Abstract
Neurogenesis during development depends on the coordinated regulation of self-renewal and differentiation of neural precursor cells. Chromatin regulation is a key step in self-renewal activity and fate decision of neural precursor cells. However, the molecular mechanism(s) of this regulation is not fully understood. Here, we demonstrate for the first time that MRG15, a chromatin regulator, is important for proliferation and neural fate decision of neural precursor cells. Neuroepithelia from Mrg15 deficient embryonic brain are much thinner than those from control, and apoptotic cells increase in this region. We isolated neural precursor cells from Mrg15 deficient and wild-type embryonic whole brains and produced neurospheres to measure the self-renewal and differentiation abilities of these cells in vitro. Neurospheres culture from Mrg15 deficient embryo grew less-efficiently than those from wild-type. Measurement of proliferation, using BrdU incorporation, revealed that Mrg15 deficient neural precursor cells have reduced proliferation ability and apoptotic cells do not increase during in vitro culture. The reduced proliferation of Mrg15 deficient neural precursor cells most likely accounts for the thinner neuroepithelia in Mrg15 deficient embryonic brain. Moreover, we also demonstrate Mrg15 deficient neural precursor cells are defective in differentiation into neurons in vitro. Our results demonstrate that MRG15 has more than one function in neurogenesis and defines a novel role for this chromatin regulator that integrates proliferation and cell-fate determination in neurogenesis during development.
Keywords: Neural precursor cell, development, chromatin, epigenetics, gene expression
INTRODUCTION
MRG15 is a chromodomain-containing nuclear protein that is evolutionarily highly conserved from yeast to human (Bertram et al. 1999; Bertram and Pereira-Smith 2001; Marin and Baker 2000). This high degree of conservation, together with the demonstrated roles of this protein in cell proliferation, regulation of gene expression and DNA repair, indicates that it is involved in fundamental processes at the cellular level. MRG15 associates with both histone acetyltransferases (HAT) and deacetylases (HDAC) and is involved in regulation of chromatin structure through association with these histone-modifying enzymes (Pardo et al. 2002). Epigenetic regulation by histone modification such as acetylation, methylation, and phosphorylation is dynamically and coordinately regulated in many physiological processes including development (Bhaumik et al. 2007).
MRG15 is present in the mammalian Tip60 HAT complex that is composed of at least sixteen subunits (Cai et al. 2005; Cai et al. 2003; Doyon et al. 2004; Hayakawa et al. 2007; Sardiu et al. 2008), and is very similar to that found in budding yeast (NuA4 complex)(Eisen et al. 2001) and Drosophila (dTip60 complex)(Kusch et al. 2004). Additionally, both dTip60 and dMrg15 have been shown to be essential for viability of Drosophila embryos and histone variant exchange during DNA double strand break repair (Downs and Cote 2005; Kusch et al. 2004).
MRG15 also associates with a mSin3/HDAC complex. Pf1 (or PHF12), which is a PHD finger containing protein, and appears to act as a scaffold protein in this complex because MRG15 and mSin3 can bind directly to Pf1 at different sites (Yochum and Ayer 2002). Although this complex has not been studied in detail in mammalian cells, in budding yeast it has been shown to be recruited to the coding regions of actively expressed genes to prevent improper transcriptional initiation. The chromodomain in Eaf3, the MRG15 ortholog of budding yeast, in association with the PHD finger in Rco1, the Pf1 ortholog, recognizes trimetylation at lysine 36 in histone H3 (H3K36me3) and inhibits transcriptional initiation at incorrect sites (Carrozza et al. 2005; Keogh et al. 2005; Li et al. 2007). Although the chromodomain of mammalian MRG15 can also recognize and bind H3K36me3 (Zhang et al. 2006), it is not known if this MRG15/mSin3/HDAC complex functions similar to that of yeast or whether it acts also as a transcriptional repressor.
Mrg15 knockout mice, which we have generated, exhibit perinatal embryonic lethality, cell growth defects and delayed development in many organ systems, including the brain (Tominaga et al. 2005). Mouse embryonic fibroblasts (MEFs) from Mrg15 deficient embryos proliferate poorly and after a very limited number of serial passages cease dividing, in part through premature induction of p21, the Cdk inhibitor (Tominaga et al. 2005).
The generation of the central nervous system, involves a tightly regulated balance between proliferation and differentiation of neural stem/progenitor cells, which we will refer to as neural precursor cells, and during embryonic development multipotent progenitors generate more restricted precursors and finally produce fully differentiated cell types such as neurons and glia (Gotz and Huttner 2005; Lledo et al. 2006; Ming and Song 2005; Pevny and Rao 2003). The fate determination processes which direct differentiation and maintenance of neural precursor cells are thought to be coordinately regulated by many intrinsic and extrinsic factors. Neural precursor cells from the brain of embryos and adult animals can be cultured and propagated in vitro as neurospheres, and require the presence of epidermal growth factor (EGF) and basic fibroblast growth factor 2 (FGF2) (Gritti et al. 1996; Reynolds et al. 1992; Reynolds and Weiss 1992; Reynolds and Weiss 1996; Tropepe et al. 1999; Vescovi et al. 1993). These neural precursor cells can be induced to differentiate in vitro into neurons, astrocytes, and oligodendrocytes following removal of mitogens. This neurosphere system has been used to determine whether defects in the brain, caused by mutation of genes, results from cell-autonomous defects in these cells or a loss of response to critical trophic factors.
In this study, we performed a histological analysis of Mrg15 null and control embryos to determine the role of MRG15 in neural precursor cell maintenance and differentiation during early development. The results indicated that in MRG15 null embryos the neural tube was much thinner than control and this decreased size was most likely a result of both the inability of neural precursor cells to enter mitosis and increased apoptosis in this cell population.
To further validate these results we used the in vitro neurosphere culture system and determined that Mrg15 deficiency leads to a decrease in the number and size of neurospheres obtained from the brain of null embryos when compared with wild-type. This is a consequence of a reduction in the number of growing cells and not an increase in apoptosis. In addition, Mrg15 deficient neural precursor cells were less efficient in differentiating into neurons when compared with wild-type cells. These data indicate that MRG15 is critical for the proper development, maintenance and differentiation of neural precursor cells.
MATERIALS AND METHODS
Tissue Preparation and Immunohistochemistry
Generation of Mrg15 null embryos has been described previously. Procedures were approved by the institutional animal care committee and were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals. We used a later generation (ten generations) of mixed background (C57BL/6J and 129SvEv) mice in this study. We could not obtain live Mrg15 null embryos later than E12.5 from this mouse colony, although we had been able to do so in the earlier generation of the mixed background. This difference most likely reflects the genetic background. Therefore, we used E10.5 embryos for the studies described here.
Embryos at E10.5 were isolated, fixed with 4% paraformaldehyde (Sigma, St. Louis, MO, USA) in PBS, pH 7.4 for 2 h, and cryoprotected in 30% sucrose overnight at 4°C. Embryos were frozen in OCT compound and cryosectioned (12-μm coronal sections). The sections were blocked in PBS-T (phosphate-buffered saline with 0.02% TritonX-100) with 5% skimmed milk and 5% normal goat sera for 5 min at room temperature, followed by incubation with the appropriate dilution of primary antibody in PBS-T containing 2% skimmed milk and 5% goat sera overnight at 4°C. The following primary antibodies used for this study: mouse anti-phospho-Ser/Thr-Pro, anti-MPM2 (1:10,000)(Upstate, Temecula, CA) and mouse anti-nestin (1:50, Developmental Studies Hybridoma Bani). Signal detection was performed with Alexa Fluor 647 conjugated goat anti-rabbit F (ab)2 fragment or Alexa Fluor 488 conjugated anti-mouse F (ab)2 fragment (1:1000) (Molecular Probes, Eugene, OR) for 45 min at 4°C. Detection of apoptotic cells was performed with In situ cell death detection kit (Roche Applied Science, Indianapolis, IN) for 1h at 37°C at the same time as secondary antibody incubation for nestin staining. The sections were examined with an LSM 5 PASCAL confocal microscope (Carl Zeiss, Deutschland, Germany).
Neural Precursor Cell (NPC) Culture
Neural precursor cells (NPCs) from embryonic brain were isolated and propagated using the methods developed by Reynolds et al and Reynolds and Weiss (Reynolds and Weiss 1992; Reynolds and Weiss 1996). Briefly, C57BL/129 mixed background mouse embryos, E10.5, were obtained and cells from the brain micro-dissected and triturated with a polished pasteur pipette. Neural cells were purified by subculturing twice in 35-mm tissue culture dishes (Corning Incorporated, Corning, NY, USA) using neural precursor cell growth medium which is Neurobasal Medium minus L-glutamine (Gibco-BRL, Gaithersburg, MD, USA) supplemented with 20 ng/ml FGF-2/basic FGF (Upstate, Temecula, CA, USA), 20ng/ml EGF (BD Bioscience, Bedford, MA, USA), 2 mM L-glutamine (Gibco-BRL, Gaithersburg, MD, USA), 1 × B27 (Gibco-BRL, Gaithersburg, MD, USA), and 50 U/ml penicillin 50 μg/ml streptomycin (Gibco-BRL, Gaithersburg, MD, USA). The cells were maintained in a humidified incubator at 37°C in 95% air/5% CO2. Four independent embryos of each genotype were obtained from two separate litters. The neurospheres were serially subcultured by trypsinization and the same number of cells from null and wild-type sources plated at each subculture.
Neurosphere Formation Assay (NSFA)
The Neurosphere Formation Assay (NSFA) was carried out according to the protocol of Reynolds and Weiss (Reynolds and Weiss 1992) and Louis (Louis et al. 2008). Briefly, primary neurospheres were dispersed with 0.025% trypsin and mechanical trituration after trypsin inactivation. Single neural precursor cells were seeded into 96-well plates (1000, 100 or 10 cells/well) and cultured for 10 days in Neurobasal Medium supplemented with the growth factor components described in the section on Primary NPC Culture. Secondary neurosphere cultures were generated by picking spheres larger than 100 μm, from the primary cultures. Single cells were prepared from the large neurospheres and seeded in 96-well plates at 1000, 100, or 10 cells per well. After 10 days the largest sphere from each culture was picked and again dispersed into single cells. These cells were seeded into 96-well plates at 500, 50, or 5 cells per well and cultured for 14 days. The spheres were counted and categorized according to sphere size at each subculture.
BrdU Incorporation by NPC and Detection by Immunocytochemistry
The effect of MRG15 on the proliferative activity of neural precursor cells was evaluated by bromodeoxyuridine (BrdU) immunocytochemistry. Single-cell suspensions from either primary cultures or cultures that had been passaged four times were prepared by 0.025% trypsin digestion followed by mechanical dissociation after trypsin inactivation. Cells were seeded on poly-L-lysine (Sigma, St. Louis, MO, USA, 20 μg/ml, pH 8.4, overnight incubation)-coated coverslips at 4 × 104 cells per 13-mm round coverslip (Nunc, Inc., Naperville, IL, USA) and grown for 36 h in neural precursor cell growth medium. The cells were pulsed with 10 μM BrdU for 4 h and fixed with ice cold 70% ethanol for 20 min. DNA was denatured by treatment with 2.5 N HCl for 20 min at room temperature followed by 0.1 M boric acid treatment to neutralize the cells. Endogenous peroxidase activity was quenched by incubating the cells with 3% H2O2 in PBS (phosphate-buffered saline) for 20 min and nonspecific binding blocked by incubating the cells with 5% non-fat milk plus 10% goat serum in PBST for 10 min at room temperature. Incorporated BrdU was detected using a mouse monoclonal anti-BrdU antibody (BD Biosciences, San Jose, CA, USA. 1:5 diluted in 2% non-fat milk and 10% goat serum) that was incubated with the cells overnight at 4°C. After 3 washes with PBST, biotinylated anti-mouse IgG antibody from VECTASTAIN ABC Reagent kit (Vector Labotories, Burlingame, CA, USA) was added, followed by VECTASTAIN ABC Reagent from the kit, according to the manufacturer's instructions. This was followed by DAB (3, 3′-diaminobenzidine) staining. The percentage of BrdU positive cells was determined by counting under a phase contrast microscope and at least 500 cells per sample were scored.
MRG15 Adenoviral Infection of Neural precursor cells
Neurospheres were dispersed into single neural precursor cells and infected with 1000 moi MRG15 adenovirus in growth medium for 24 h. One week later, the cells were infected again with the same titer of the virus. Two days after the second infection, cells were subcultured onto poly-L-lysine-coated coverslips and grown for two more days. The cells were pulsed with 10 μM BrdU for 4 h and positive cells detected using the immunocytochemistry method described above. The same titer of a GFP expressing adenovirus was used as the control for each infection.
Nestin Immunocytochemistry and TUNEL Assay Analyses of NPC Cultures
Nestin expression was determined using immunocytochemistry. Briefly, the same number of primary neural precursor cells from both knockout and wild-type embryos were plated on poly-L-lysine-coated coverslips, and cultured for 5 days with Neurobasal Medium supplemented with stem cell growth factors. The cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Fixed cells were washed with PBS and permeabilized with PBST for 10 min at room temperature. Nonspecific binding was blocked with 10% goat serum in PBST for 30 min at room temperature. Cells were incubated with mouse anti-nestin monoclonal antibody (Chemicon, USA, 1:20 diluted in 1% BSA) overnight at 4°C. Cells were washed three times with PBST, then incubated with Texas Red-conjugated goat anti-mouse IgG antibody (Molecular Probes, USA, 1:1000 dilution) for 1 h at 37°C. After washing the cells were counterstained with 0.1 μg/ml DAPI (4′, 6-Diamidino-2-phenylindole, Sigma, St. Louis, MO, USA). Coverslips were mounted onto Superfrost® Microscope Slides (Fisher Scientific, USA) with Fluoro-Gel (Electron Microscope Science, Hatfield, PA, USA), and results observed under a fluorescence microscope (Axiovert 200M microscope with X-Cite™ 120 fluorescence illumination unit and AxioCam MR, Carl Zeiss Inc., Germany).
Similar cultures were used for TUNEL Assay using the In Situ Cell Death detection kit (Roche, Mannheim, Germany), according to the manufacturer's instructions.
β-III Tubulin (Tuj1) and Glial Fibrillary Acidic Protein (GFAP) Immunocytochemistry Analyses of Differentiating NPC Cultures
Approximately, 7 times passaged neurospheres were collected, the cells dispersed and plated on poly-L-lysine-coated coverslips and grown for 7 days in differentiation medium (Neurobasal Medium with all supplements for neural stem cell growth except FGF and EGF and supplied with 10% fetal bovine serum). The cells were fixed with 4% paraformaldehyde for 10 min at room temperature. After washing with PBS, the cells were permeabilized with PBST, and nonspecific binding blocked with 5% milk and 10% goat serum in PBST. The cells were incubated with the neural differentiation marker β-III tubulin antibody (Rabbit anti-beta III Tubulin Polyclonal Antibody, Covance, 1:7000 dilution in 10% goat serum with 2% milk PBST), and mouse anti-glial fibrillary acidic protein (GFAP) monoclonal antibody (Chemicon, USA, 1:200 dilution in 10% goat serum with 2% milk PBST) at 4°C overnight. Following a wash with PBST, the cells were incubated with Alexa Fluor 488™-conjugated goat anti-rabbit IgG antibody (Invitrogen, USA, 1:1000 dilution in 10% goat serum with 2% milk PBST) and Alexa Fluor 594™-conjugated goat anti-mouse IgG antibody (Invitrogen, USA, 1:1000 dilution in 10% goat serum with 2% milk PBST). After washing with PBS, cells were counterstained with 0.1μg/ml of DAPI for nuclear staining and mounted onto Superfrost® Microscope Slides with Fluoro-Gel. Results were observed under a fluorescence microscope and percentage of Tuj1 (for neurons) or GFAP (for glia) positive cells scored.
RESULTS
Mrg15 is required for neural precursor cell division and maintenance in vivo
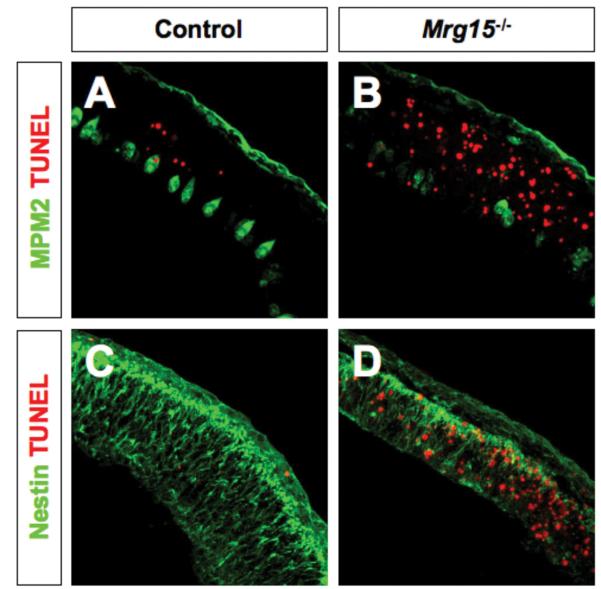
To determine the role of Mrg15 in neural development, we immunostained sections from the brain of Mrg15 null embryos and controls with markers for neural progenitors and differentiated neuronal cells. The most striking effect of loss of MRG15 expression was a general thinning of the neural tube that we observed in null embryos (Fig. 1 and data not shown). This indicated that lack of MRG15 results in a decrease in the number of progenitors and postmitotic neurons in the developing neural tube. To assess whether this was the result of decreased cell division or increased apoptosis, we immunostained coronal sections from embryonic day 10.5 embryos with MMP2, which detects cells in mitosis. In the forebrain, for example, there were fewer cells positive for MMP2 in null embryos, suggesting defects in completion of the cell cycle in some precursor cells (Fig. 1A, 1B). When we analyzed for apoptosis by the TUNEL assay there was increased TUNEL positive staining in the MRG15 null embryonic forebrain. Thus it appears that apoptosis also contributes to the thinning of the neural tube that was observed. Interestingly, the surviving precursor cells were nestin positive, suggesting that they would be effective in self renewal (Fig. 1C, 1D), and differentiated neurons could be detected by Tuj1 staining (data not shown), further indicating that surviving cells were capable of differentiation
Figure 1. Expression of progenitor and mitotic markers in E10.5 Mrg15 neural tube.
A, B: MPM2 (green)/TUNEL (red) double-staining of coronal section of E10.5 embryonic forebrain from Mrg15 heterozygote (A) and Mrg15 null (B). C, D: Nestin (green)/TUNEL (red) double-staining of coronal section of E10.5 embryonic forebrain from Mrg15 heterozygote (C) and Mrg15 null (D).
Mouse Mrg15 Null Neural precursor cells Exhibit Reduced Self-Renewing Capacity
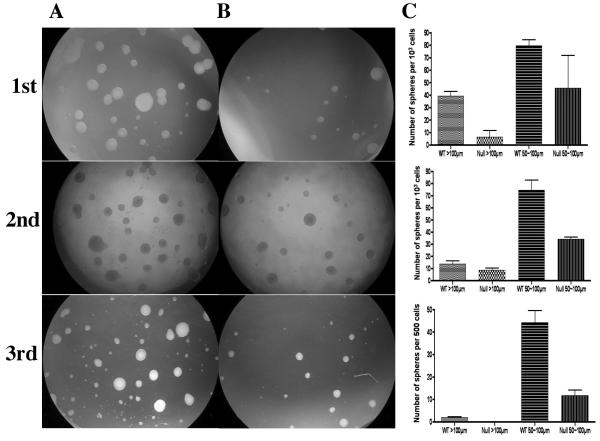
We extended these studies to analyses of neural precursor cells in vitro, to bypass the many factors that affect cell behavior in vivo, and also to determine their response when induced to proliferate and differentiate. We isolated neural precursor cell cultures from E10.5 Mrg15 null and wild-type embryo brain and it was immediately obvious that fewer cells were obtained from the brain of null embryos. This was not surprising in view of the increased apoptosis observed in the histological studies. We then performed the neurosphere formation assay (NSFA). Single neural precursor cells were dispersed from primary neurospheres of Mrg15 null and wild-type cultures and equal numbers seeded into 96-well plates. We seeded the cells at three different densities, using at least two wells per dilution, to account for variation in cell preparation and number actually seeded. We found that fewer large neurospheres (>50 μm) were formed in Mrg15 null cell cultures in the same growth period when compared with wild-type cells (Fig. 2A, 2B). Since the cells we dissociated were a mixture of neural stem cells and neural progenitor cells, one possibility was that the different cell types might proliferate at different rates in culture. To eliminate this, spheres of the same size (> 50 μm) were collected from each genotype to prepare single cell suspensions and serially subcultured. Once again, the size of the majority of the spheres derived from Mrg15 null cells were smaller than that of wild-type (Fig. 2C) and though neurosphere formation ability decreased with serial passage in both wild-type and null cultures, the sphere size was always smaller in the null. Additionally, the number of times the null cells could be passaged was fewer than wild-type (9 versus 25 passages).
Figure 2. Decreased neurosphere formation in Mrg15 null neural precursor cells.
A, B: Representative pictures of primary (upper row), secondary (middle row), and tertiary (lower) neurosphere cultures from wild-type (panel A) and Mrg15 null (panel B) cells. Single neural precursor cells were seeded into 96-well plate at 103, 102 and 10 cells per well cultured in Neurobasal Medium supplemented with EGF and FGF2 for the neurosphere formation assay for 10 days (Primary and Secondary). The same size spheres from both wild-type and null cell that were larger than 100 μm were collected and treated with trypsin to make a single cell suspension for secondary NSFA. Single cell suspension made from one of the largest spheres from each genotype was seeded into a 96-well plate at 500, 50, 5 cells per well and cultured for 14 days (Tertiary). C: Quantitative analyses of neurosphere number and size. After defined time points, sphere size and number were measured under the microscope. Y-axis indicates sphere numbers per 1000 or 500 cells which were initially seeded.
Neural Precursor Cells Derived from Mrg15 Null Embryonic Mouse Brain Exhibit Decreased Proliferation and No Increased Apoptosis
The decreased neurosphere size observed in neural precursor cell cultures of Mrg15 null compared with wild-type could be due to either a decrease in proliferative capacity or increased apoptosis. To determine whether one or both mechanisms were causing the phenotype, we used BrdU incorporation to determine the number of cells entering the S phase of the cell cycle and a cell death detection kit to measure apoptosis.
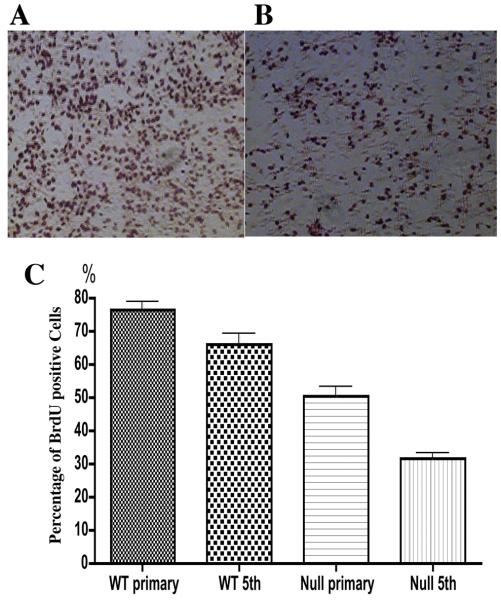
We pulsed primary cultures, or cells that had been passaged four times, with BrdU and determined that the percentage of positive cells was lower in null cultures (p<0.001)(Fig. 3A, 3B). Though BrdU incorporation into wild-type neural precursor cell decreased with the serial passage (P<0.05), the percent positive null cells was always lower than wild-type at later passages (Fig. 3C).
Figure 3. Reduced cell proliferation in Mrg15 null neural precursor cells.
A, B: Representative BrdU immunostaining in wild-type (A) and Mrg15 null (B) cells at fifth passage (x4 magnification). Single cells were plated on poly-L-lysine-coated coverslips, grown for 36 h, and incubated with 10 μM BrdU for 4 h. Incorporated BrdU was detected by mouse anti-BrdU antibody followed by incubation with biotinilyted anti-mouse IgG and positive cells demonstrated by DAB staining. C: Quantitative analyses of BrdU positive cells. Percentage of positive cells is shown. BrdU positive cells in Mrg15 null population were fewer than those in wild-type population both in primary culture and at fifth subculture (P<0.001).
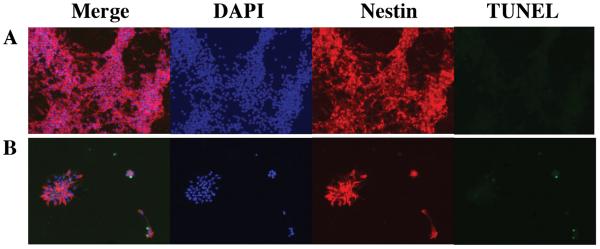
TUNEL assay to detect apoptosis indicated that there were a few dead cells in both Mrg15 null and wild-type cell cultures (Fig. 4). Interestingly, nestin-positive neural precursor cells from Mrg15 null embryos did not spread into monolayers as did wild-type, but rather tended to aggregate on the coated coverslips.
Figure 4. Apoptosis in Mrg15 null neural precursor cells does not increase in neurosphere culture.
Single cells from wild-type and Mrg15 null neural precursor cells were seeded on poly-L-lysine-coated coverslips and cultured in Neurobasal Medium supplemented with stem cell growth component for 5 days. The cells on coverslips were fixed with 4% paraformaldehyde and TUNEL assay (green) used to detect apoptotic cells. We co-stained these cells with anti-nestin antibody (red) to detect the neural precursor cell population. Cells were counterstained with 0.1 μg/ml of DAPI (blue) for nuclear staining. Wild-type (A) and Mrg15 null (B) neural precursor cells are shown.
Introduction of Exogenous MRG15 Rescues the Proliferation Defect of Mrg15 Null NPCs
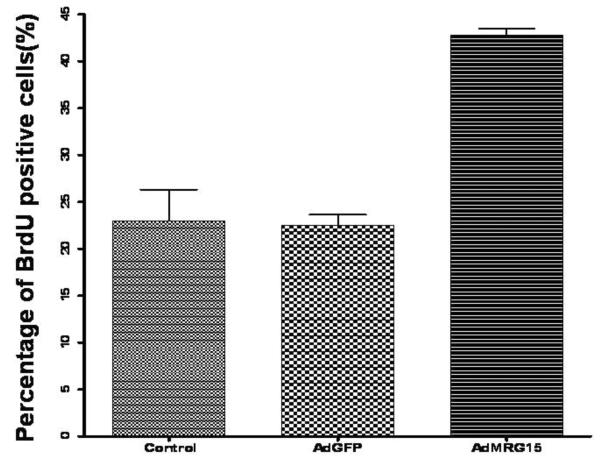
To determine whether the proliferation defect of Mrg15 null NPCs was the result of lack of MRG15, we infected the Mrg15 null cells with an adenovirus construct expressing MRG15. Since high titers of adenovirus caused some toxicity, we used lower titers that were nontoxic. This resulted in about 40% infection of cells as indicated by the GFP adenovirus control. To obtain about 85% infection, we re-infected the cells 7 days later, and then measured BrdU incorporation. Infected Mrg15 null neural precursor cells had a higher BrdU incorporation rate when compared with GFP infected control cells (Fig. 5).
Figure 5. Re-expression of MRG15 by adenovirus can rescue the cell proliferation defect in Mrg15 null neural precursor cells.
Mrg15 null neural precursor cells were infected with adenovirus expressing MRG15 or EGFP at 1000 moi twice at a one week interval. Four days after the second infection, cells were incubated with 10 μM BrdU for 4 h and BrdU positive cells detected by immunostaining. The percentage of BrdU positive cells in adenovirus MRG15 infected Mrg15 null cells significantly increased compared with Mrg15 null control cells (P<0.01), although the control adenovirus EGFP did not have any effect on BrdU incorporation in these cells.
Mrg15 Null NPCs Exhibit a Decreased Ability to Differentiate into Neurons
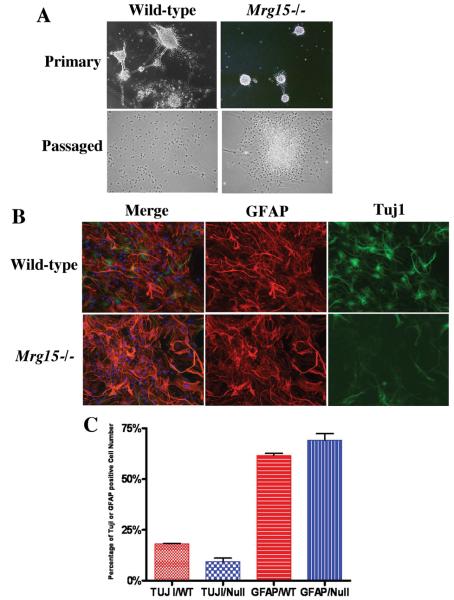
Neural stem cells are capable of self-renewal and differentiation into neural and glial lineages. To determine if MRG15 has any influence on neural stem cell differentiation, primary or neurospheres that had been passaged approximately 7 times were plated on poly-L-lysine-coated coverslips and cultured in differentiation medium for 7 to 10 days. The morphology of wild-type cells began to change on day 3 after culture in differentiation medium. Neurospheres lost their spherical shape and flattened to form a monolayer (Fig. 6A). Attached monolayer cells changed their shape to the morphology of bipolar cells or cells with fine extended processes. In contrast, the morphology of primary Mrg15 null cells showed minimal change in only a subset of colonies even after 10 days in differentiation medium (Fig. 6A). The majority of the Mrg15 null cells remained as cell aggregrates, very different from that of wild-type cells. In the case of cells that had been passaged 7 times, after 7 days in differentiation conditions, neurons or glia were easily detected in wild-type cultures using the neuronal differentiation marker Neuronal Class III β-tubulin (Tuj1) and the astrocyte/glial cell marker Glial Fibrillary Acidic Protein (GFAP)(Fig. 6B). In the case of Mrg15 null cultures, though the number of differentiated glial cells was similar to that in wild-type cultures, the number of differentiated neurons in Mrg15 null cells was clearly fewer than wild-type (P<0.05)(Fig. 6B).
Figure 6. MRG15 is required for differentiation into neural lineages.
A: Representative pictures of primary wild-type and Mrg15 null cells 10 days after exposure to differentiation conditions (upper row) and passaged cells 7 days after differentiation condition (lower row). Wild-type cells (left panel) changed morphology and appeared to be differentiated under this condition, but most of the Mrg15 null cells (right panel) remained as colony-like aggregates. B: Reduced Tuj1 immunoresposive cell number of differentiated cells from passaged Mrg15 null neural precursor cells (lower row) compared with wild-type (upper row). Neurospheres were plated on poly-L-lysine-coated coverslips and cultured under differentiation condition for 7 days. Cells were fixed with 4% paraformaldehyde and stained with antibody against Tuj1 (neurons, green), GFAP (glial, red), followed by incubation with fluorescent conjugated secondary antibodies. Cells were counterstained with 0.1 μg/ml of DAPI (blue). C: Quantitative analyses of Tuj1 and GFAP positive cells. The percentage of positive cells is shown. Tuj1 positive cells in the Mrg15 null population were fewer than those in the wild-type population (P<0.05).
DISCUSSION
In this report, we demonstrate that expression of MRG15, a chromatin regulator, is required for both proliferation and differentiation of neural precursor cells. We have found that the number of these primary stem/progenitor cells is lower in the initial isolation of cells from the embryonic null brain, most likely because of the increased apoptosis observed in vivo in the histological analyses. We observed that the overall neural tube is thinner in Mrg15 null embryos and this is most likely due to the presence of fewer neural precursor cells. As a result when the cells are cultured in vitro the number of large spheres is decreased in cell cultures derived from Mrg15 deficient embryonic brain. Cells derived from the largest neurospheres, when subcultured, continue to exhibit defects in number of spheres greater than 50 μm at each subsequent passage. BrdU incorporation in Mrg15 deficient cells is reduced when compared with wild-type, however, apoptosis is not affected indicating that in vitro defects in neural precursor proliferation is the result of reduced growth rate and long term growth potential, but not increased cell death. The difference in results between in vivo and in vitro studies is not surprising as the milieu of cells in culture is very different from that in the embryo. For example, overlying non-neuronal tissues may provide extrinsic or non-cell autonomous signals that are necessary for cell survival in vivo. However, the final results are similar in that cell cycle progression and completion are affected in both cases, with a mitotic defect contributing to a decreased number of precursor cells in vivo. The fact that infection with an adenovirus expressing MRG15 causes increased BrdU incorporation in null cells in vitro, demonstrates that it is the deficiency in MRG15 that causes the proliferation defect we observed.
Differentiation into neurons was also affected in Mrg15 deficient neural precursor cells in vitro. They did not attach to the tissue culture dishes as well as wild-type and many of the cells remained in aggregates in differentiation media. This suggests that the abnormalities observed in the developing brain of Mrg15 deficient embryos are a result of cell-autonomous defects in these neural precursor cells. Abnormalities observed in many other tissues of Mrg15 deficient embryo may be caused by a similar molecular mechanism(s) as that seen in brain tissue, and the data we have regarding proliferative defects in MEFs derived from null and wild-type embryos supports this possibility.
MRG15 associates in complexes with the HAT Tip60 (Cai et al. 2005; Cai et al. 2003; Doyon et al. 2004; Hayakawa et al. 2007; Sardiu et al. 2008) and also mSin3/HDAC (Doyon et al. 2004; Hayakawa et al. 2007; Yochum and Ayer 2002) and is thereby involved in the regulation of gene expression by changing the acetylation status of histones surrounding target genes. Recently, Fazzio et al. have reported that the Tip60-p400 complex is important for maintenance of embryonic stem cell (ESC) identity (Fazzio et al. 2008). Knockdown of either of these components of the complex in ESCs resulted in reduced growth rate, flattened cell morphology, changes in gene expression, and loss of ESC markers eg: alkaline phosphatase activity was decreased and embryoid body formation less efficient. Gene expression analyses demonstrated that cell cycle regulators and cell division related genes were down-regulated and differentiation and embryonic development related genes were up-regulated following knockdown of these genes. However, interestingly, MRG15 knockdown in ESCs did not have a significant phenotype. One possibility is that MRGX, a mammalian homolog of MRG15, may compensate for MRG15 function in these cells, because MRGX can also associate in complexes with Tip60 (Cai et al. 2003; Hayakawa et al. 2007; Lerin et al. 2006; Sardiu et al. 2008). Since Tip60 HAT is a multi-subunit complex, another possibility is that MRG15 in this complex is not required for ESCs but is important for tissue precursor cells, such as the neural precursor cells we have studied in this report.
In our study we have found that Mrg15 deficient neural precursor cells exhibit differentiation defects in addition to growth defects. Mrg15 deficient neural precursor cells appear to be maintained as stem-like aggregates in differentiation medium and differentiate into neurons less-efficiently than wild-type cells. It is known that hematopoietic competence is a rare property of neural stem cells and epigenetic alterations can cause fate switching (Morshead et al. 2002). Thus, treatment of neurospheres with trichostatin A (an HDAC inhibitor) and 5-aza-2′-deoxycytidine (a DNA methyltransferase inhibitor) can yield a transplantable hematopoietic population (Schmittwolf et al. 2005). Additionally, Mbd3, which is a component of the nucleosome remodeling and histone deacetylation (NuRD) complex, is essential for commitment to developmental lineages in ESCs (Kaji et al. 2006). The NuRD complex contains at least seven subunits and HDAC1 and HDAC2 are catalytic subunits of this complex (Wade et al. 1999; Zhang et al. 1999). Mbd3 deficient ESCs are viable but fail to silence genes which are important for maintenance of ESCs, such as Oct4, Nanog, and Rex1, under differentiation conditions (without LIF). Mbd3 deficient ESCs also cannot form neuroectoderm in culture. Normal ESCs lose Oct4 expression and express markers of neural progenitors (nestin-positive) and postmitotic neurons (Tuj1-positive) after 10 days in differentiation conditions. However, the majority of Mbd3 deficient cells continue to express Oct4 under these conditions and retain a stem cell-like growth (Kaji et al. 2006). It is also known that inhibition of HDAC activities in neural progenitors induce neural differentiation but inhibit glial differentiation (Hsieh and Gage 2004; Hsieh et al. 2004). Taken together these data suggest that co-repressor complexes involving HDACs are also important for cell-fate determination and differentiation of stem/precursor cells.
MRG15 is also a component of HDAC1 and HDAC2 containing complexes. Although it is known that this complex acts to suppress spurious intragenic transcription in budding yeast, the function in mammalian cells is still unclear. MRG15 containing HDAC complex(es) may also work as a repressor of expression of genes required for stem cells to maintain their stem cells status in addition to inhibiting incorrect transcription initiation. The defects in self-renewal and differentiation, we have observed in Mrg15 deficient neural precursor cells may therefore be the result of inactivation of two or more independent MRG15 containing complexes.
We here present evidence for a role of MRG15 in neural cell proliferation. In an earlier study using an antibody against the chromodomain of MRG15 protein, we had analyzed hippocampal tissue samples from histopathologically confirmed Alzheimer's disease (AD) and non-AD age matched controls. We observed specific labeling of large pyramidal neurons only in AD cases, and that there was significant overlap of immunostaining of this protein and phosphorylated tau (Raina et al. 2001). Age matched normal controls showed no immunoreactivity. The presence of a positive chromatin remodeling, transcriptionally controlling protein in association with intraneuronal neurofibrillary pathology is consistent with the multiple gene expression changes that have been observed in such regions. It also provides support for the idea that neurons in AD re-enter the cell cycle. Interestingly, the expression of microRNAs (miRNAs), that have been implicated in brain development and neuronal specification, have recently been demonstrated to be altered in AD brain suggesting functional deficits occur at various stages of the disease (Cogswell et al. 2008). Thus, chromatin remodeling and the resulting gene expression modifications could well be a contributing factor to the initiation and progression of AD.
CONCLUSIONS
Our study demonstrates a critical role for the chromatin regulator MRG15 in proliferation and differentiation into neurons of neural precursor cells in vivo and in vitro. An understanding of the molecular mechanisms that act to establish a functional nervous system during development is essential and future studies involving identification of the regulatory complexes that are affected by loss of Mrg15, and their gene targets should contribute to a better understanding of this developmental process.
ACKNOWLEDGEMENTS
We thank Dr. Mei Qiang for technical advice regarding isolation of neural precursor cells and cultivation of neurospheres. We also thank Drs. James L. Roberts and Randy Strong for helpful discussion. This study was supported by funding from The Ellison Medical Foundation and NIH/NIA RO1AG032134 to OMPS, the UTSA-STARS Award to GG, and an ERC grant from The University of Texas Health Science Center at San Antonio to KT.
Grant information: Ellison Medical Foundation
National Institutes of Health/National Institute on Aging
University of Texas at San Antonio-STARS Award
University of Texas Health Science Center at San Antonio-ERC grant
REFERENCES
- Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, Clark LJ, Parkinson EK, Barrett JC, Smith JR, Pereira-Smith OM. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of cording regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Downs JA, Cote J. Dynamics of chromatin during the repair of DNA double-strand breaks. Cell Cycle. 2005;4:1373–1376. doi: 10.4161/cc.4.10.2108. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem. 2001;276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nature Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gritti A, Parati EA, Cova L, Frolichsthal P, Galli R, Wanke E, Faravelli L, Morassutti DJ, Roisen F, Nickel DD, Vescovi AL. Multipotential stem cells from the adult mouse brain proliferate and self-renew in response to basic fibroblast growth factor. J Neurosci. 1996;16:1091–1100. doi: 10.1523/JNEUROSCI.16-03-01091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Kurdistani SK, Morrus SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, MacDonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim S, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1a. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and Chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Lledo P-M, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Louis SA, Rietze RL, Deleyrolle L, Wagey RE, Thomas TE, Eaves AC, Reynolds BA. Enumeration of neural stem and progenitor cells in the neural colony-forming cell assay. Stem Cells. 2008;26:988–996. doi: 10.1634/stemcells.2007-0867. [DOI] [PubMed] [Google Scholar]
- Marin I, Baker BS. Origin and evolution of the regulatory gene male-specific lethal-3. Mol Biol Evol. 2000;17:1240–1250. doi: 10.1093/oxfordjournals.molbev.a026407. [DOI] [PubMed] [Google Scholar]
- Ming G-l, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- Morshead CM, Benveniste P, Iscove NN, van der Kooy D. Hematopoietic competence is a rare property of neural stem cells that may depend on genetic and epigenetic alterations. Nat Med. 2002;8:268–273. doi: 10.1038/nm0302-268. [DOI] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15 a novel chromodomain protein is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- Pevny L, Rao MS. The stem-cell menagerie. Trends Neurosci. 2003;26:351–359. doi: 10.1016/S0166-2236(03)00169-3. [DOI] [PubMed] [Google Scholar]
- Raina AK, Pardo P, Rottkamp CA, Zhu X, Pereira-Smith OM, Smith MA. Neurons in Alzheimer disease emerge from senescence. Mech Ageing Dev. 2001;123:3–9. doi: 10.1016/s0047-6374(01)00333-5. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci USA. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittwolf C, Kirchhof N, Jauch A, Durr M, Harder F, Zenke M, Muller AM. In vivo haematopoietic activity is induced in neurosphere cells by chromatin-modifying agents. EMBO J. 2005;24:554–566. doi: 10.1038/sj.emboj.7600546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev Biol. 1999;208:166–188. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
- Vescovi AL, Reynolds BA, Fraser DD, Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng H-H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]