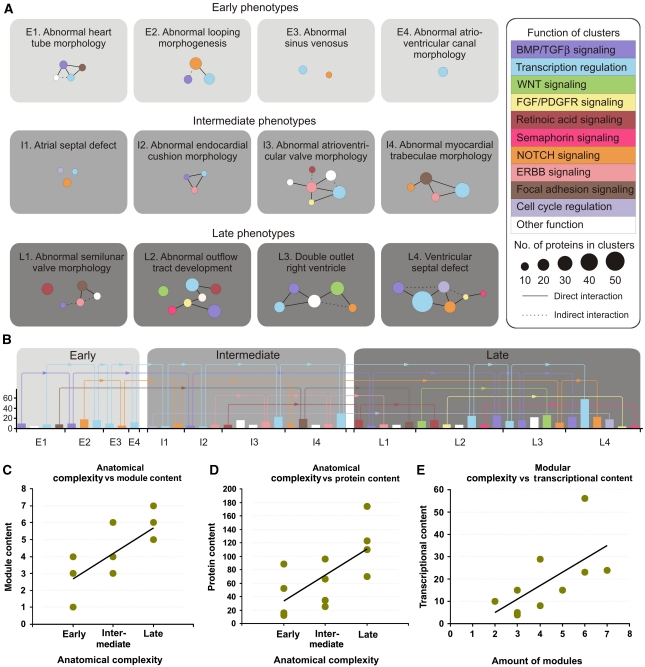
To get insight into how the modularity of heart development is organized across spatio-temporal morphological stages, we created module maps of the different networks and grouped them according to temporal development (i.e. early, intermediate, and late developmental stages of organogenesis; Figure 2). Here, the modular design of the functional networks becomes clear across developmental stages and anatomical structures. Surprisingly, although the networks in some instances contain hundreds of proteins, they consist of relatively few protein modules that are extensively recycled across developmental stages (Figure 2A and B). Moreover, each network consists of a combinatorial unique module pattern. Although modularity is known to be a core feature of the organization of organisms (Wagner et al, 2007), to our knowledge, this concept has not been shown at the level of protein networks in organ development before. This organizational concept allows for the formation of a very complex organ using relatively simple building blocks and suggests how mutations in the same genes and modules can lead to very diverse phenotypes. For example, NOTCH signaling modules are present in the networks representing atrial septal defects (ASD; Supplementary Figure S2A) and double outlet right ventricle (DORV; Supplementary Figure S3C) in line with the observation that mutations in NOTCH1 may lead to ASDs in one individual, and DORV in another (Garg et al, 2005).
Figure 2.
An overview of the modular organization of heart development. (A) Protein interaction networks are plotted at the resolution of functional modules. Each module is color coded according to functional assignment as determined by literature curation. The amount of proteins in each module is proportional to the area of its corresponding node. Edges indicate direct (lines) or indirect (dotted lines) interactions between proteins from the relevant modules. (B) Recycling of functional modules during heart development. The bars represent functional modules and recycling is indicated by arrows. The bars follow the color code of (A) and the height of the bars represent the number of proteins in each module, as shown on the y axis (left). (C–E) Correlations between anatomical, modular, and transcriptional complexity in organ developmental networks. We plotted network complexity along an axis of increasing anatomical complexity as defined by the early, late, and intermediate phenotypes (C, D), and observe a significant correlation. Also, modular and transcriptional complexity correlate significantly during the traversing of organ developmental programs and stages (E). In a given network, module content is the amount of modules, protein content is the amount of proteins, and transcriptional content is the amount of proteins directly involved in transcriptional regulation.
Development of the human heart starts ∼2 weeks after fertilization, with the formation of the cardiac crescent and the subsequent formation and looping of the primitive heart tube. At this stage, the heart is an anatomically simple structure associated with the ‘early phenotype' networks in Figure 2. Looping is followed by extensive tissue remodeling, which includes septation of the atrium and ventricles, and development of trabeculae within the ventricles. Defects at this stage results in ‘intermediate phenotypes.' The last stages of heart development include construction of the heart valves and separation of the outflow tract, as determined by ‘late phenotypes.' Throughout this transformation, the organ, along with the embryo, becomes an anatomically much more elaborate structure (Srivastava, 2006), which remarkably is mirrored in the complexity of the functional networks we have identified as drivers of these processes.
We have quantified network complexity based on (1) the number of distinct functional modules present in each network and (2) the total amount of proteins in each network. The amount of modules in networks associated with ‘early phenotypes' is on average 2.5, which increases to an average of 5.8 for ‘late phenotypes' (Figure 2C; Spearman ρ=0.76, P=0.010). A similar observation can be made for the total amount of proteins in each network that increases from an average of 42 to an average of 119 (Figure 2D; Spearman ρ=0.72, P=0.016). Although the stages of heart development are broadly defined, there is a clear trend across all networks for later phenotypes to be associated with more complex and functionally diverse networks.
Thus, our analysis of the networks strongly suggest that increased morphological complexity of the heart is correlated with increased signaling complexity at the molecular level, which support a model predicting that the four-chambered heart has evolved by addition of new autonomous anatomical structures (Moorman and Christoffels, 2003; Olson, 2006). Analysis of functional data at the systems level suggests that this evolution in part relies on recycling and shuffling of existing functional modules, to create combinatorial unique functional networks that drive the formation of new anatomical structures. Furthermore, our results mirror the findings of evolution modeling and algorithms, which show that modularity in networks can spontaneously arise under changing environments (Lipson et al, 2002; Kashtan and Alon, 2005), a principle that allows for rapid organism adaptation to new demands (Kashtan and Alon, 2005). Importantly, we couple these findings to the anatomical development of organs and together these studies give insight into the forces that advance structural simplicity in biological networks underlying organ development.
Formation of organs depends on highly conserved sets of transcription factors (Satou and Satoh, 2006) of which spatio-temporal regulation is critical to achieve correct patterning (Pourquie, 2003). During development, series of transcription factors function hierarchically to regulate specific developmental programs (Kim et al, 2001; Maduro and Rothman, 2002; Skeath and Thor, 2003). These observations raise the question of how transcriptional programs are linked to the timing of developmental processes and the relationship between the transcriptional programs, cellular networks, diseases, and the developing organ.
GATA4, NKX2-5, and TBX5 are known to be involved in many stages of heart development, and defects in these genes have been established as the cause of familial CHD (Basson et al, 1997; Schott et al, 1998; Garg et al, 2003). As expected, we observe these transcription factors participating in most of the networks and across almost all stages of heart development, stressing their importance (Supplementary Figures S1, S2, S3 and S4). In addition to GATA4, NKX2-5, and TBX5, the networks also contain a large amount of other proteins directly involved in transcriptional control either as transcription factors, or by participating in transcription initiation complexes and networks (Figure 2; Supplementary Figures S1, S2, S3 and S4).
Two transcriptional concepts have been observed in organ development. Some regulators are only active for a brief period of time and usually produce a uniform response in the expressing cells (Kornberg and Tabata, 1993; Maduro and Rothman, 2002; Skeath and Thor, 2003). Other regulators such as GATA4, NKX2-5, and TBX5 are continuously expressed, but activate different sets of genes at different developmental stages, suggesting they are parts of more heterogeneous and complex transcriptional programs (Weatherbee et al, 1998; Bergstrom et al, 2002; Gaudet and Mango, 2002; Gaudet et al, 2004). The latter type of regulators exert their specific function by exploiting promoter affinity gradients, and through complicated patterns of promoter elements that scaffold sets of transcriptional proteins (Gaudet and Mango, 2002; Gaudet et al, 2004). Our data show that GATA4, NKX2-5, and TBX5 participate in most of the transcriptional modules throughout heart development as expected (Supplementary Figures S1, S2, S3 and S4), but interestingly, the modules vary widely in complexity and in the specific composition of the participating proteins. Thus, on the level of transcriptional protein networks, we observe combinatorial regulation, which provides the organism with a high degree of flexibility for GATA4, NKX2-5, and TBX5, and enables them to have a broad function during heart development. This is consistent with the remarkable variability of phenotypic outcome that can be the result of mutations in each of these genes.
Interestingly, the amount of transcriptional proteins in the networks increases from an average of 11 in the networks associated with ‘early phenotypes,' to 32 in the networks associated with ‘late phenotypes.' Moreover, there is a significant correlation between the amount of modules in each network and the amount of proteins directly involved in transcriptional control in the same networks (Figure 2E; Spearman ρ=0.69, P=0.035). Thus, our results show a direct relationship between anatomical, modular, and transcriptional complexity during the traversing of organ developmental programs and support the concept of combinatorial regulation at the protein level.
To experimentally test the biological accuracy of the module maps, we systematically identified 49 novel heart developmental proteins from the modules (Supplementary Tables S2 and S3). These candidates were interacting significantly with the CD set, but were not in the literature associated with heart developmental processes (the procedure for scoring and identifying the final 49 candidates is described in detail in Supplementary Information and Supplementary Figures S5). Twelve of these candidates were selected for immunohistochemistry (IH) analyses to test whether they were expressed at the time and place determined by the functional networks in which they participate and the specific morphological groups associated with those networks (the procedure and criteria for choosing the tested proteins is described in detail in Supplementary Information and Supplementary Figures S5 and S6). Immunohistochemical analyses were systematically performed on a total of 382 tissue sections from 19 developing human hearts in a blinded manner, and a semi-quantitative measure for the expression level of each protein was determined in at least six anatomical structures or tissues, and across six developmental time points (Supplementary Table S4; Supplementary Figures S7–S13).
For all 12 proteins, we see strong evidence of heart developmental function (Supplementary Table S4). Importantly, 11 of the proteins (SNX9, DLL1 (DELTA), NOTCH3, JAG2, PTGS2 (COX-2), CAV3, SRC, MAPK3 (ERK1), MAPK8 (JNK1), BMX, and PTK2B) are specifically expressed in the anatomical structures associated with the morphological grouping of the functional network in which they participate (Supplementary Table S5 and Supplementary Figures S7–S13).
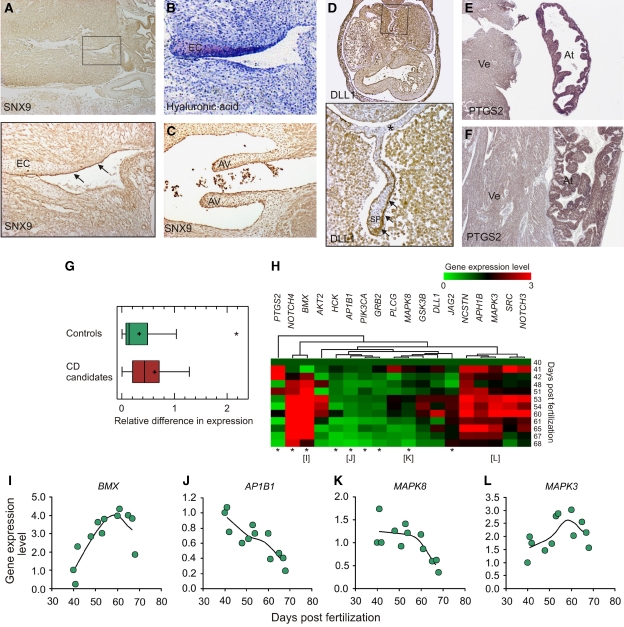
As a standardized estimate of the network-based prediction signal, we calculated the precision of our predictions as the true functional predictions among all functional predictions (or the amount of true positives among all positives). Using a conservative estimate of a true functional prediction, the precision is 0.72 (Supplementary Information; Supplementary Table S5), meaning that the morphological groups associated networks in which a candidate emerges, correlate with the candidates being specifically, and highly, expressed in relevant tissues at relevant time points in 72% of the cases. For example, SNX9, which is not associated with heart development in the literature, emerges in several networks associated with valve development (Supplementary Figures S2C and S3A). We confirmed this function of SNX9 by observing that it is highly expressed specifically in the cell populations driving the development of the endocardial cushions, which are anatomical precursors of the heart valves, and aortic valves (AVs; Figure 3A–C). Analogously, DLL1 and PTGS2 are predicted to be involved in the development of the atrial septum (Supplementary Figure S2A), and cardiac myocardium (Supplementary Figure S4A–E), respectively. These predictions are confirmed by their specific expression patterns in the relevant structures of the developing heart at the correct developmental time points (Figure 3D–F). For enlargements of IH pictures, many more details, examples of antibody specificity, and a more thorough discussion of the functional roles of these candidates see Supplementary Information and Supplementary Figures S7–S14.
Figure 3.
Examples of functional validations of candidates emerging from the networks. (A–F) The functional validations rely on a total of 382 tissue sections of which only a very small subset (6/382 or <2%) are shown in this figure. (A) SNX9 is located in clusters of unknown function involved in the development of the cardiac valves (Supplementary Figures S2C and S3A), which correlates with its specific expression in endothelial cells of the endocardial cushions (EC, precursors of heart valves) in a 9-week-old human heart. Note that strong expression of SNX9 is confined to the endothelial cells lining the developing portion of the EC (section below (A)). (B) Toluidine blue staining of hyaluronic acid, a marker for epithelial-mesenchymal transformation in the endocardial cushions correlates with the expression pattern of SNX9, hereby confirming its expression specifically in the developing parts of the EC. (C) SNX9 expression in endothelial cells of the developing aortic valves (AVs) in a 9-week-old human heart. Strong expression is shown in endothelial cells lining the valves and in cells within the valves. (D) DLL1 expression in the leading edge of septum primum (SP) in a 6-week-old human heart. DLL1 is located in a NOTCH signaling cluster involved in the development of the atrial septum (Supplementary Figure S2A). Note the stronger expression in the migrating and developing part of the SP (arrows) compared with endothelial cells lining the inner surface of the atrial wall (asterisk). (E, F) Expression in cardiomyocytes of the ventricle and atrium of PTGS2 in an 18-week-old human heart. PTGS2 is located in several clusters involved in myocardial growth and organization (Supplementary Figure S4A–E). Note the stronger expression of PTGS2 in the atrium (At) compared with the ventricle (Vt). (G–L) Validating a larger set of 49 candidates by real-time quantitative RT–PCR. (G) The gene expression level of the candidate genes and 29 control genes were measured in two hearts collected from embryos at ages 46 days and 67 days, respectively. Controls were genes corresponding to randomly chosen proteins that did not significantly interact with the 255 CD proteins, but were represented in our interaction data set. The data distribution is displayed by a box-and-whisker plot. A single outlier data point in the control group is shown with an asterisk. (H) Heatmap showing the relative level of gene expression of 18 representative candidates (level at day 40=1). The gene expression level of each of the 18 candidate genes was analyzed in hearts collected from 12 human embryos or fetuses of the indicated stages of development (between days 40–68 post-fertilization). The data were sorted in four groups according to expression pattern of the genes using hierarchical clustering. Statistical significant correlation between expression value and days post-fertilization is marked with an asterisk. (I–L) Representative plots of gene expression within the four groups. A trend line representing the average value of the data in four groups of three data points is shown as a smoothed line. Expression levels were measured by QPCR, and the data were normalized using the average value of six housekeeping genes (GAPDH, COX4A, B2M, ATP6A, HPRT, and RPL13).
To further validate the network data, we carried out expression profiling of the larger set of 49 candidates across different developmental stages using quantitative real-time RT–PCR on RNA extracted from 14 embryonic human hearts (Figure 3G–L). The candidates were significantly differentially expressed during heart development, compared with a set of controls (Figure 3G; Mann–Whitney (Wilcoxon) W test, P<0.006; Supplementary Table S6), and significantly higher expressed in heart tissues than random controls (P=0.016; Supplementary Figure S15; Supplementary Information). To investigate this trend in more detail, we analyzed the relative expression levels of a subset of the candidates in 12 additional hearts at 12 different time points between 40 and 67 days post-fertilization (Supplementary Table S7). This analysis showed that half of the candidates were significantly differentially expressed across these 12 time points further supporting their function in human heart development (Figure 3H–L; Supplementary Table S7). Together with the IH results, these data strongly establish the biological signal in our network data, and the high accuracy of the module maps.
Conclusion
We present a framework for gaining new insights into the systems biology of the protein networks driving organ development and related polygenic human disease phenotypes, exemplified here with heart development and CHD. Our analysis is the first example of large-scale integration of phenotypic data from targeted mice mutants with high-confidence experimental proteomics data and represents the most comprehensive characterization and analysis of the functional protein networks underlying the development of an organ system to date. A strength of our approach is that it immediately puts new candidates in the functional context of other, more well-characterized, network components.
We have shown that analysis of organ development at the systems level can be used to discover new developmental modules, gain insight into the evolution of organs, and understand the biology of highly polygenic disorders associated with aberrant organ development. A weakness of the method is the lack of cellular resolution of the networks due to use of macroscopic phenotypes as the starting point of the analysis. However, the morphological subgroups associated with the networks, and the IH data (which has the resolution of individual cells), strongly suggests in which cell populations the individual networks are active. The networks generated here can be used as a community resource for addressing major questions in developmental and cardiac biology, and we have made a database of the relevant network data available at http://www.cbs.dtu.dk/suppl/dgf/.
In principle, the framework can be applied to any organ, to widen our understanding of the functional architecture of protein networks that drive the formation of organs. In addition, they can facilitate the evolution of novel approaches in regenerative medicine, because a thorough characterization and understanding of the genes, proteins, pathways, and concepts underlying organ developmental programs will be necessary for the successful manipulation of these systems in tissue-engineering technologies and stem cell therapy. Finally, the networks can be used as a functional scaffold for understanding combinatorial effects of gene–gene and gene–environment interactions in complex heart phenotypes.
Materials and methods
Generating a functional network
A network is generated by determining the first- and second-order interactions of CD proteins associated with a given morphological subgroup in a human protein interaction network consisting of refined experimental proteomics data. This network is described in high detail in Lage et al (2007, 2008), and online (http://www.cbs.dtu.dk/suppl/dgf/). The full network (InWeb 29) can be downloaded from (http://www.cbs.dtu.dk/suppl/dgf/). Interactions of the CD proteins are integrated into a network by always including direct interactions between CD proteins, and only including indirect interactions mediated through proteins with Q percent of its interactions to the CD set. Various thresholds for Q are iteratively tested and value of Q for the final network is chosen based on which value gives the optimal network significance, this procedure is described in detail in Bergholdt et al (2007) and D'Hertog et al (2007). The method for determining network significances can be seen below. Detailed views of the networks can be seen in Supplementary Figures S1, S2, S3 and S4.
Determining network significances
The significance of each of the generated 19 networks was determined by randomization testing as described in detail earlier (Bergholdt et al, 2007; D'Hertog et al, 2007). Specifically, for an input set of Ninput proteins yielding an interaction network (connected component) with G input proteins and T total proteins, a network score (NSinput) was determined. This network score is the fraction of input proteins of all proteins in the network (G/T). We then determined the significance of the network score by empirically estimating the probability of observing a similar or better network score in networks generated by using 10 000 random input sets of size Ninput. The random gene sets were chosen so the degree distribution of proteins in the random sets approximate the input set. As each query generates a varying number of networks (connected components) the probability estimates can be calculated from the total amount of networks produced by all 10 000 randomizations that have a network score > NSinput. For this reason, network P-values can be lower than the amount of random queries. All network P-values can be seen in Supplementary Figures S1, S2, S3 and S4 below the title of the network. To rule out the chance of functional bias in the CD set, we analyzed the set for bias as discussed in Supplementary Information.
Identifying candidates for IH and expression analyses
A set of raw candidates were determined by querying all proteins in our interaction network for the amount of interactions to the CD set and determining the hypergeometric probability of this interaction profile. Out of all proteins in the proteome, 49 novel candidates had a significant interaction profile to the CD proteins after adjustment for multiple testing (described in detail in Supplementary Information and Supplementary Figure S5). We then used the functional networks assigned to each morphological subgroup to determine the most likely developmental function of the candidates. This was done by identifying the specific subnetworks to which the interactions of the candidates were most significant (as described in Supplementary Information and Supplementary Figure S6). In all, 12 of the 49 candidates were chosen for IH analysis based on the overlap between morphological subgroups and the developmental stages present in our panel of embryonic hearts available for validation experiments.
Human embryonic and fetal heart tissues
Human embryonic tissues were collected from legal abortions, according to the Helsinki Declaration II, and their use was approved by the local science ethics committee. Embryonic or fetal age was based on measurement of crown-rump length. Immediately after dissection, the samples were snap frozen in liquid nitrogen or treated with RNA later according to the manufacturer's instructions (Ambion, Austin, TX). Samples for IH were dissected into appropriate tissue blocks and fixed for 12–24 h at 4 °C in either 10% neutral-buffered formalin, 4% Formol-Calcium, Lillie's or Bouin's fixatives. The specimens were dehydrated with graded alcohols, cleared in xylene and paraffin embedded. Serial sections, 3–5 μm thick, were cut in transverse, sagittal or horizontal planes and placed on silanized slides.
Immunohistochemical analysis
Sections were deparaffinized and rehydrated in xylene followed by a series of graded alcohols according to the established procedures. The sections were treated with a fresh 0.5% solution of hydrogen peroxide in methanol for 15 min to quench endogenous peroxidase and then rinsed in TRIS-buffered saline (TBS, 5 mM Tris–HCl, 146 mM NaCl, pH 7.6). Non-specific binding was inhibited by incubation for 30 min with blocking buffer (ChemMate antibody diluent S2022, DakoCytomation, Glostrup, Denmark) at room temperature. The sections were then incubated overnight at 4°C with the primary antibody in blocking buffer (ChemMate antibody diluent S2022, DakoCytomation). The sections were washed with TBS and then incubated for 30 min with a peroxidase-labeled secondary antibody. The sections were washed with TBS, followed by incubation for 10 min with 3,3′-diamino-benzidine chromogen solution. Positive staining was recognized as a brown color. The sections were dehydrated in graded alcohols followed by xylene and coverslipped with DPX mounting media. Non-immune rabbit IgG1 (X0936) was used as negative control. Specificity of the antibodies were determined by their ability to stain specific cell populations in the tissue sections (examples are shown in Supplementary Figures S7–S12).
The following primary antibodies were used: anti-PTGS2 (35-8200, Invitrogen), anti-MAPK8 (SC-6254, Santa Cruz Biotechnology, Santa Cruz, CA), anti-CAV3 (610421, BD transduction laboratories, Franklin Lakes, NJ), anti-MAPK3 (SC-7383, Santa Cruz Biotechnology), anti-SRC (AT-7016, MBL International, Woborn, MA), anti-JAG2 (SC-8157, Santa Cruz Biotechnology), anti-DLL1 (SC-9102, Santa Cruz Biotechnology), anti-NOTCH3 (SC-7474, Santa Cruz Biotechnology), anti-NOTCH4 (SC-5594, Santa Cruz Biotechnology), anti-BMX (ab73887, Abcam, Cambridge, UK), anti-PTK2B (ab78119, Abcam), anti-BMP4 (ab31165, Abcam ), Anti-EGFR (#2232, Cell Signaling Technology, Boston, MA).
Real-time quantitative RT–PCR (QPCR)
We chose quantitative real-time quantitative RT–PCR for this analysis because it is considered to be the most accurate and sensitive method for detecting RNA differences also at very small amounts (Lang et al, 2009). Total RNA was isolated from tissues using TRIzol Reagent (Invitrogen, Taastrup, Denmark) and cDNA synthesized with SuperScript II (RNase H−) reverse transcriptase (Invitrogen) according to the manufacturer's instructions. QPCR analysis was carried out on an ABI 7500 Fast real-time PCR system using a LightCycler FastStart DNA MasterPLUS SYBR GreenI kit (Roche, Hvidovre, Denmark). Primer sequences used for QPCR analysis are available on request. To exclude that polymorphic gene expression between the individual developing hearts could account for the observed differential expression trends reported by QPCR, we also used polony multiplex analysis of gene expression (Kim et al, 2007) to measure the expression of the 49 candidates in right ventricular outflow tract from TOF patients at the time of primary surgical repair and left ventricle collected from patients with either heart failure or diabetic cardiomyopathy. The expression levels of the candidates were compared with the expression levels of a different set of 49 randomly chosen controls after normalizing both gene sets against gene expression in glioblastoma tissue. Here, the heart developmental candidates were significantly higher expressed in heart tissue than the controls (P=0.016) (see Supplementary Information and Supplementary Figure S13).
Supplementary Material
Supplementary text
Acknowledgments
We thank Christine Kocks, Barbara Pober, Chris Cotsapas, Josh Korn, Soumya Raychaudhuri, Ben Voight, Gerasimos Sykiotis, and Lars Juhl Jensen for critical discussions. We are also grateful to the Charles Lee laboratory for discussions and input on the project and results. This work was supported by The Danish Heart Foundation, the Villum Kann Rasmussen Foundation, and The Novo Nordisk Foundation. Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation. KL is supported by a grant from ‘Forskningsrådet for Sundhed og Sygdom'; and KL and PKD are supported by NICHD RO1 grant HD055150-03. We thank Lillian Rasmussen and Kirsten Winther for technical assistance.
Footnotes
The authors declare that they have no conflict of interest.
References
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE (1997) Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15: 30–35 [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD (1999) Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 104: 1567–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholdt R, Storling ZM, Lage K, Karlberg EO, Olason PI, Aalund M, Nerup J, Brunak S, Workman CT, Pociot F (2007) Integrative analysis for finding genes and networks involved in diabetes and other complex diseases. Genome Biol 8: R253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9: 587–600 [DOI] [PubMed] [Google Scholar]
- Bruneau BG (2008) The developmental genetics of congenital heart disease. Nature 451: 943–948 [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA (2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res 36: D724–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien KR, Domian IJ, Parker KK (2008) Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science (New York, NY) 322: 1494–1497 [DOI] [PubMed] [Google Scholar]
- D'Hertog W, Overbergh L, Lage K, Ferreira GB, Maris M, Gysemans C, Flamez D, Cardozo AK, Van den Bergh G, Schoofs L, Arckens L, Moreau Y, Hansen DA, Eizirik DL, Waelkens E, Mathieu C (2007) Proteomics analysis of cytokine-induced dysfunction and death in insulin-producing INS-1E cells: new insights into the pathways involved. Mol Cell Proteomics 6: 2180–2199 [DOI] [PubMed] [Google Scholar]
- Fishman MC, Olson EN (1997) Parsing the heart: genetic modules for organ assembly. Cell 91: 153–156 [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447 [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D (2005) Mutations in NOTCH1 cause aortic valve disease. Nature 437: 270–274 [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE (2002) Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science (New York, NY) 295: 821–825 [DOI] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE (2004) Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol 2: e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Perez-Pomares JM, de la Pompa JL (2007) Notch signaling is essential for ventricular chamber development. Dev Cell 12: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, Huynen MA, de Vlieg J, Groenen PM (2006) Benchmarking ortholog identification methods using functional genomics data. Genome Biol 7: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Alon U (2005) Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci USA 102: 13773–13778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS (2001) A gene expression map for Caenorhabditis elegans. Science (New York, NY) 293: 2087–2092 [DOI] [PubMed] [Google Scholar]
- Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG (2007) Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science 316: 1481–1484. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Tabata T (1993) Segmentation of the Drosophila embryo. Curr Opin Genet Dev 3: 585–594 [DOI] [PubMed] [Google Scholar]
- Lage K, Hansen NT, Karlberg EO, Eklund AC, Roque FS, Donahoe PK, Szallasi Z, Jensen TS, Brunak S (2008) A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci USA 105: 20870–20875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, Moreau Y, Brunak S (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotech 25: 309–316 [DOI] [PubMed] [Google Scholar]
- Lang JE, Magbanua MJ, Scott JH, Makrigiorgos GM, Wang G, Federman S, Esserman LJ, Park JW, Haqq CM (2009) A comparison of RNA amplification techniques at sub-nanogram input concentration. BMC Genomics 10: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson H, Pollack JB, Suh NP (2002) On the origin of modular variation. Evolution 56: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH (2002) Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev Biol 246: 68–85 [DOI] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM (2003) Cardiac chamber formation: development, genes, and evolution. Physiol Rev 83:1223–1267. [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL (2002) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33 Database Issue: D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN (2004) A decade of discoveries in cardiac biology. Nat Med 10: 467–474. [DOI] [PubMed] [Google Scholar]
- Olson EN (2006) Gene regulatory networks in the evolution and development of the heart. Science 313: 1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O (2003) The segmentation clock: converting embryonic time into spatial pattern. Science (New York, NY) 301: 328–330 [DOI] [PubMed] [Google Scholar]
- Satou Y, Satoh N (2006) Gene regulatory networks for the development and evolution of the chordate heart. Genes Dev 20: 2634–2638 [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG (1998) Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science (New York, NY) 281: 108–111 [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S (2003) Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol 13: 8–15 [DOI] [PubMed] [Google Scholar]
- Srivastava D (2006) Making or breaking the heart: from lineage determination to morphogenesis. Cell 126: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM (2007) The road to modularity. Nat Rev Genet 8: 921–931 [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N et al. (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science (New York, NY) 320: 539–543 [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S (1998) Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev 12: 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text