Abstract
Background Genetic variants in 15q25 have been identified as potential risk markers for lung cancer (LC), but controversy exists as to whether this is a direct association, or whether the 15q variant is simply a proxy for increased exposure to tobacco carcinogens.
Methods We performed a detailed analysis of one 15q single nucleotide polymorphism (SNP) (rs16969968) with smoking behaviour and cancer risk in a total of 17 300 subjects from five LC studies and four upper aerodigestive tract (UADT) cancer studies.
Results Subjects with one minor allele smoked on average 0.3 cigarettes per day (CPD) more, whereas subjects with the homozygous minor AA genotype smoked on average 1.2 CPD more than subjects with a GG genotype (P < 0.001). The variant was associated with heavy smoking (>20 CPD) [odds ratio (OR) = 1.13, 95% confidence interval (CI) 0.96–1.34, P = 0.13 for heterozygotes and 1.81, 95% CI 1.39–2.35 for homozygotes, P < 0.0001]. The strong association between the variant and LC risk (OR = 1.30, 95% CI 1.23–1.38, P = 1 × 10–18), was virtually unchanged after adjusting for this smoking association (smoking adjusted OR = 1.27, 95% CI 1.19–1.35, P = 5 × 10–13). Furthermore, we found an association between the variant allele and an earlier age of LC onset (P = 0.02). The association was also noted in UADT cancers (OR = 1.08, 95% CI 1.01–1.15, P = 0.02). Genome wide association (GWA) analysis of over 300 000 SNPs on 11 219 subjects did not identify any additional variants related to smoking behaviour.
Conclusions This study confirms the strong association between 15q gene variants and LC and shows an independent association with smoking quantity, as well as an association with UADT cancers.
Keywords: Lung cancer, nicotine dependence, smoking quantity, UADT cancer
Introduction
We and others have recently identified an association between chromosome 15q variants and risk of lung cancer (LC; in particular rs16969968 and rs8034191) using a genome wide association (GWA) approach.1–3 The susceptibility region contains three cholinergic nicotine receptor genes (CHRNA3, CHRNA5 and CHRNB4), encoding nicotine receptors in neuronal and other tissues. Amos et al.1 and our study2 identified the variants directly via its association with the risk of LC, whereas the study by Thorgeirsson et al.3 identified an association with the same genetic region and smoking quantity, and concluded that the variant increases LC risk indirectly through smoking. In our initial study we did not observe an association between the genetic variant and nicotine dependence, whereas Amos et al. noted only a weak association, but a much stronger direct association with LC. In addition to LC, the variant allele was also associated with peripheral arterial disease3 and chronic obstructive pulmonary disease.4
Although all three initial GWA studies1–3 reported almost identical associations between the 15q variant and LC risk [an allelic odds ratio (OR) between 1.30 and 1.32], they differed as to whether this was a direct association, or whether the 15q variant was simply a proxy for increased exposure to tobacco carcinogens.5 However, all three concluded that the association with cigarettes smoked per day (CPD) did not explain the observed LC risk totally. In addition to these initial studies, a variety of other genome wide and candidate gene studies showed an association of 15q25 single nucleotide polymorphisms (SNPs) and smoking behaviour.6–11
To clarify the relation between the 15q variant, smoking behaviour and LC, we have extended our previous LC studies. We additionally included 3968 upper aerodigestive tract (UADT) cancer cases (comprising oral cavity, oropharynx, hypopharynx, larynx and oesophagus) to determine the association of the genotype with other cancers strongly associated with smoking. With over 7000 subjects, this is one of the largest studies performed so far for CHRN gene variants, smoking behaviour and cancer risk. Finally, based on genome-wide data on more than 11 000 subjects we have attempted to identify additional genetic variants associated with smoking behaviour.
Methods
Study characteristics
The studies included in this article were five LC studies and four UADT cancer studies. The LC studies were Central Europe (Czech Republic, Hungary, Poland, Romania, Russia and Slovakia), Toronto (Canada), EPIC (Sweden, The Netherlands, UK, France, Germany, Spain, Italy, Greece, Norway), Liverpool (UK) and Hunt/Tromsø (Norway). The LC studies participated in our initial LC GWA study and details about each study have been described.2 However, for the current study there were 1334 additional subjects from the EPIC study available. The UADT studies were Central Europe (Romania, Poland, Russia, Slovakia, Czech Republic), ARCAGE (Czech Republic, Greece, Italy, Norway, UK, Spain, Croatia, France), Latin America (Cuba, Brazil, Argentina) and Rome (Italy) and have been previously described.12 The Central Europe UADT study and the ARCAGE study were already analysed for rs16969968 in our previous GWA study.2 In the Central Europe study the same controls were used for the LC and UADT cancer comparisons. After quality control, we had valid genotypes on rs16969968 for 3898 LC cases, 3968 UADT cancer cases and 9434 controls from 10 different studies. In comparison with our previous study on 15q variants, there were 1334 samples from the EPIC LC study (397 cases and 937 controls) and 3018 new samples from the UADT cancer studies (Latin America: 1429 cases, 1093 controls; Rome: 267 cases, 229 controls). Table 1 shows an overview of the different study populations.
Table 1.
Characteristics of study populations
| Study | n | Males | Females | Mean age (SD) | Never | Ever | Former | Current | MAFa |
|---|---|---|---|---|---|---|---|---|---|
| LC studies | |||||||||
| Central Europe lung casesb | 1790 | 1393 | 397 | 60.29 (8.73) | 136 | 1654 | 352 | 1298 | 0.40 |
| Central Europe Controlsb,c | 2362 | 1720 | 642 | 59.61(9.77) | 836 | 1524 | 608 | 909 | 0.34 |
| Toronto casesb | 329 | 157 | 172 | 63.75 (11.5) | 77 | 193 | 93 | 91 | 0.40 |
| Toronto controlsb | 462 | 160 | 302 | 51.85 (15.55) | 186 | 235 | 126 | 77 | 0.35 |
| EPIC casesd | 1176 | 705 | 471 | 63.16 (7.89) | 101 | 1075 | 307 | 754 | 0.42 |
| EPIC controlsd | 2515 | 1467 | 1048 | 65.5 (7.43) | 932 | 1583 | 878 | 670 | 0.36 |
| Liverpool casesb | 389 | 234 | 155 | 66.92 (8.8) | 16 | 372 | 153 | 216 | 0.36 |
| Liverpool controls | 812 | 498 | 314 | 64.71 (8.62) | 230 | 582 | 395 | 145 | 0.32 |
| Hunt/Tromso casesb | 214 | 129 | 85 | 62.95 (10.67) | 12 | 199 | 43 | 150 | 0.41 |
| Hunt/Tromso controlsb | 326 | 194 | 132 | 63.94 (11.15) | 97 | 207 | 121 | 84 | 0.31 |
| UADT cancer studies | |||||||||
| Central Europe UADT casesb | 719 | 636 | 83 | 58.46 (9.42) | 51 | 668 | 90 | 578 | 0.35 |
| ARCAGE casesb | 1553 | 1256 | 297 | 59.15 (10.19) | 144 | 1409 | 357 | 1052 | 0.39 |
| ARCAGE controlsb | 1635 | 1235 | 400 | 59.16 (11.62) | 544 | 1091 | 554 | 537 | 0.37 |
| Latin America casesd | 1429 | 1219 | 210 | 58.15 (10.39) | 87 | 1342 | 309 | 1031 | 0.28 |
| Latin America controlsd | 1093 | 858 | 235 | 56.87 (11.5) | 318 | 775 | 362 | 413 | 0.26 |
| Rome casesd | 267 | 46 | 221 | 63.36 (11.19) | 33 | 234 | 127 | 107 | 0.45 |
| Rome controlsd | 229 | 91 | 138 | 63.72 (13.5) | 130 | 99 | 50 | 49 | 0.39 |
| Total candidate gene study | 17 300 | 11 998 | 5302 | 60.86 (10.43) | 3930 | 13 242 | 4925 | 8161 | 0.36 |
| Extra studies GWAS | |||||||||
| Estonia casesb | 109 | 87 | 22 | 64.50 (10.26) | 14 | 95 | 1 | 94 | e |
| Estonia controlsb | 875 | 473 | 402 | 42.99 (16.08) | 428 | 446 | 149 | 297 | e |
| Paris casesb | 135 | 126 | 9 | 58.53 (9.90) | 0 | 135 | 35 | 100 | e |
| Paris controlsb | 146 | 139 | 7 | 55.09 (10.73) | 0 | 146 | 44 | 102 | e |
aMinor allele frequency for rs16969968.
bThese studies contributed data for the GWA analysis.
cFor the Central Europe study the same controls were used for lung and head and neck cancer cases.
dContributed new samples to the current study (compared with Hung et al.).
eOnly GWA data.
SD = standard deviation.
MAF = minor allele frequency.
Smoking phenotypes
Smoking status (never/ever/current/former) was available for all studies. Ever smokers are current and former smokers, while never smokers had smoked less than 100 cigarettes during their lifetime or had never smoked regularly. Former smokers had quit smoking for ≥2 years at the time of diagnosis/interview. Smoking quantity data were available for 12 310 subjects as the average number of CPD. Smokers were asked at what age they started smoking regularly. Former smokers were asked their age at the time of quitting. Light smoking was defined as 1–10 CPD, whereas heavy smoking was defined as >20 CPD. Smoking quantity, age of initiation and age of cessation were not available for the Rome study.
Genotyping
Genotyping for rs16969968 was performed using the 5′-exonuclease assay (TaqMan, Applied Biosystems, Foster city, CA, USA) and was centrally performed at the International Agency for Research on Cancer (Lyon, France) for all participating studies. We did not include data from the GWA study for the rs16969968 analyses in this article. Cases and controls were randomly mixed when genotyped and laboratory personnel were blinded to case/control status. A randomly selected 10% of the study subjects (both cases and controls) were re-genotyped to examine the reliability of the genotyping assays. Internal duplicate concordance was >99.9% and genotyping success rate was 97.9%.
Statistical analysis
All analyses were performed in SAS 9.1. For quantitative variables (CPD, age of initiation of smoking, age of cessation, age of onset of disease) linear regression was performed on log transformed data and adjusted mean values and 95% confidence intervals (CIs) are presented. For discrete variables ORs and 95% CIs were calculated using logistic regression. Multiplicative (AA vs GA vs GG: trend) and genotype specific (defined as AA + GA vs AA and AA vs GA + GG) models were computed and compared using the likelihood ratio test. All analyses were adjusted for age, sex and country, by including the variables in logistic or linear regression models. If cases and controls were analysed together, case/control status was included as a covariate. Lung and UADT cancer risk were both calculated without and with adjustment for smoking quantity (CPD). Only current and former smokers were included in the analyses involving the smoking phenotypes with the exception of smoking initiation, which included never smokers as well. Chi-squared tests for heterogeneity were performed.
GWA analysis
We attempted to identify additional genes associated with smoking behaviour by analysis of genome wide data from a total of 11 219 participants with smoking phenotypes (5687 non-cancer participants and 5532 cancer participants). Studies that provided genome-wide data are identified in Table 1. In addition to these data, the Estonia study and the Paris study contributed extra data for the GWA analysis only. Genome-wide data were available from the Illumina platform as previously described.2,13 GWA study analysis was performed for all studies centrally at the Centre National de Genotypage (Evry, France). Briefly, genotyping was conducted using Illumina Sentrix HumanHap 300 BeadChips. We excluded variants with a call rate of <95% or whose allele distributions deviated strongly from Hardy–Weinberg equilibrium among controls (P < 10–7). We also excluded subjects with a completion rate of <95% or whose reported sex did not match with the inferred sex based on the heterozygosity rate from the X chromosomes. Unexpected duplicates and unexpected first-degree relatives were also excluded from the analysis. Population outliers were detected using STRUCTURE with HapMap subjects as internal controls, and were subsequently excluded from the analysis.
Phenotypes for the analysis in this study were smoking quantity (CPD) as a log transformed continuous variable, smoking initiation (ever/never; dichotomous), smoking cessation (current/former; dichotomous), heavy smoking (heavy/light; dichotomous) and age of smoking initiation (log transformed continuous variable). In a subset of the ARACAGE study (1504 subjects) participants were asked questions relating to tobacco addiction based on the Fagerstrom tolerance questionnaire.14 Two of these questions (‘time to first cigarette’ and ‘numbers of CPD’) have been shown to be particularly strongly associated with nicotine dependence and responses to both questions result in a heaviness of smoking index (HSI).15 We performed GWA analysis in the ARCAGE study for the HSI (score 0–6). For all GWA analyses logistic and linear regression were performed in PLINK assuming a co-dominant genetic model.16 Adjustment was performed for age, sex, country and case/control status.
Results
Smoking quantity
We investigated if the rs16969968 variant allele was related to smoking quantity, using the amount of CPD as a quantitative variable. The analysis was performed stratified for cases and controls, per study and combined. In the controls subjects with one minor variant allele smoked on average 0.3 CPD more, whereas subjects with the homozygous minor AA genotype smoked on average 1.2 CPD more than subjects with a GG genotype (P-trend = 0.01) (Table 2). We determined the association both under a genotype specific and a multiplicative model. As they gave comparable fits, we used the multiplicative model for further analysis. The difference in CPD between the two homozygous genotypes was 0.9 CPD (P = 0.03) among LC cases and 1.3 CPD (P = 0.04) for UADT cancer cases. If all cases and controls were combined (after adjustment for case/control status), the adjusted mean difference between the two homozygote genotypes was 1.2 CPD (P < 0.0001). However, there was considerable heterogeneity between the studies (P-heterogeneity = 0.01 for the controls, P = 0.06 for the combined analysis). The association between smoking quantity and rs16966968 genotype was similar for former and current smokers (data not shown).
Table 2.
Association of rs16969968 with smoking quantity in CPD
|
Controls |
LC cases |
UADT cancer cases |
Cases and controls combined |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Genotype | n | Mean | 95% CI | P-trend | n | Mean | 95% CI | P-trend | n | Mean | 95% CI | P-trend | n | Mean | 95% CI | P-trend |
| All | GG | 2470 | 12.3 | 11.9–12.8 | 1183 | 15.7 | 15.2–16.2 | 1489 | 16.2 | 15.5–17 | 5142 | 14.1 | 13.8–14.4 | ||||
| GA | 2493 | 12.6 | 12.1–13 | 1560 | 15.9 | 15.4–16.3 | 1520 | 16.6 | 15.8–17.3 | 5573 | 14.4 | 14.1–14.7 | |||||
| AA | 644 | 13.5 | 12.7–14.3 | 0.01 | 563 | 16.6 | 15.9–17.4 | 0.03 | 388 | 17.5 | 16.3–18.7 | 0.04 | 1595 | 15.3 | 14.8–15.8 | <.0001 | |
| Abs. diff. | 1.2 | 0.9 | 1.3 | 1.2 | |||||||||||||
| Central Europe | GG | 643 | 12.4 | 11.7–13.2 | 580 | 16 | 15.3–16.7 | 280 | 15.9 | 14.6–17.2 | 1503 | 14.1 | 13.7–14.6 | ||||
| GA | 691 | 12.6 | 11.9–13.3 | 799 | 16.3 | 15.7–16.9 | 305 | 15.6 | 14.5–16.8 | 1795 | 14.3 | 13.8–14.7 | |||||
| AA | 180 | 12.5 | 11.4–13.7 | 0.81 | 270 | 16.4 | 15.5–17.3 | 0.38 | 76 | 17 | 15.1–19 | 0.51 | 526 | 14.5 | 13.8–15.2 | 0.32 | |
| Abs. diff. | 0.1 | 0.4 | 1.1 | 0.4 | |||||||||||||
| Toronto | GG | 87 | 17.1 | 14.3–20.4 | 64 | 17.6 | 14.7–21.1 | 151 | 17.5 | 15.4–19.8 | |||||||
| GA | 88 | 15.2 | 12.7–18 | 92 | 18.4 | 15.8–21.3 | 180 | 16.6 | 14.8–18.6 | ||||||||
| AA | 25 | 14.3 | 10.4–19.8 | 0.25 | 25 | 26.4 | 19.8–35.1 | 0.05 | 50 | 19.2 | 15.5–23.9 | 0.72 | |||||
| Abs. diff. | −2.8 | 8.8 | 1.7 | ||||||||||||||
| EPIC | GG | 532 | 10.6 | 10–11.4 | 309 | 15.5 | 14.5–16.5 | 841 | 12.8 | 12.2–13.4 | |||||||
| GA | 591 | 11 | 10.3–11.7 | 427 | 15.6 | 14.7–16.5 | 1018 | 13 | 12.5–13.6) | ||||||||
| AA | 169 | 11.6 | 10.4–13 | 0.16 | 184 | 16 | 14.7–17.4 | 0.52 | 353 | 13.6 | 12.7–14.6 | 0.14 | |||||
| Abs. diff. | 1 | 0.5 | 0.8 | ||||||||||||||
| Liverpool | GG | 269 | 11.6 | 10.5–12.8 | 160 | 17.6 | 16–19.4 | 429 | 14.3 | 13.3–15.3 | |||||||
| GA | 248 | 12.6 | 11.3–14 | 154 | 17.4 | 15.8–19.1 | 402 | 14.9 | 13.8–16.1 | ||||||||
| AA | 53 | 14.2 | 11.4–17.7 | 0.08 | 56 | 20.2 | 17.2–23.6 | 0.28 | 109 | 16.8 | 14.6–19.2 | 0.05 | |||||
| Abs. diff. | 2.6 | 2.6 | 2.5 | ||||||||||||||
| Hunt/Tromso | GG | 78 | 7.8 | 6.8–8.8 | 70 | 12 | 10.6–13.5 | 148 | 9.6 | 8.8–10.5 | |||||||
| GA | 79 | 10.1 | 8.8–11.5 | 88 | 12 | 10.8–13.3 | 167 | 10.9 | 10.1–11.9 | ||||||||
| AA | 14 | 11.5 | 8.5–15.6 | 0.002 | 28 | 12.4 | 10.3–14.9 | 0.83 | 42 | 11.6 | 9.8–13.7 | 0.01 | |||||
| Abs. diff. | 3.7 | 0.4 | 2 | ||||||||||||||
| ARCAGE | GG | 446 | 14.4 | 13–15.8 | 512 | 17 | 15.8–18.2 | 958 | 15.6 | 14.7–16.5 | |||||||
| GA | 504 | 13.7 | 12.6–15 | 690 | 17 | 15.9–18.1 | 1194 | 15.3 | 14.5–16.1 | ||||||||
| AA | 140 | 15 | 13–17.3 | 0.95 | 199 | 17.4 | 15.8–19.2 | 0.68 | 339 | 16 | 14.8–17.4 | 0.82 | |||||
| Abs. diff. | 0.6 | 0.4 | 0.4 | ||||||||||||||
| Latin America | GG | 415 | 12.3 | 11–13.9 | 697 | 16.6 | 15.3–17.9 | 1112 | 14.5 | 13.5–15.5 | |||||||
| GA | 292 | 12.9 | 11.3–14.7 | 525 | 17.4 | 16.1–18.9 | 817 | 15.3 | 14.2–16.4 | ||||||||
| AA | 63 | 16.9 | 13.6–21 | 0.02 | 113 | 19.1 | 16.7–21.9 | 0.03 | 176 | 17.8 | 15.9–20 | 0.0007 | |||||
| Abs. diff. | 4.6 | 2.5 | 3.3 | ||||||||||||||
| P-heterogeneity between studies | 0.01 | 0.45 | 0.33 | 0.06 | |||||||||||||
| Males | GG | 1878 | 14.8 | 14.2–15.5 | 864 | 18.3 | 17.6–18.9 | 1301 | 19.4 | 18.6–20.3 | 4043 | 16.8 | 16.4–17.2 | ||||
| GA | 1890 | 15.2 | 14.6–15.8 | 1139 | 18.2 | 17.6–18.7 | 1323 | 20 | 19.2–20.9 | 4352 | 17.1 | 16.7–17.5 | |||||
| AA | 507 | 16 | 15–17.1 | 0.04 | 392 | 18.5 | 17.6–19.5 | 0.78 | 331 | 20.9 | 19.5–22.3 | 0.04 | 1230 | 17.7 | 17.1–18.4 | 0.01 | |
| Abs. diff.a | 1.2 | 0.2 | 1.5 | 0.9 | |||||||||||||
| Females | GG | 592 | 9.9 | 9–10.8 | 319 | 12.5 | 11.6–13.5 | 188 | 14.6 | 12.7–16.8 | 1099 | 11.4 | 10.8–12 | ||||
| GA | 603 | 9.8 | 9–10.7 | 421 | 13.3 | 12.4–14.2 | 197 | 13.9 | 12.2–15.8 | 1221 | 11.5 | 10.9–12.1 | |||||
| AA | 137 | 10.9 | 9.5–12.5 | 0.32 | 171 | 15.1 | 13.8–16.6 | 0.0004 | 57 | 16 | 12.9–19.7 | 0.75 | 365 | 13.2 | 12.2–14.2 | 0.003 | |
| Abs. diff.a | 1 | 2.6 | 1.4 | 1.8 | |||||||||||||
| P-heterogeneity between sex | 0.94 | 0.003 | 0.75 | 0.13 | |||||||||||||
Linear regression analysis of CPD was performed. Means are adjusted for age, sex, country and case/control status for the combined analysis.
aDifference in CPD between both homozygous variant (AA) and homozygous (GG) genotypes.
We determined the association of rs16969968 with smoking quantity after stratifying by sex. The effect of the variant on smoking quantity was slightly higher for women than men (overall difference between homozygotes was 1.8 CPD for women, vs 0.9 CPD for men) (Table 2) (P-heterogeneity = 0.13). This association was not consistent between cases and controls. In the LC cases women showed a 2.6 CPD increase in the homozygous variant cases, whereas we did not detect any association in men (P-heterogeneity = 0.003). However, in the controls men showed a larger association than women. The UADT cancer cases did not show a different association between rs16969968 and smoking quantity for sexes.
Smoking initiation, cessation and heavy smoking
We determined subsequently if smoking initiation or cessation was modified by the 15q variant, among 13 242 ever smokers and 3930 never smokers. We observed no association with smoking initiation in controls alone, or cases alone or combined (OR = 1.01, 95% CI 0.94–1.08, P = 0.83 among controls and OR = 1.03, 95% CI 0.97–1.09, P = 0.36 among all subjects) (Table 3). Similarly, we did not identify an association between the 15q variant and quitting smoking (OR = 1.00, 95% CI 0.92–1.09, P = 1.00 among controls; and OR = 1.00, 95% CI 0.94–1.06, P = 0.91 for all subjects) (Table 3).
Table 3.
Association between smoking initiation, cessation and heavy smoking with rs16969968 genotype
|
Ever vs never smokers |
Current vs former smokers |
Heavy vs light current smokers |
Heavy vs light former smokers |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ever | Never | OR | 95% CI | P-value | Current | Former | OR | 95% CI | P-value | Heavy | Light | OR | 95% CI | P-value | Heavy | Light | OR | 95% CI | P-value | |
| Controls | ||||||||||||||||||||
| GG | 2666 | 1432 | 1.00 | Ref. | – | 1253 | 1356 | 1.00 | Ref. | – | 273 | 332 | 1.00 | Ref. | – | 301 | 477 | 1.00 | Ref. | – |
| GA | 2720 | 1454 | 1.02 | 0.93–1.12 | 0.71 | 1295 | 1378 | 1.01 | 0.90–1.13 | 0.89 | 264 | 319 | 0.98 | 0.76–1.27 | 0.87 | 320 | 439 | 1.20 | (0.95–1.50) | 0.12 |
| AA | 710 | 387 | 1.00 | 0.87–1.16 | 0.95 | 336 | 360 | 0.99 | 0.83–1.19 | 0.93 | 78 | 53 | 1.60 | 1.04–2.46 | 0.03 | 80 | 112 | 1.21 | (0.85–1.73) | 0.28 |
| OR trend | – | – | 1.01 | 0.94–1.08 | 0.83 | – | – | 1.00 | 0.92–1.09 | 1.00 | – | – | 1.15 | 0.96–1.39 | 0.14 | – | – | 1.13 | (0.97–1.33) | 0.13 |
| LC cases | ||||||||||||||||||||
| GG | 1247 | 133 | 1.00 | Ref. | – | 890 | 342 | 1.00 | Ref. | – | 253 | 115 | 1.00 | Ref. | – | 97 | 75 | 1.00 | Ref. | – |
| GA | 1649 | 155 | 1.18 | 0.90–1.56 | 0.23 | 1179 | 452 | 0.94 | 0.79–1.12 | 0.48 | 366 | 145 | 1.10 | 0.78–1.53 | 0.59 | 122 | 90 | 1.30 | (0.81–2.09) | 0.27 |
| AA | 597 | 54 | 1.27 | 0.88–1.84 | 0.20 | 440 | 154 | 0.98 | 0.77–1.24 | 0.88 | 140 | 48 | 1.27 | 0.81–1.99 | 0.30 | 50 | 26 | 2.07 | (1.08–3.96) | 0.03 |
| OR trend | – | – | 1.14 | 0.95–1.36 | 0.15 | – | – | 0.98 | 0.88–1.10 | 0.74 | – | – | 1.12 | 0.90–1.39 | 0.30 | – | – | 1.41 | (1.03–1.92) | 0.03 |
| UADT cancer cases | ||||||||||||||||||||
| GG | 1569 | 132 | 1.00 | Ref. | – | 1187 | 380 | 1.00 | Ref. | – | 393 | 189 | 1.00 | Ref. | – | 126 | 72 | 1.00 | Ref. | – |
| GA | 1649 | 141 | 1.13 | 0.87–1.47 | 0.35 | 1261 | 388 | 1.13 | 0.94–1.35 | 0.18 | 460 | 153 | 1.37 | 1.03–1.82 | 0.03 | 128 | 68 | 1.11 | (0.68–1.79) | 0.68 |
| AA | 435 | 42 | 1.14 | 0.77–1.68 | 0.52 | 320 | 115 | 0.99 | 0.76–1.29 | 0.93 | 136 | 22 | 3.09 | 1.81–5.29 | <0.0001 | 31 | 23 | 0.77 | (0.37–1.60) | 0.49 |
| OR trend | – | – | 1.09 | 0.90–1.30 | 0.38 | – | – | 1.03 | 0.91–1.17 | 0.61 | – | – | 1.57 | 1.27–1.95 | <0.0001 | – | – | 0.94 | (0.67–1.32) | 0.73 |
| Cases and controls combined | ||||||||||||||||||||
| GG | 5482 | 1697 | 1.00 | Ref. | – | 3330 | 2078 | 1.00 | Ref. | – | 919 | 636 | 1.00 | Ref. | – | 524 | 624 | 1.00 | Ref. | – |
| GA | 6018 | 1750 | 1.04 | 0.96–1.13 | 0.34 | 3735 | 2218 | 1.01 | 0.93–1.10 | 0.79 | 1090 | 617 | 1.13 | 0.96–1.34 | 0.13 | 570 | 597 | 1.17 | (0.98–1.41) | 0.09 |
| AA | 1742 | 483 | 1.04 | 0.92–1.19 | 0.51 | 1096 | 629 | 0.98 | 0.87–1.11 | 0.77 | 354 | 123 | 1.81 | 1.39–2.35 | <0.0001 | 161 | 161 | 1.25 | (0.95–1.65) | 0.12 |
| OR trend | – | – | 1.03 | 0.97–1.09 | 0.36 | – | – | 1.00 | 0.94–1.06 | 0.91 | – | – | 1.27 | 1.13–1.43 | <0.0001 | – | – | 1.13 | (1.00–1.29) | 0.05 |
ORs are obtained by logistic regression, comparing GA and AA phenotype with GG phenotype. Adjusted for age, sex, country and case/control status for the combined analysis.
Ref. = reference.
We next examined if rs16969968 genotype was associated with a heavy smoking phenotype. Heavy smoking was defined as >20 CPD and light smoking was defined as 1–10 CPD. If cases and controls were combined the overall OR for a heavy smoking phenotype was 1.81 (95% CI 1.30–2.13, P < 0.001) for the AA genotype. Current smoking controls with the AA genotype had a 60% increased OR of being a heavy smoker (OR = 1.60, 95% CI 1.04–2.46, P = 0.03) (Table 3). The LC cases only showed no association. However, in UADT cancer cases only the OR was 3.09 (95% CI 1.81–5.29, P < 0.001) for the AA genotype. Former smokers had slightly lower ORs in controls and all subjects combined (OR = 1.21, 95% CI 0.85–1.73, P = 0.28; and OR = 1.25, 95% CI, 0.95–1.65, P = 0.12). No heterogeneity between studies was observed (P-heterogeneity = 0.13 and 0.14, respectively, for current and former smokers in the combined analysis).
We also examined if the variant allele modified age of smoking initiation or age of smoking cessation. Adjusted mean age of initiation was not different between the genotypes, neither was age of cessation (Table 4).
Table 4.
Age of onset smoking and age of cessation by rs16969968 genotype
|
Age of smoking initiation |
Age of smoking cessation |
|||||||
|---|---|---|---|---|---|---|---|---|
| n | Mean age | 95% CI | P-trend | n | Mean age | 95% CI | P-trend | |
| Controls | ||||||||
| GG | 2588 | 18.5 | 18.2–18.7 | 1320 | 41.7 | 40.8–42.7 | ||
| GA | 2623 | 18.4 | 18.2–18.7 | 1332 | 41 | 40.1–42 | ||
| AA | 680 | 18.5 | 18.1–19 | 0.86 | 349 | 40.6 | 39.2–42.1 | 0.08 |
| LC cases | ||||||||
| GG | 1232 | 18.2 | 17.9–18.5 | 330 | 48.9 | 47.4–50.4 | ||
| GA | 1629 | 18 | 17.8–18.3 | 445 | 49.3 | 48–50.7 | ||
| AA | 588 | 18 | 17.7–18.4 | 0.42 | 153 | 49.5 | 47.6–51.6 | 0.50 |
| UADT cancer cases | ||||||||
| GG | 1499 | 17.2 | 16.8–17.5 | 340 | 48.5 | 46.3–50.8 | ||
| GA | 1525 | 17.4 | 17.1–17.8 | 326 | 50.3 | 48.2–52.6 | ||
| AA | 390 | 17.3 | 16.8–17.9 | 0.30 | 90 | 48 | 44.9–51.2 | 0.61 |
| Cases and controls combined | ||||||||
| GG | 5319 | 18 | 17.8–18.2 | 1990 | 45 | 44.2–45.8 | ||
| GA | 5777 | 18 | 17.9–18.2 | 2103 | 44.8 | 44.1–45.6 | ||
| AA | 1658 | 18 | 17.8–18.3 | 0.77 | 592 | 44.4 | 43.3–45.6 | 0.35 |
Linear regression of age of onset and age of cessation was performed. Means are adjusted for age, sex and country and case/control status for the combined analysis.
rs16969968 and risk of smoking-related cancers
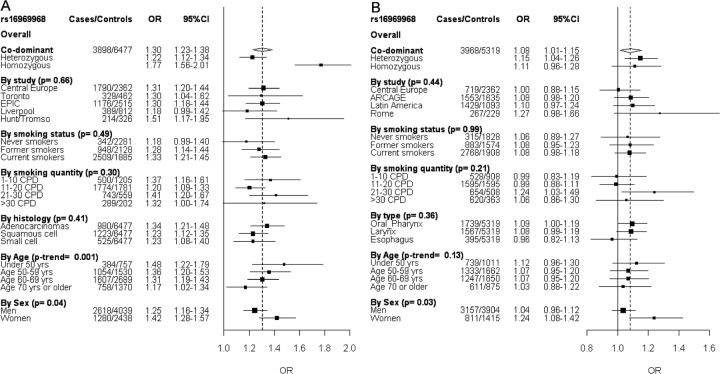
Figure 1A shows the association between rs16969968 and LC risk, stratified by study, smoking phenotype, histology, age and sex. This is an extension of our previous analysis. Under a co-dominant model the OR for LC was 1.30 (95% CI 1.23–1.38, P = 1 × 10–18), after adjustment for age, sex and country. There was no heterogeneity observed between studies, smoking status or histology. rs16969968 genotype was associated with LC in former and current smokers (OR = 1.28, 95% CI 1.14–1.44, P < 0.0001; and OR = 1.33, 95% CI 1.21–1.45, P < 0.0001, respectively), and a trend was observed in never smokers (OR = 1.18, 95% CI 0.99–1.40, P = 0.07) (P-heterogeneity = 0.49). Younger subjects showed a higher OR than older subjects (P-trend = 0.001). Heterogeneity was observed for sex (P-heterogeneity = 0.04), with a higher OR for women (OR = 1.42, 95% CI 1.28–1.57, P < 0.0001) than men (OR = 1.25, 95% CI 1.16–1.34, P < 0.0001). When we adjusted for smoking quantity (CPD) the association was only slightly changed (overall OR = 1.27, 95% CI 1.19–1.35, P < 0.0001).
Figure 1.
Forest plots representing LC (A) and UADT cancer (B) risk and rs16969968 genotype. Unless specified, the ORs and 95% CIs are derived from the per allele model including age, sex and country. The overall OR is shown by the broken vertical line. P-values are from heterogeneity tests, unless the P-trend for the age effect
Figure 1B shows stratified results for the association between UADT cancer risk and rs16969968 genotypes. We observed an overall OR for UADT cancer of 1.08 (1.01–1.15 95% CI, P = 0.02) under a co-dominant model. No heterogeneity was observed between studies, smoking status, organ subtype or age. The OR ratio for UADT cancer was higher in subjects smoking 21–30 CPD (OR = 1.23, 95% CI 1.03–1.49, P = 0.01) than the other smoking categories. The association between UADT cancer and rs16969968 genotype was different between sexes (P-heterogeneity = 0.03); we did not detect an association in men, whereas women with the variant allele had an OR of 1.24 (95% CI 1.08–1.42, P = 0.003). After adjusting for smoking quantity (CPD) the overall OR was 1.05 (95% CI 0.98–1.13, P = 0.15).
Finally, adjusted mean age of LC diagnosis was <1.1 years for homozygous AA vs homozygous GG (P = 0.02) (Table 5). There was evidence of heterogeneity between the studies (P = 0.02). For UADT cancer adjusted mean age of diagnosis was <0.9 years for homozygote variant genotypes, but this difference did not reach statistical significance (P = 0.10). Adjustment for smoking quantity (CPD) did not influence the results (data not shown).
Table 5.
Association between age of onset of LC and UADT cancer with rs16969968 genotype
| LC | UADT cancer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Adj. mean | 95% CI | P-trend | n | Adj. mean | 95% CI | P-trend | ||||
| Overall | GG | 1405 | 61.8 | 61.2–62.3 | Overall | GG | 1701 | 58.5 | 57.8–59.2 | ||
| GA | 1828 | 61.5 | 61–62 | GA | 1790 | 58.2 | 57.5–58.9 | ||||
| AA | 665 | 60.7 | 59.9–61.4 | 0.02 | AA | 477 | 57.6 | 56.6–58.6 | 0.10 | ||
| Diff. in yearsa | 1.1 | Diff. in yearsa | 0.9 | ||||||||
| Central Europe | GG | 640 | 59.4 | 58.6–60.2 | Central Europe | GG | 305 | 57.6 | 56.1–59.2 | ||
| GA | 854 | 59.3 | 58.6–60 | GA | 331 | 56.9 | 55.5–58.4 | ||||
| AA | 296 | 59.2 | 58.1–60.3 | 0.80 | AA | 83 | 56.9 | 54.7–59.2 | 0.38 | ||
| Diff. in yearsa | 0.2 | Diff. in yearsa | 0.7 | ||||||||
| Toronto | GG | 122 | 62 | 59.8–64.2 | ARCAGE | GG | 576 | 58.4 | 57.3–59.5 | ||
| GA | 154 | 63.1 | 61.1–65.1 | GA | 757 | 58.4 | 57.4–59.5 | ||||
| AA | 53 | 62.8 | 59.5–66.4 | 0.56 | AA | 220 | 56.8 | 55.3–58.4 | 0.14 | ||
| Diff. in yearsa | –0.8 | Diff. in yearsa | 1.6 | ||||||||
| EPIC | GG | 399 | 62.6 | 61.7–63.4 | Latin America | GG | 743 | 59.4 | 58.2–60.6 | ||
| GA | 555 | 62.5 | 61.8–63.2 | GA | 561 | 59.3 | 58.1–60.6 | ||||
| AA | 222 | 61 | 60–62 | 0.02 | AA | 125 | 59.6 | 57.5–61.7 | 0.96 | ||
| Diff. in yearsa | 1.6 | Diff. in yearsa | –0.2 | ||||||||
| Liverpool | GG | 168 | 67.7 | 66.3–69.1 | Rome | GG | 77 | 64.9 | 61.9–68 | ||
| GA | 165 | 65.6 | 64.2–67 | GA | 141 | 61.8 | 59.4–64.2 | ||||
| AA | 56 | 64.5 | 62.2–66.8 | 0.008 | AA | 49 | 62.4 | 58.9–66.2 | 0.20 | ||
| Diff. in yearsa | 3.2 | Diff. in yearsa | 2.5 | ||||||||
| Hunt/Tromso | GG | 76 | 64 | 61.4–66.6 | P-heterogeneity between studies | 0.58 | |||||
| GA | 100 | 62.2 | 60.1–64.4 | ||||||||
| AA | 38 | 57.7 | 54.5–61 | 0.006 | |||||||
| Diff. in yearsa | 6.3 | ||||||||||
| P-heterogeneity between studies | 0.02 | ||||||||||
Linear regression of age of onset was performed. Means are adjusted for sex and country.
Difference in years between homozygous variant (AA) and homozygous (GG) genotypes.
GWA analysis to identify other smoking-associated genes
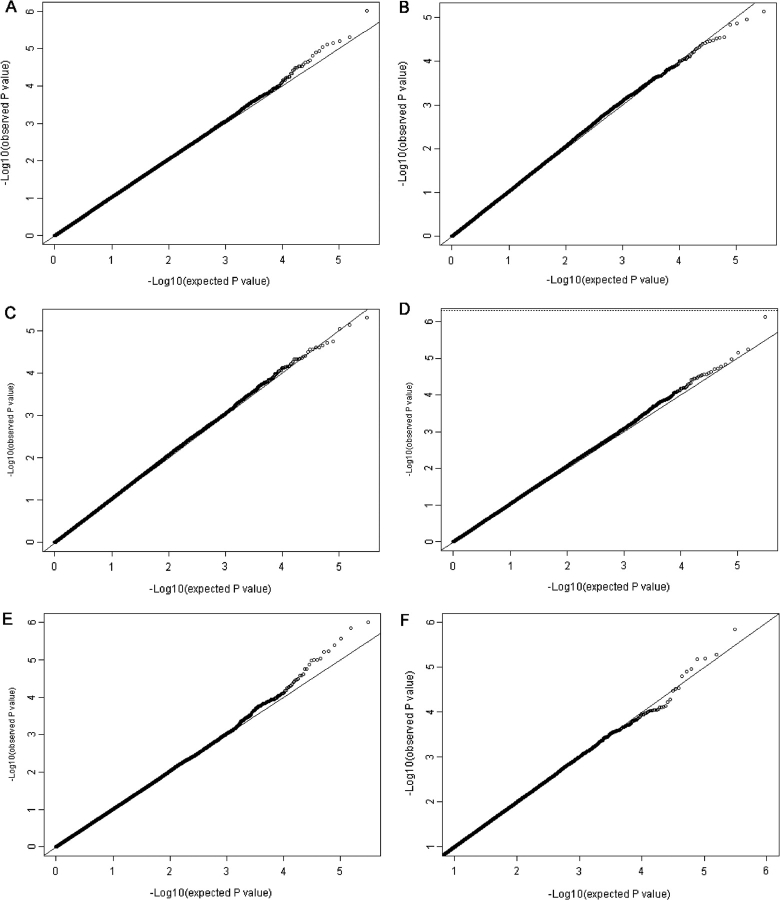
To identify additional genetic regions than 15q25 involved in smoking behaviour, we performed a GWA study of 11 219 subjects, using GWA methods as described. We analysed 317 139 SNPs for the following phenotypes: smoking quantity (CPD), smoking initiation, smoking cessation, heavy smoking, age at smoking initiation. We also looked in a subset of data from the ARCAGE study at the HSI, an index deduced from the Fagerstrom test for nicotine dependence.15 Adjustment was performed for country, age, sex and case/control status. Figure 2 shows quantile–quantile plots for these analyses. No clear inflation of the observed and expected P-values was observed, neither were particular outliers pointing on significant SNPs. No SNPs with a P-value <5 × 10–7 were observed. For each analysis, the 100 SNPs with lowest P-values are shown in the supplementary table available as supplementary data at IJE online.
Figure 2.
Quantile–quantile plot from the GWA study. The observed P-values (Y-axis) are plotted against the expected P-values (X-axis) for the various smoking phenotypes: smoking quantity (CPD) (A), smoking initiation (ever vs never smokers) (B), smoking cessation (current vs former smokers) (C), heavy smoking (D), age of smoking initiation (E), heaviness of smoking index (F)
Discussion
In this study we performed a detailed analysis between the 15q25 LC risk locus, smoking behaviour and related cancers, using data on patients and controls from five LC studies and four UADT cancer studies. We have previously reported that the variant was associated with LC and that this association was largely independent of smoking.2 In our extended analysis, we confirm our previous observations and show in 13 242 ever smokers a small association between the locus and smoking quantity. We used the non-synonymous SNP rs16969968 as the marker to characterize the locus. Overall, although there was no effect on prevalence of smoking, we found that smokers with two copies of the allele associated with increased risk of LC for this SNP, smoked on average 1.2 CPD more, and people with one copy of the risk allele 0.3 CPD more, than individuals in whom this allele was absent. We also found that the risk allele increased the risk for a heavy smoker phenotype. This relationship with heavy smoking is compatible with the quantitative effect on tobacco consumption. Several studies reported an association between 15q variants and nicotine dependence or smoking quantity.3,6,9 Saccone et al. found a potential recessive effect of the rs16969968 on nicotine dependence.9 We found both evidence for a multiplicative and a genotypic model, indicating a potential recessive effect. Thorgeirsson et al. found a 1 CPD increase in their discovery set and a 0.74 CPD increase in their validation set for each copy of the variant allele.3 The latter would lead to a 1.5 CPD increase for two variant alleles in the replication set, similar to our overall estimate of 1.2 CPD.
In a recent publication, Bierut et al. described a high variability in rs16969968 minor allele frequencies between different populations, with percentages ranging from 0% in African populations to 37% in Europeans,7 and postulated that different allele frequencies can lead to differences in the prevalence of nicotine dependence. In our study, we found that the frequency of the risk allele varied in controls between 26% (Latin America) and 39% (Rome). On the other hand, we found weak evidence for heterogeneity of the association with smoking quantity between the studies included in our investigation, with some showing a null or marginal association, whereas others showed strong association, (P = 0.06 for heterogeneity).
Weiss et al. have recently reported that specific CHRNA5-A3-B4 haplotypes are associated with age-dependent nicotine addiction11 and a study by Schaepfer et al. showed that CHRNA5-A3-B4 variations might influence behaviours that promote early alcohol and tobacco initiation.17 Therefore, we examined the possibility of an association with age in our data. We found no association between age at initiation of smoking, or age at quitting in former smokers. We should mention here that numbers were small for age at quitting in the lung and UADT cancer cases. As these people are not likely to stop smoking, this resulted in low sample numbers. Interestingly, however, the mean age of LC onset was <1.1 years for homozygous AA subjects (P = 0.02) and the OR for LC increased with younger age of disease onset (P = 0.001). This age-dependent association was not previously reported, and supports the notion of a more prominent genetic effect in early onset compared with late onset LC.18
The strong association observed with LC risk in our data is not accounted by the relationship of the chromosome 15q locus with smoking quantity, as was already observed in previous LC GWA studies.1–3 We now attempted to formalize this conclusion by using a model developed by Doll and Peto, who described a dose–response relationship among smoking and LC in the British doctors cohort study.19 They found that, amongst male current smokers aged 40–79 years who started smoking between ages 16 and 25 years and who smoked ≤40 CPD, the annual LC incidence was proportional to (CPD + 6)2 × (age – 22.5)4,5, where age – 22.5 was used as a proxy for smoking duration. Given that the vast majority of smokers in our study started between ages of 16 and 25 years, and smoked <40 CPD, we applied this model to our data and calculated the increase in LC risk that is associated with smoking 1.2 CPD extra (the increase in CPD associated with the homozygous variant genotype). For a subject smoking 20 CPD, an increase of tobacco consumption with 1.2 CPD would lead to a 9% increase in LC risk. This is substantially lower than the observed direct association between rs16969968 and LC: the homozygous variant genotype is associated with a 77% increase in LC risk (in fact, an extra 8.6 CPD are required to increase risk by 77%). The corresponding risk increase of 1.2 extra CPD for someone smoking either 10 or 30 CPD is 13 and 6%, respectively. If the variant genotype would increase LC risk by ∼80% solely through its association with smoking quantity, this would imply that the assessment of smoking quantity by our questionnaires is subject to extreme levels of misclassification, which would be in contradiction with the strong relationship between smoking and LC observed in our studies.20 Adjustment for smoking quantity had little effect on the estimation of the LC risk associated with the locus. Moreover, in our data we found an increased LC risk not only in present or former smokers, but also in never smokers. However, it is possible that actual nicotine exposure is not fully captured by smoking quantity. Depth of inhalation, smoking to the end of the butt are, among others, contributing to nicotine exposure. Cotinine measurement, which is a more appropriate method for nicotine exposure, was weakly correlated to smoking quantity in the EPIC study (correlation coefficient = 0.2866). Unfortunately, cotinine levels were not available for the other studies, so only smoking quantity could be used for large-scale analysis.
It is unlikely that the association of the SNP with LC is the result of inflation by chance (Winner's Curse). When we consider the four replication studies (Toronto, Epic, Liverpool, Hunt) they all show the same association of the minor allele with LC as the discovery dataset (Central Europe study) (Figure 1A), also after adjusting for smoking quantity.
Therefore, we conclude that a more plausible explanation is that the variant allele is indeed associated with LC independent of or in addition to an association with tobacco smoking, presumably through a direct effect on the bronchial epithelium. Previous studies have assessed that nicotine receptor genes are expressed in LC cells and might play a role in lung carcinogenesis.21–23 The rs16969968 SNP leads to a substitution of D to N at position 398 of the CHRNA5 gene, which is a highly conserved region between species.2 Functional studies by Bierut et al.7 demonstrated that the risk allele decreased response to a nicotine agonist. However, the functional consequences of the D398N alteration and its possible role in lung carcinogenesis remain to be established.
A recent study by LeMarchand24 found that carriers of these variants extract a greater amount of toxic substances per cigarette than non-carriers, resulting in an increased risk for LC. So this study also provides evidence that the variant is influencing other aspects of smoking behaviour.
It should be noted that the studies by Amos et al. and Spitz et al. did not identify an association of the locus with LC risk in never smokers.1,25 However, the number of never smokers in all three studies is small, and further research to clarify these differences is required. As discussed above, Spitz et al.1,25 found an association between the minor allele and smoking quantity that is equivalent to that reported here, and which similarly does not appear to be sufficient to explain alone the increased risk of LC.
In our study, we also found an association between the locus and UADT cancer (OR = 1.08) that was marginally stronger in women compared with men (P-heterogeneity = 0.03), but much smaller than the association with LC. Like in the LC studies, the OR diminishes after adjusting for smoking. This suggests that the relationship between this locus and UADT cancer could be mediated through effects on smoking quantity. Larger UADT cancer studies should confirm if the association between rs16969968 and UADT cancer is direct or mediated through smoking.
In an attempt to find other smoking-related genes, we analysed GWA data on over 11 000 subjects for six different smoking phenotypes. However, this analysis did not identify any SNPs clearly related to smoking behaviour. This finding is in accordance with the other large GWA of smoking behaviour3 that identified an association between 15q25 variants and smoking behaviour and indicated that additional common variants with a similar or greater effect are unlikely to exist. Comparison of our top gene lists with data from other published GWA studies6,8,26–28 on smoking quantity or nicotine dependence did not result in any overlap. It is also of interest that in our GWA 15q variant rs8034191 shows a P-value of 0.02 (rs16969968 was not on the Illumina array), which is in the same order of magnitude as findings from our candidate gene approach on rs16969968.
This study has some limitations. First, many comparisons were made and therefore some associations might be due to chance. However, for most analyses sample numbers were sufficiently large to detect real associations. Secondly, samples were obtained from cancer studies. Samples in most studies investigating smoking behaviour are obtained from population-based cohorts. In this way there may be a selection bias in study subject, even when we adjust for case/control status. As most studies were case–control studies it is slightly more difficult to obtain the OR for LC for the general population. For such a purpose cohort meta-analysis of cohort studies only is more appropriate. Finally, GWA data were not available for two studies (Latin America and Rome) and we were therefore unable to identify individuals of mixed ethnicity for these two studies. Any potential population stratification resulting from the inclusion of these two studies is however likely to be minimal.
In conclusion, the results of this study confirm that the rs16969968 gene variant is associated with both nicotine dependence and LC risk. The modest association with cigarette smoking would indicate that the major association with LC cannot be explained by the variant's effect on smoking quantity. The association between rs16969968 and age at onset of LC and the difference between sexes in both lung and UADT cancers risk need to be confirmed in independent validation series.
Supplementary data
Supplementary data are available at IJE online.
Funding
NCI grants ‘Genetics of tobacco and alcohol related cancers: R01CA092039-07’ and ‘International lung cancer consortium: R03CA133939-01’.
KEY MESSAGES.
This study shows that the association between rs16969968 and LC is strong and largely independent from smoking quantity.
A small association between rs16969968 and smoking quantity was observed.
For the first time an association between the rs16969968 SNP and UADT cancer risk was shown.
Rs16969968 was associated with an earlier age of LC onset and the association between the minor allele and LC was more pronounced in women than in men.
GWA analysis on 11 219 subjects did not identify any additional variants related to smoking behaviour.
Supplementary Material
Acknowledgements
T.V.M. partly worked on this study while at the University of Manchester. The authors acknowledge the help of Prof. Gary J Macfarlane, Dr Anne-Marie Biggs, Dr Richard Oliver and Prof. Martin Tickle in study conduct at the Manchester centre, and Prof. Phil Sloan and Prof. Nalin Thakker who, in addition, coordinated sample collection and processing for all the UK centres.
Conflict of interest: None declared.
Appendix
Paolo Vineis,1,2 Francoise Clavel-Chapelon,3 Domenico Palli,4 RosarioTumino,5 Vittorio Krogh,6 Salvatore Panico,7 Carlos A González,8 José Ramón Quirós,9 Carmen Martínez,10 Carmen Navarro,11,12 Eva Ardanaz,13 Nerea Larrañaga,14 Kay Tee Khaw,15 Timothy Key,16 H Bas Bueno-de-Mesquita,17 Petra HM Peeters,18 Antonia Trichopoulou,19 Jakob Linseisen,20 Heiner Boeing,21 Göran Hallmans,22 Kim Overvad,23 Anne Tjønneland,24 Merethe Kumle25 and Elio Riboli2
1Servizio di Epidemiologia dei Tumori, Università di Torino and CPO-Piemonte, Turin, Italy.
2Department of Epidemiology and Public Health, Imperial College, London, UK.
3INSERM, E3N-EPIC Group Institut Gustave Roussy, Villejuif.
4Molecular and Nutrional Epidemiology Unit, Center for Cancer Research and Prevention Scientific Institute of Tuscany, Florence, Italy.
5Cancer Registry and Histopathology Unit, Azienda Ospedaliera "Civile M.P.Arezzo", Ragusa, Italy
6Istituto Nazionale dei Tumori, Milan, Italy.
7Dipartimento di Medicina Clinica e Sperimentale, Universita di Napoli, Federico II, Naples, Italy.
8Servicio de Epidemiología y registro del Cáncer, Instituto Catalán de Oncología, Barcelona, Spain.
9Jefe Sección Información Sanitaria, Consejería de Servicios Sociales, Principado de Asturias, Oviedo, Spain.
10Escuela Andaluza de Salud Pública, Granada, Spain.
11Department of Epidemiology, Health Council of Murcia, Murcia, Spain.
12CIBER Epidemiologia y Salud Publica (CIBERESP), Spain.
13Registro de Cáncer de Navarra, Instituto de Salud Pública,Gobierno de Navarra, Pamplona, Spain.
14Subdirección de Salud Pública de Gipuzkoa, Gobierno Vasco, San Sebastian, Spain.
15MRC Dunn Human Nutrition Unit, Cambridge, UK.
16Cancer Research UK, University of Oxford, Oxford, UK.
17Centre for Food and Health, National Institute of Public Health and the Environment (RIVM), Bilthoven, The Netherlands.
18Julius Center for Health Sciences and Primary Care, Department of Epidemiology, University of Utrecht, Utrecht, The Netherlands.
19Department of Hygiene and Epidemiology, University of Athens, Athens, Greece.
20Division of Clinical Epidemiology, German Cancer Research Centre, Heidelberg, Germany.
21Department of Epidemiology, Deutsches Institut für Ernährungsforschung, Potsdam-Rehbrücke, Germany.
22Department of Public Health and Clinical Medicine, University of Umeå, Umeå, Sweden.
23Department of Epidemiology and Social Medicine, Aarhus University, Aarhus, Denmark.
24The Danish Cancer Society, Institute of Cancer Epidemiology, Copenhagen, Denmark.
25Institute of Community Medicine, University of Tromsø, Tromsø, Norway.
References
- 1.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–37. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanock SJ, Hunter DJ. Genomics: when the smoke clears. Nature. 2008;452:537–38. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 6.Berrettini W, Yuan X, Tozzi F, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens VL, Bierut LJ, Talbot JT, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17:3517–25. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashibe M, McKay JD, Curado MP, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40:707–9. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- 13.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fagerstrom KO, Schneider NG. Measuring nicotine dependence: a review of the Fagerstrom Tolerance Questionnaire. J Behav Med. 1989;12:159–82. doi: 10.1007/BF00846549. [DOI] [PubMed] [Google Scholar]
- 15.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom test for nicotine dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–27. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlaepfer IR, Hoft NR, Collins AC, et al. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–46. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cassidy A, Myles JP, Duffy SW, Liloglou T, Field JK. Family history and risk of lung cancer: age-at-diagnosis in cases and first-degree relatives. Br J Cancer. 2006;95:1288–90. doi: 10.1038/sj.bjc.6603386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doll R, Peto R. Cigarette smoking and bronchial carcinoma: dose and time relationships among regular smokers and lifelong non-smokers. J Epidemiol Community Health. 1978;32:303–13. doi: 10.1136/jech.32.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan P, Crispo A, Zaridze D, et al. High cumulative risk of lung cancer death among smokers and nonsmokers in Central and Eastern Europe. Am J Epidemiol. 2006;164:1233–41. doi: 10.1093/aje/kwj340. [DOI] [PubMed] [Google Scholar]
- 21.Song P, Sekhon HS, Fu XW, et al. Activated cholinergic signaling provides a target in squamous cell lung carcinoma. Cancer Res. 2008;68:4693–700. doi: 10.1158/0008-5472.CAN-08-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam DC, Girard L, Ramirez R, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638–47. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- 23.Minna JD. Nicotine exposure and bronchial epithelial cell nicotinic acetylcholine receptor expression in the pathogenesis of lung cancer. J Clin Invest. 2003;111:31–33. doi: 10.1172/JCI17492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le ML, Derby KS, Murphy SE, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–40. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–56. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drgon T, Montoya I, Johnson C, et al. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15:21–27. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.