Abstract
We previously reported up-regulation of aryl hydrocarbon receptor (AhR) expression as a mechanism by which overexpression of Cu/Zn-superoxide dismutase (SOD) and/or catalase accelerates benzo(a)pyrene (BaP) detoxification in mouse aorta endothelial cells (MAECs). The objective of this study was to investigate the regulatory role of specificity protein-1 (Sp1) in AhR expression in MAECs that overexpress Cu/Zn-SOD and/or catalase. Our data demonstrated comparable levels of nuclear Sp1 protein in the transgenic and wild-type MAECs; however, binding of Sp1 protein to the AhR promoter region was more than 2-fold higher in MAECs overexpressing Cu/Zn-SOD and/or catalase than in wild-type cells. Inhibition of Sp1 binding to the AhR promoter by mithramycin A reduced AhR expression and eliminated the differences between wild-type MAECs, and three lines of transgenic cells. Functional promoter analysis indicated that AhR promoter activity was significantly higher in MAECs overexpressing catalase than in wild-type cells. Mutation of an AhR promoter Sp1-binding site or addition of hydrogen peroxide to the culture medium reduced AhR promoter activity, and decreased the differences between wild-type MAECs and transgenic cells overexpressing catalase. These results suggest that increased Sp1 binding to the AhR promoter region is an underlying mechanism for up-regulation of AhR expression in MAECs that overexpress Cu/Zn-SOD and/or catalase.
Keywords: Specific protein-1, aryl hydrocarbon receptor, endothelial cells, catalase, Cu/Zn-superoxide dismutase
INTRODUCTION
We recently demonstrated that overexpression of Cu/Zn-superoxide dismutase (SOD) and/or catalase inhibited benzo(a)pyrene (BaP)-induced atherosclerosis in hypercholesterolemic mice [1], accelerated BaP detoxification, and up-regulated aryl hydrocarbon receptor (AhR) expression in mouse aorta endothelial cells (MAECs) [2]. AhR is a ligand-activated transcription factor that regulates the expression of phase I and phase II xenobiotic-metabolizing enzymes [3-5]. It is known that phase I enzymes, i.e., cytochrome P450 (CYP) enzymes, metabolize BaP into reactive intermediates, whereas phase II enzymes detoxify BaP reactive intermediates and other xenobiotic chemicals, usually by addition of a conjugate to the functional group resulting in the conversion of relatively hydrophobic chemicals to hydrophilic products that are more readily excreted [6]. Phase I and phase II enzymes therefore work together to detoxify toxins such as BaP. Our previous studies demonstrated that overexpression of Cu/Zn-SOD and/or catalase up-regulated the expression of phase I and phase II enzymes, such as CYP1A1 and glutathione S-transferase (GST), and that knockdown of AhR reduced CYP1A1 and GST expression and decreased the beneficial effect of Cu/Zn-SOD and/or catalase overexpression on BaP detoxification [2]. Taken together, these findings suggest that up-regulation of AhR expression is a causal mechanism underlying the increase in xenobiotic-metabolizing enzyme expression and BaP detoxification in endothelial cells that overexpress Cu/Zn-SOD and/or catalase [2].
The expression of AhR is regulated by a number of genetic and epigenetic factors. Specifically, DNA hypermethylation [7] and histone deacetylation [8] have been shown to down-regulate AhR expression, whereas activation of transcription factor specificity protein-1 (Sp1) increases AhR transcription [9]. Sp transcription factors form a family of four C2H2 zinc-finger DNA binding proteins (Sp1-4) that regulate basal and inducible transcription of many genes via binding to GC boxes (GGGCGG), GT motifs (GGGTGTGGC), or CT elements (CCTCCTCCTCCTCGGCCTCCTCCCC) in the promoter region of target genes [10]. Sp1, Sp2, and Sp4 often function as activators of promoter activity, whereas Sp3 can function as either a transcriptional activator or repressor. Within the Sp-family of transcription factors, Sp1 is ubiquitously expressed, and is particularly important for regulation of the TATA-less genes [10,11]. Nucleotide sequence analysis revealed that the mouse AhR gene lacks a TATA box and contains four Sp1-binding sites within a highly GC-rich region [12]. Furthermore, functional analysis of the AhR promoter region demonstrated that some of the Sp1-binding sites are involved in regulation of AhR expression [9,13].
In the present study, we examined the regulatory role of Sp1 in AhR expression in MAECs that overexpress Cu/Zn-SOD and/or catalase. Our data suggest that increased binding of Sp1 to the AhR promoter is a mechanism by which overexpression of Cu/Zn-SOD and/or catalase upregulates AhR expression. These findings contribute to the larger goal of understanding the molecular mechanisms underlying the protective role of Cu/Zn-SOD and/or catalase overexpression against BaP-induced atherosclerosis.
MATERIALS AND METHODS
Isolation and culture of mouse aortic endothelial cells
Transgenic mice overexpressing human Cu/Zn-SOD (hsod1Tg), human catalase (hcatTg), or both Cu/Zn-SOD and catalase (Sod/CatTg) were generated as described previously [14]. For the present study, endothelial cells were obtained from the aortas of these transgenic mice and their wild-type littermates using an outgrowth technique, and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin at 37°C in a 95% air and 5% CO2 atmosphere. These cells displayed a cobblestone-like monolayer and expressed von Willebrand factor and platelet-endothelial cell molecule-1 (CD31), characteristics of freshly isolated endothelial cells. In addition, endothelial cells from hSod1Tg, hCatTg, and Sod/CatTg mice had approximately 2.5-fold higher activities of Cu/Zn-SOD and/or catalase compared with cells obtained from their wild-type littermates [15]. The 8th and 9th passages of cells were used for experiments.
Western blot analysis
MAECs grown to confluence in 100-mm culture dishes were made quiescent by culture in serum-free DMEM for 12 h. In experiments involving mithramycin A, MAECs were incubated for 24 h with 50-500 nM mithramycin A or culture medium alone as a control. For whole-cell protein extraction, cells were lysed in M-PER mammalian protein extraction reagent (Thermo Scientific, Rockford, IL) and cell lysates were subjected to sonication and centrifugation to remove cellular debris. For nuclear extracts, nuclei were isolated using a nuclear isolation kit, and lysed in nuclear extraction reagent according to the manufacturer’s protocol (Thermo Scientific). Final protein concentrations were determined using a BCA protein assay kit (Thermo Scientific). Samples containing 5 μg (for detection of β-actin) or 35 μg (for detection of AhR and Sp1) protein were separated on a 10% SDS-polyacrylamide gel, and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). After blocking with 5% fat-free milk, the membranes were incubated with antibodies against β-actin, AhR, or Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA), and then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology). Immunoreactive bands were visualized using ECL-plus chemiluminescence reagent (GE Healthcare Healthcare-Amersham, Piscataway, NJ), and analyzed with a GS-700 Imaging Densitometer (Bio-Rad, Hercules, CA).
Quantitative real-time RT-PCR assay
Total RNA was extracted using RNAeasy plus mini kits (Qiagen, Valencia, CA). Reverse transcription was performed using a SuperScript first-strand synthesis system with random hexamers as primers (Applied Biosystems, Foster City, CA). The resulting cDNAs were subjected to quantitative real-time PCR with an iCycler system (Bio-Bad) using specific primers provided by Qiagen for amplification of Sp1 and AhR. Reactions were performed in a final volume of 20 μl with reagents from an ABsolute blue QPCR SYBR Green Fluorescein kit (ABgene, Rochester, NY). A standard curve of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was generated by 10-fold serial dilutions of total RNA. The expression levels of target mRNAs were normalized to the GAPDH mRNA level [2].
Chromatin immunoprecipitation (ChIP) analysis
MAECs were grown to confluence in 100 mm tissue culture dishes. In experiments involving H2O2, MAECs were incubated with 25-100 μM H2O2 for 2 h or with culture medium alone as a control. In experiments involving mithramycin A, MAECs were incubated for 24 h with 500 nM mithramycin A or culture medium alone as a control. Cells were cross-linked with 1% formaldehyde at room temperature. After washing three times with ice-cold PBS, cell pellets were resuspended in 500 μl cell lysis buffer (5 mM Pipes, 85 mM KCl, 0.5% Nonidet P-40, 1 mM PMSF, and protease inhibitor cocktail, pH 8.0) for 20 min on ice and centrifuged at 5000 rpm for 5 min. The pellet containing nuclei was homogenized in 1 ml lysis buffer (50 mM Tris–HCl, 1% SDS, 10 mM EDTA, and protease inhibitor cocktail, pH 8.1). The suspension was sonicated until cross-linked chromatin was sheared to an average DNA fragment length of 0.3-1.0 kb, and then centrifuged at 14,000 rpm for 10 min. Five microliters of supernatant was used as an input control, and the remaining supernatant was diluted 10-fold with ChIP dilution buffer (16.7 mM Tris–HCl, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, and protease inhibitor cocktail, pH 8.1), and precleaned with salmon sperm DNA/protein A-agarose (Santa Cruz Biotechnology). The precleaned sample was incubated overnight at 4°C with Sp1 antibody or rabbit IgG as a non-specific binding control. The reactants were then incubated with salmon sperm DNA/protein A-agarose for another 3 h. The bound protein-DNA complex was eluted with a solution containing 0.1 M NaHCO3 and 1% SDS. After reversion of the protein-DNA cross-links, DNA fragments in the eluate and the input controls were purified using a QIAquick PCR purification kit (Qiagen). PCR amplification of a 192-bp fragment containing Sp1-binding sites 2-4 of the AhR promoter region (Fig. 1) was performed with the following primers: 5′-AACAGGTACTGGGCGAACAC-3′ (forward); and 5′-CTCCGCAGCCGCGACCCCGC-3′ (reverse). The PCR signals obtained from the IgG-immunoprecipitated DNA were subtracted from those obtained from the anti-Sp1-immunoprecipitated DNA. The DNA-binding activity of Sp1 was expressed as the ratio of the PCR product from the anti-Sp1 immunoprecipitation versus that from the input controls [2].
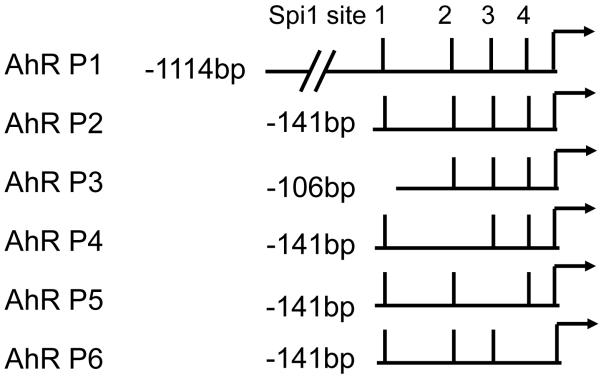
Fig. 1. Chimeric Ahr promoter constructs.
The four Sp1 binding sites in the AhR promoter region are indicated by solid bars. The arrow indicates the major transcription initiation site. For simplification, the bar furthest upstream of the transcription initiation site is designated as the first Sp1-binding site, and others are numbered consecutively towards the initiation site.
Recombinant plasmid construction and cell transfection
Four putative Sp1 binding sites have been described in the AhR promoter region, spanning nucleotides −12 to −140 upstream of the major transcription initiation site [12]. For functional analysis of the AhR promoter, six AhR promoter fragments (AhR P1-6) (Fig. 1) were generated by PCR using AccuPrime™ GC-Rich DNA Polymerase (Invitrogen, Carlsbad, CA) and C57BL/6J mouse genomic DNA as a template. The reverse primer for these fragments starts at nucleotide 88 downstream from the transcription start site of the AhR gene. The forward primers for AhR P1, P2, and P3 start at nucleotides −1114, −141, and −106 upstream from the transcription start site of the AhR gene, respectively. AhR P4, P5, and P6 were generated from AhR P2 as a template using two rounds of PCR [16]. Specifically, AhR P4 was generated using the PCR primers 5′-TAGAATCCTCTTCGCAGCACG-3′ (reverse), and 5′-CGTGCTGCGAAGAGGATTCTACCCTCCTGGGAACCGTGA-3′ (forward) to mutate the second Sp1 binding site in the AhR promoter; AhR P5 was generated using PCR primers 5′-GTCTCACGGTTCCCAGGAGG-3′ (reverse), and 5′-CCTCCTGGGAACCGTGAGACTACAAGCCGAGCAGGTGGGGC-3′ (forward) to mutate the third Sp1 binding site in the AhR promoter; and AhR P6 was generated with primers 5′-ATTACCTGCTCGGCCCCG-3′ (reverse) and 5′-CGGGGCCGAGCAGGTAATGCGGGGCTGGAGC-3′ (forward) to mutate the fourth Sp1-binding site in the AhR promoter region. PCR products were cloned into the XhoI/HindIII sites of the pGL2 luciferase vector (Promega, Madison, WI).
The recombinant plasmid constructs and a β-galactosidase expression plasmid pCMV-SPORT-βgal (Promega, Madison, WI) were co-transfected into MAECs using a lipofectamine 2000 kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Briefly, MAECs grown in 6-well plates to 60-70% confluence were incubated with 2 ml serum-free DMEM containing 10 μl lipofectamine 2000, 1 μg β-galactosidase expression plasmid, and 4 μg AhR promoter-luciferase reporter plasmid or 4 μg empty pGL2 luciferase vector. After 6 h transfection, cells were replenished with fresh medium containing 10% FBS and cultured for an additional 48 h prior to analysis by luciferase and β-galactosidase assays. In experiments involving H2O2, the indicated concentrations of H2O2 were added to the culture medium 18 h before the reporter enzyme assays.
Luciferase and β-galactosidase assays
Luciferase and β-galactosidase activities were determined using two commercial assay kits according to the manufacturer’s instructions (Luciferase Assay System, Promega, Madison, WI; and Galacto-Star™ System, Applied Biosystems, Bedford, MA, respectively). For luciferase assays, transfected cells were washed twice with PBS and lysed with 200 μl reporter lysis buffer. A 20 μl aliquot of the lysate was incubated with 100 μl luciferase assay reagent in a 96-well plate at room temperature for 4 min. For β-galactosidase assays, cells were lysed with 100 μl lysis buffer and 10 μl aliquots of the lysate were incubated with 100 μl reaction buffer in a 96-well plate at room temperature for 60 min. Luminescence derived from the luciferase and β-galactosidase assays was detected using a luminometer (BL10000 LumiCount, Packard BioScience, Meriden, CT). Luciferase activities were expressed as the luminosity ratio of the luciferase assay relative to that of the β-galactosidase assay. The AhR promoter-induced luciferase activity was expressed as the difference between cells transfected with the AhR promoter-luciferase reporter plasmid and the empty pGL2 luciferase vector.
Statistical analysis
For experiments using the microplate reader, the mean value for each experiment was calculated from triplicate wells in the same plate. Data were reported as mean ± standard error of the mean from at least five independent experiments. The differences between wild-type MAECs and three lines of transgenic cells were analyzed by one-way or multiple factor analysis of variance and student’s unpaired t-test. Differences were considered significant at a P value <0.05. Statistix software (Statistix, Tallahassee, FL) was used for statistical analysis of data.
RESULTS
The effect of overexpressing Cu/Zn-SOD and/or catalase on Sp1 expression
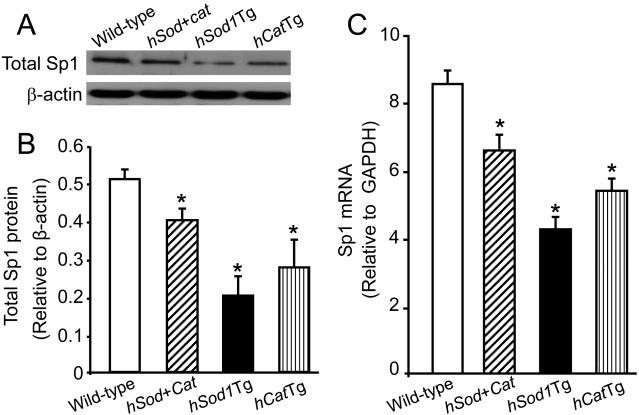
We previously reported that the AhR protein level in the hCatTg, Sod/CatTg and hSod1Tg MAECs was approximately 19-, 20-, and 18-fold higher, respectively, than in wild-type cells, and that this was mainly due to elevated expression of AhR mRNA [2]. Transcription factor Sp1 has been reported to enhance AhR expression [9]. Therefore, to investigate whether the increased AhR expression in MAECs that overexpress Cu/Zn-SOD and/or catalase is associated with increased Sp1 expression, we measured the Sp1 protein level in whole-cell lysates of hCatTg, Sod/CatTg, hSod1Tg, and wild-type MAECs. As shown in Fig. 2A-B, the Sp1 protein level in hCatTg, Sod/CatTg, and hSod1Tg MAECs was significantly lower than that in wild-type cells. Consistent with reduced Sp1 protein expression, the Sp1 mRNA level was also decreased in these transgenic MAECs: specifically, the Sp1 mRNA level in Sod/CatTg, hSod1Tg, and hCatTg MAECs was approximately 24%, 50%, and 31% lower, respectively, than in wild-type cells (Fig. 2C). Thus, overexpression of Cu/Zn-SOD and/or catalase does not up-regulate, but rather down-regulates, Sp1 expression in MAECs. These findings indicate that the previously observed up-regulation of AhR expression in hCatTg, Sod/CatTg, hSod1Tg MAECs is not the result of elevated Sp1 expression.
Fig. 2. The effect of overexpressing Cu/Zn-SOD and/or catalase on Sp1 expression.
MAECs were obtained from hSod1Tg, hCatTg, hSod+CatTg mice and their wild-type littermates. The Sp1 protein level (A and B) in whole-cell lysates was determined by immunoblotting and expressed relative to the immunoblot intensity of β-actin. The Sp1 mRNA level (C) was determined by quantitative real-time RT-PCR and expressed relative to the GAPDH mRNA level. Values represent the mean ± SEM of five separate experiments in which MAECs from four mice were pooled. *P<0.05 vs. wild-type cells.
The effect of overexpressing Cu/Zn-SOD and/or catalase on Sp1 binding to the AhR promoter
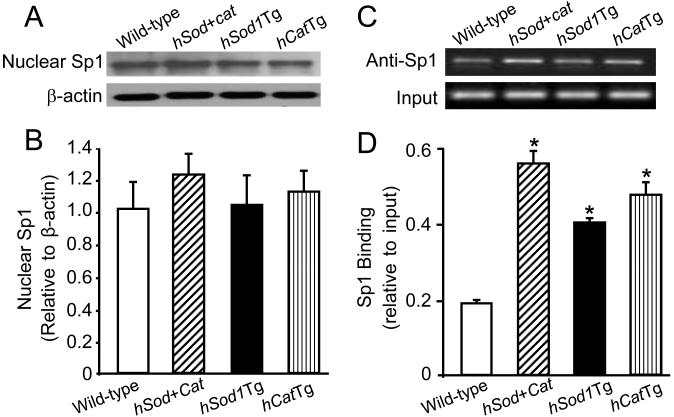
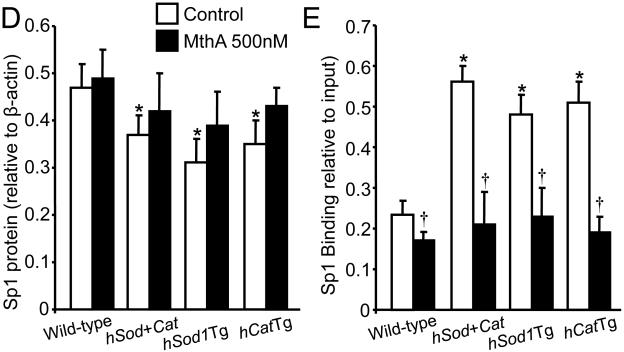
Sp1 translocates into the nucleus and enhances transcription via binding to the promoter region of its target genes [9,13]. We therefore determined the effect of overexpressing Cu/Zn-SOD and/or catalase on the level of nuclear Sp1 protein and the amount of Sp1 bound to the AhR promoter. As shown in Fig. 3A-B, the level of nuclear Sp1 was comparable between wild-type MAECs and transgenic cells overexpressing Cu/Zn-SOD and/or catalase. However, ChIP analysis indicated that the amount of AhR promoter fragments co-immunoprecipitated by anti-Sp1 antibody was approximately 2.8-, 2.0-, and 2.2- fold higher in Sod/CatTg, hSod1Tg, and hCatTg MAECs, respectively, than in wild-type cells (Fig. 3C-D). These results suggest that the amount of Sp1 protein bound to the AhR promoter is higher in transgenic cells that overexpress Cu/Zn-SOD and/or catalase than in wild-type cells.
Fig. 3. The effect of overexpressing Cu/Zn-SOD and/or catalase on nuclear Sp1 protein level and Sp1 DNA-binding activity.
The Sp1 protein level (A and B) in the nuclei of MAECs from hSod1Tg, hCatTg, and hSod+CatTg mice and their wild-type littermates was determined by immunoblotting and expressed relative to the immunoblot intensity of β-actin. The amount of Sp1 bound to the AhR promoter was determined using ChIP analysis, and expressed as the ratio of input controls. Values represent the mean ± SEM of five independent experiments in which MAECs from four mice were pooled. *P<0.05 vs. wild-type cells.
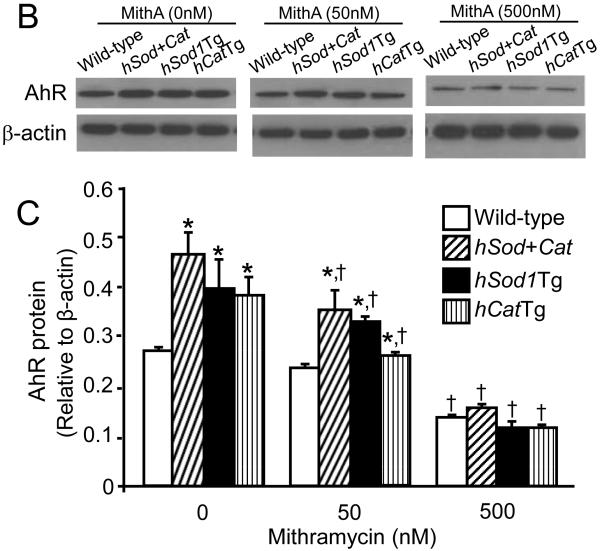
The effect of mithramycin A on Sp1 and AhR expression, and Sp1 binding to the AhR promoter
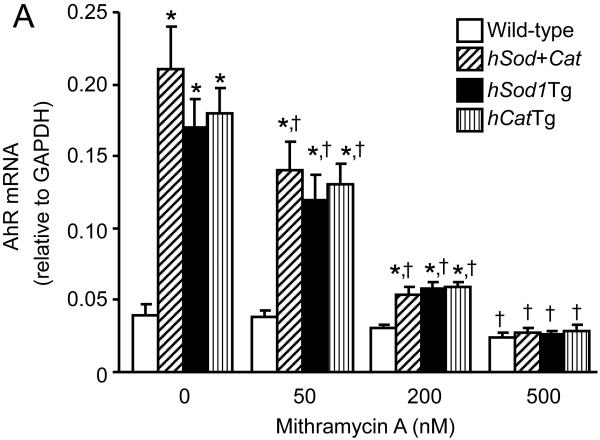
Mithramycin A is a chemotherapeutic drug that binds to GC-rich DNA sequences thereby blocking the binding of transcription factors such as Sp1 to GC-specific regions of DNA [17]. A wide range of concentrations of mithramycin A (10 nM - 1 μM) have been used to inhibit the transcriptional activity of Sp1 in various types of cells, including human bronchial epithelial cells and multiple myeloma cells [17,18]. Data from the present study showed that mithramycin A did not alter the Sp1 protein level (Fig. 4D), but significantly reduced the amount of Sp1 bound to the AhR promoter in both wild-type MAECs and transgenic cells overexpressing Cu/Zn-SOD and/or catalase, with a greater extent in transgenic cells (Fig. 4E). Correspondingly, wild-type and transgenic MAECs also showed different responses to mithramycin A with respect to AhR expression (Fig. 4A-C). Specifically, low concentrations (50 nM) of mithramycin A significantly reduced AhR mRNA (Fig. 4A) and protein (Fig. 4B-C) levels in Sod/CatTg, hSod1Tg, and hCatTg MAECs, but not in wild-type cells. However, a higher concentration of mithramycin A (500 nM) significantly reduced AhR mRNA and protein levels in wild-type MAECs and transgenic cells overexpressing Cu/Zn-SOD and/or catalase, and eliminated the differences in AhR expression between these cells (Fig. 4-C). Thus, the AhR expression level was comparable in wild-type and transgenic MAECs treated with high concentrations of mithramycin A. Together with the observation that more Sp1 protein bound to the AhR promoter region in Sod/CatTg, hSod1Tg, and hCatTg MAECs than in wild-type cells (Fig. 3C-D), these results suggest that increased Sp1 binding to the AhR promoter is a mechanism by which overexpression of Cu/Zn-SOD and/or catalase upregulates AhR expression.
Fig. 4. The effect of mithramycin A on Sp1 and AhR expression, and Sp1 binding to the AhR promoter.
MAECs obtained from hSod1Tg, hCatTg, hSod+Cat mice and their wild-type littermates were treated with the indicated concentrations of mithramycin A for 24 h. The Sp1 mRNA level (A) was determined by quantitative real-time RT-PCR and expressed relative to the GAPDH mRNA level. The AhR and Sp1 protein levels (B-D) were determined by western blot analysis and expressed relative to the immunoblot intensity of β-actin. The amount of Sp1 bound to the AhR promoter (E) was determined using ChIP analysis, and expressed as the ratio of input controls. Values represent means ± SEM of five separate experiments in which MAECs were pooled from four mice. *P<0.05 vs. wild-type cells under the same culture conditions; †P<0.05 vs. the same genotype cells without mithramycin A treatment.
The effect of catalase overexpression on AhR promoter activity
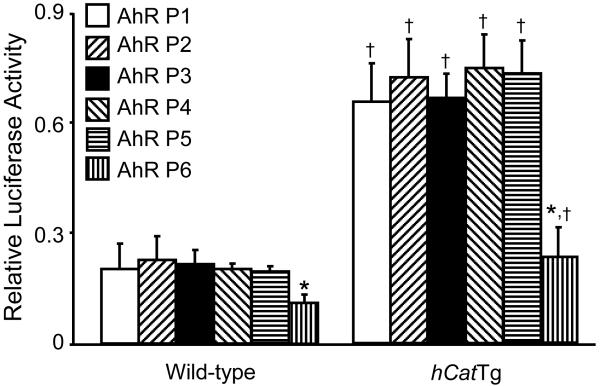
Basal transcription of the AhR gene largely depends on the Sp1-binding sites in the proximal promoter region, and deletion of as much as 5 kb of DNA sequence upstream of the multiple Sp1-binding sites does not affect AhR expression [13]. However, the capability of the Sp1-binding sites in the AhR promoter region to drive AhR transcription varies greatly. Specifically, the third and fourth Sp1-binding sites have been shown to be required for efficient basal AhR transcription in a number of cell lines, whereas the activity of the first and the second Sp1-binding sites appears to be cell-type specific and deletion or mutation of these sites reduces AhR promoter activity in some cell types but not in others [13,19]. The present study is the first to examine the transcription activity of these AhR promoter Sp1-binding sites in mouse endothelial cells by transfection of a series of AhR promoter-luciferase reporter chimeric constructs (Fig. 1). As shown in Fig. 5, the luciferase activity of all the chimeric constructs was significantly higher in hCatTg than in wild-type MAECs. Deletion of approximately 1 kb of DNA sequence upstream of the multiple Sp1-binding sites (AhR-P2 vs. AhR-P1) did not significantly affect promoter activity in wild-type or hCatTg MAECs. DNA sequence analysis reveals a number of functional motifs in this deleted fragment, including two AP1 binding sites, two AP2 binding sites, one GRE binding site, one E2A binding site, one YY1 binding site, one NF-E1 binding site, and one DRE binding site [12]. It therefore appears that these motifs are not responsible for the increased AhR promoter activity in hCatTg MAECs.
Fig. 5. Functional analysis of the Ahr promoter chimeric constructs.
AhR promoter-luciferase reporter plasmids were constructed and co-transfected into hCatTg and wild-type MAECs with a β-galactosidase expression plasmid as described in Materials and Methods. Luciferase activity was measured using a luminescence assay and expressed relative to the luminosity of the β-galactosidase assay. Values denote means ± SEM of five independent transfection experiments. *P <0.01 vs. the same genotype cells transfected with AhR-P2; †P <0.01 vs. wild type cells transfected with the same plasmid construct.
Data in Fig. 5 also show that luciferase activity was not affected by selective deletion of the first Sp1-binding site (AhR-P3), or mutation of the second (AhR-P4) or third (AhR-P5) Sp1-binding site. In contrast, mutation of the fourth Sp1-binding site (AhR-P6) significantly reduced luciferase activity in both wild-type and hCatTg MAECs, suggesting that this proximal Sp1-binding site is essential for regulation of basal expression of AhR in mouse endothelial cells. Moreover, the inhibitory effect of Sp1-binding site mutation on luciferase activity was greater in hCatTg than in wild-type MAECs: mutation of the fourth Sp1-binding site reduced luciferase activity approximately 3-fold in hCatTg and 2-fold in wild-type MAECs (Fig. 5). These findings indicate that the fourth Sp1 binding site in the AhR promoter region plays a role in the increased AhR promoter activity in MAECs that overexpress catalase.
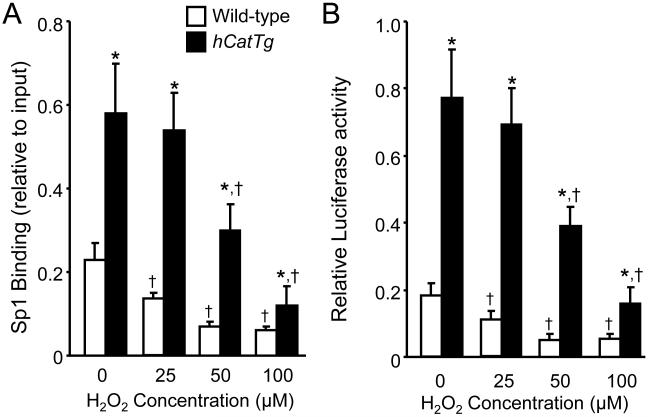
The effect of H2O2 on Sp1 binding and AhR promoter activity
Previous studies have shown that overexpression of Cu/Zn-SOD and/or catalase reduces cellular O2−· and H2O2 levels in mouse endothelial cells [2,15]. Here, we studied the effect of H2O2 on Sp1 binding and AhR promoter activity in wild-type and hCatTg MAECs. As shown in Fig. 6, low concentrations (25 μM) of H2O2 significantly reduced Sp1 binding and AhR promoter activity in wild-type cells but not in hCatTg cells, whereas higher concentrations (50 and 100 μM) of H2O2 reduced Sp1 binding and AhR promoter activity in both wild-type and hCatTg MAECs, and decreased the difference between these cells. We also observed that 200 μM H2O2 induced a further reduction in Sp1 binding and AhR promoter activity in hCatTg cells, and reduced viability of wild-type cells (data not shown). These observations provide evidence that overexpression of catalase increased Sp1 binding to the AhR promoter and enhanced AhR promoter activity via reduction of intracellular H2O2 levels. When the scavenging capability of catalase is overwhelmed by exogenously added H2O2, both the amount of Sp1 bound to the AhR promoter region and the activity of the AhR promoter are reduced.
Fig. 6. The effect of H2O2 on Sp1 DNA-binding activity and AhR promoter activity.
Panel A: Wild-type and hCatTg MAECs were treated with the indicated concentrations of H2O2 for 2 h. The level of Sp1 bound to the AhR promoter region was detected using ChIP analysis and expressed as the ratio of input controls. Panel B: Wild-type and hCatTg MAECs were co-transfected with an AhR promoter (AhR P2)-luciferase reporter plasmid and a β-galactosidase expression plasmid, and treated with the indicated concentrations of H2O2 for 18 h. The luciferase activity was measured using a luminescence assay and expressed relative to the luminosity of the β-galactosidase assay. Values represent the mean ± SEM of five independent experiments in which MAECs from four mice were pooled. *P<0.01 vs. wild-type cells treated with same concentration of H2O2, †P <0.05 vs. the same genotype cells without H2O2 treatment.
DISCUSSION
Data from this study identified the Sp1-binding site immediately upstream of the transcription initiation site, designated the fourth binding site, as a functional element of the AhR promoter. Specifically, introduction of mutations into this element significantly reduced the promoter activity of the AhR gene. In contrast, separate deletion or mutation of other three Sp1-binding sites did not significantly affect promoter activity. In addition, our data demonstrated that block of Sp1 binding to the AhR promoter by mithramycin A reduced AhR expression in both wild-type MAECs and transgenic cells that overexpress Cu/Zn-SOD and/or catalase. This effect was more pronounced in transgenic cells, and significantly diminished the difference in AhR expression between wild-type and transgenic MAECs. This finding implies that binding of Sp1 to the promoter region, i.e., the fourth Sp1-binding site, is essential for regulation of AhR transcription, and at least partially responsible for the increased AhR expression in cells that overexpress Cu/Zn-SOD and/or catalase.
Another important finding from this study is that the level of Sp1 protein in the nucleus was comparable between wild-type MAECs and transgenic cells overexpressing Cu/Zn-SOD and/or catalase, despite significantly increased Sp1 binding to the AhR promoter region in transgenic cells. These data suggest that overexpression of Cu/Zn-SOD and/or catalase increased the activity of Sp1 bound to the AhR promoter region. Cu/Zn-SOD is a protein that converts O2−· to H2O2, while catalase destroys H2O2 by converting it to molecular oxygen and water. Our previous studies consistently demonstrated that overexpression of catalase slightly reduced the basal level and markedly reduced the oxidant-induced level of H2O2 in mouse vascular cells, and that overexpression of Cu/Zn-SOD reduced cellular O2−·, but did not increase H2O2 level [2,14,15]. Taken together, it appears that both endogenously generated O2−· and H2O2 inhibit Sp1 binding to the AhR promoter, and down-regulate AhR expression. The inhibitory role of H2O2 is directly evidenced by the differential responses of wild-type and hCatTg MAECs to exogenous H2O2. Specifically, at low concentrations, H2O2 reduced Sp1 binding and repressed AhR promoter activity in wild-type MAECs but not in hCatTg cells. However, H2O2 at higher concentrations reduced Sp1 binding and AhR promoter activity in both wild-type and transgenic cells, and decreased the difference between these cells. This report did not directly examine the regulatory role of exogenous O2−· because of lacking reagents that exclusively elevate cellular O2−·. Considering the factor that overexpression of Cu/Zn-SOD reduced cellular O2−·, the hSod1Tg MAECs would be predicted to be less sensitive to O2−·-reduced Sp1 binding and AhR promoter activity, as compared with wild-type cells.
There is great interest in understanding the regulatory effect of cellular redox status on the DNA binding activity of Sp1. For example, it has been shown that purified Sp1 treated with H2O2 loses the ability to bind to its DNA cis-element, and that DNA binding efficiency is fully restored after incubation with the thiol reducing reagent dithiothreitol [20]. Further studies demonstrated that treatment of cells with diamide, a thiol oxidant, reduced Sp1 binding to the promoter region of its target genes, and repressed the promoter activity and transcription of these genes [19]. These findings suggest that oxidation of the cysteine sulfhydryl group is a mechanism by which oxidants such as H2O2 and diamide decrease the DNA-binding efficiency of Sp1 [19,20]. It is highly likely that reduction in cellular O2−· and H2O2 by overexpression of Cu/Zn-SOD and/or catalase protects Sp1 protein from oxidation, and therefore increases its ability to bind to the AhR promoter and up-regulates AhR expression.
In summary, the present study showed that the binding efficiency of Sp1 to the AhR promoter region is higher in endothelial cells that overexpress Cu/Zn-SOD and/or catalase than in wild-type cells, and that chemical inhibition of Sp1 binding to DNA by mithramycin A eliminated the difference in AhR expression between wild-type MAECs and the three lines of transgenic cells. Moreover, direct mutation of the fourth Sp1-binding site or addition of H2O2 to the culture medium repressed AhR promoter activity in both hCatTg and wild-type cells, with a greater effect in transgenic cells. These observations suggest that increased Sp1 binding to the AhR promoter region is a mechanism by which overexpression of Cu/Zn-SOD and/or catalase up-regulates AhR expression, and provide an insight into the molecular mechanisms underlying the protective role of these antioxidant enzymes against BaP in vascular cells [2] and atherosclerosis mouse models [1]. It is known that other TATA-less genes, particularly those that lack TATA elements but contain conserved Sp1-binding sites, are similarly redox-regulated [19,20], and it is highly likely that overexpression of antioxidant enzymes would affect the expression of those genes. Further studies are required to investigate the role of these Sp1 target genes in the beneficial effect of Cu/Zn-SOD and/or catalase overexpression. In addition, the Sp1 protein level in the whole-cell lysates was significantly lower in transgenic cells overexpressing Cu/Zn-SOD and/or catalase than in wild-type cells. It appears that increased Sp1 activity in a reduced redox status induces a compensatory down-regulation of Sp1 expression, which is consistent with a previous report showing that oxidative stress increases Sp1 expression [21]. Further studies are necessary to address this issue.
Acknowledgments
This study was supported by NIH grants G12RR003032 (Hong Yang), and R01ES014471 and R01HL089382 (ZhongMao Guo).
Footnotes
The authors have no conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yang H, Zhou L, Wang Z, Roberts LJ, Lin X, Zhao Y, Guo Z. Overexpression of antioxidant enzymes in ApoE-deficient mice suppresses Benzo(a)pyrene-accelerated atherosclerosis. Atherosclerosis. 2009;207:51–58. doi: 10.1016/j.atherosclerosis.2009.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang Z, Yang H, Ramesh A, Roberts LJ, Zhou L, Lin X, Zhao Y, Guo Z. Overexpression of Cu/Zn-superoxide dismutase and/or catalase accelerates benzo(a)pyrene detoxification by upregulation of the aryl hydrocarbon receptor in mouse endothelial cells. Free Radic. Biol. Med. 2009;47:1221–1229. doi: 10.1016/j.freeradbiomed.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fujii-Kuriyama Y, Mimura J. Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 2005;338:311–317. doi: 10.1016/j.bbrc.2005.08.162. [DOI] [PubMed] [Google Scholar]
- [4].Nebert DW. Drug-metabolizing enzymes in ligand-modulated transcription. Biochem. Pharmacol. 1994;47:25–37. doi: 10.1016/0006-2952(94)90434-0. [DOI] [PubMed] [Google Scholar]
- [5].Nebert DW, Puga A, Vasiliou V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann. N. Y. Acad. Sci. 1993;685:624–640. doi: 10.1111/j.1749-6632.1993.tb35928.x. [DOI] [PubMed] [Google Scholar]
- [6].Gelboin HV. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- [7].Mulero-Navarro S, Carvajal-Gonzalez JM, Herranz M, Ballestar E, Fraga MF, Ropero S, Esteller M, Fernandez-Salguero PM. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006;27:1099–1104. doi: 10.1093/carcin/bgi344. [DOI] [PubMed] [Google Scholar]
- [8].Garrison PM, Rogers JM, Brackney WR, Denison MS. Effects of histone deacetylase inhibitors on the Ah receptor gene promoter. Arch. Biochem. Biophys. 2000;374:161–171. doi: 10.1006/abbi.1999.1620. [DOI] [PubMed] [Google Scholar]
- [9].Fitzgerald CT, Nebert DW, Puga A. Regulation of mouse Ah receptor (Ahr) gene basal expression by members of the Sp family of transcription factors. DNA Cell Biol. 1998;17:811–822. doi: 10.1089/dna.1998.17.811. [DOI] [PubMed] [Google Scholar]
- [10].Yu JH, Schwartzbauer G, Kazlman A, Menon RK. Role of the Sp family of transcription factors in the ontogeny of growth hormone receptor gene expression. J. Biol. Chem. 1999;274:34327–34336. doi: 10.1074/jbc.274.48.34327. [DOI] [PubMed] [Google Scholar]
- [11].Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- [12].Garrison PM, Denison MS. Analysis of the murine AhR gene promoter. J. Biochem. Mol. Toxicol. 2000;14:1–10. doi: 10.1002/(sici)1099-0461(2000)14:1<1::aid-jbt1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- [13].Fitzgerald CT, Fernandez-Salguero P, Gonzalez FJ, Nebert DW, Puga A. Differential regulation of mouse Ah receptor gene expression in cell lines of different tissue origins. Arch. Biochem. Biophys. 1996;333:170–178. doi: 10.1006/abbi.1996.0378. [DOI] [PubMed] [Google Scholar]
- [14].Guo ZM, Van Remmen H, Yang H, Chen XL, Mele J, Vijg J, Epstein CJ, Ho Y-S, Richardson A. Changes in expression of antioxidant enzymes affect cell-mediated LDL oxidation and oxLDL-induced apoptosis in mouse aorta cells. Arterioscler. Thromb. Vasc. Biol. 2001;21:1131–1138. doi: 10.1161/hq0701.092092. [DOI] [PubMed] [Google Scholar]
- [15].Yang H, Shi MJ, Richardson A, Vijg J, Guo ZM. Attenuation of leukocyte-endothelium interaction by antioxidant enzymes. Free Radic. Biol. Med. 2003;35:266–276. doi: 10.1016/s0891-5849(03)00277-6. [DOI] [PubMed] [Google Scholar]
- [16].Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J. Clin. Invest. 1991;88:1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reamon-Buettner SM, Borlak J. Epigenetic silencing of cell adhesion molecule 1 in different cancer progenitor cells of transgenic c-Myc and c-Raf mouse lung tumors. Cancer Res. 2008;68:7587–7596. doi: 10.1158/0008-5472.CAN-08-0967. [DOI] [PubMed] [Google Scholar]
- [19].Wu X, Bishopric NH, Discher DJ, Murphy BJ, Webster KA. Physical and functional sensitivity of zinc finger transcription factors to redox change. Mol. Cell Biol. 1996;16:1035–1046. doi: 10.1128/mcb.16.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ammendola R, Mesuraca M, Russo T, Cimino F. The DNA-binding efficiency of Sp1 is affected by redox changes. Eur. J. Biochem. 1994;225:483–489. doi: 10.1111/j.1432-1033.1994.t01-1-00483.x. [DOI] [PubMed] [Google Scholar]
- [21].Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, Ratan RR. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]