SUMMARY
Telomerase ribonucleoprotein complexes copy an internal RNA template to synthesize DNA repeats. DNA-interacting subunits other than telomerase reverse transcriptase (TERT) and telomerase RNA (TER) have been hypothesized to account for high repeat addition processivity of telomerase holoenzyme compared to the minimal catalytic RNP. Here, we present the identification of three additional subunits of Tetrahymena thermophila telomerase holoenzyme. Each of seven telomerase proteins is required for telomere maintenance and copurifies active RNP. The catalytic core (p65-TER-TERT) is assembled with a three-protein subcomplex (p75-p45-p19) and two peripheral subunits (p82 and p50). Remarkably, only a p82-enriched subset of the total holoenzyme population is capable of high repeat addition processivity, as shown by p82 immunodepletion and add-back. The RPA-like p82 subunit binds sequence specifically to multiple telomeric repeats. These discoveries establish the existence of a telomerase holoenzyme processivity subunit with sequence-specific DNA binding.
INTRODUCTION
Telomeric repeats nucleate the formation of specialized chromatin that allows chromosome ends to persist as DNA breaks (Gilson and Geli, 2007; Palm and de Lange, 2008). Each terminal repeat array is maintained in a dynamic equilibrium of repeat loss and new repeat synthesis. Telomerase elongates the guanosine-rich (G-rich) strand of telomeric repeats, T2AG3 in vertebrates or T2G4 in the ciliate Tetrahymena thermophila, using an active site within telomerase reverse transcriptase (TERT) and a template within telomerase RNA (TER). TERT and TER from some species can be combined in a heterologous cell extract to reconstitute telomeric repeat synthesis activity (Collins, 2009). Telomerase biogenesis and function in vivo require additional proteins that remain associated with TERT and TER in a telomerase holoenzyme (Gallardo and Chartrand, 2008; Collins, 2008). The catalytic features of recombinant TERT+TER can differ from the features of holoenzyme (Hardy et al., 2001; Errington et al., 2008). For example, for the relatively well-studied T. thermophila enzymes, the recombinant telomerase catalytic core differs from endogenous holoenzyme in the mechanism and extent of repeat addition processivity (RAP).
The current inventory of telomerase-associated factors includes proteins that interact with TER, TERT, and/or catalytically active holoenzyme. Among these factors, two types of holoenzyme protein have been most amenable to functional characterization. The first class is comprised of TER-binding proteins that promote accumulation of biologically stable, mature RNP (Teixeira and Gilson, 2007; Collins, 2009). These proteins are relatively straightforward to define by interaction assays and the in vivo depletion phenotype of reduced TER. Ciliates possess a telomerase-specific protein with a La motif that folds and protects RNA Pol III-transcribed TER. RNA Pol II-transcribed TERs of vertebrates or yeasts instead assemble with proteins shared by small nuclear or small nucleolar RNPs. A second class of holoenzyme protein acts to allow telomerase engagement with the substrate single-stranded DNA (ssDNA) at chromosome termini (Gilson and Geli, 2007). The best-characterized members of this group are budding yeast Est1 and Est3, which act in a coordinated, cell cycle-dependent fashion to activate the RNP catalytic core through a mechanism involving Est1 association with the telomeric ssDNA-binding protein Cdc13 (Pennock et al., 2001; Osterhage et al., 2006; Chan et al., 2008). Additional proteins in this category have been shown to affect telomerase subunit localization and association with telomeric chromatin (Fisher et al., 2004; Gallardo et al., 2008; Venteicher et al., 2009).
Ciliates provide a favorable model system for telomerase holoenzyme characterization due to the relative abundance of active telomerase. We have focused on T. thermophila to exploit the useful combination of telomerase abundance and robust molecular genetic methods. Previous work discovered and characterized the T. thermophila telomerase holoenzyme proteins p65, p45, and p75 encoded by TAP65, TAP45, and TAP75 (Witkin and Collins, 2004; Witkin et al., 2007). In vivo depletion of the La motif-containing p65, but not p45 or p75, reduced the biological accumulation of TERT and TER. Biochemical and biophysical assays revealed that p65 binds and folds TER to promote subsequent TERT-TER interaction, producing a conformationally homogeneous p65-TER-TERT ternary complex (Prathapam et al., 2005; O'Connor and Collins, 2006; Stone et al., 2007). The in vitro assembly specificity of this RNP catalytic core parallels that of TERT and TER accumulation as stable RNP in vivo. On the other hand, it remains unclear how p45 and p75 contribute to telomerase function. At a genetic level, these proteins have roles comparable to yeast Est1 and Est3 because protein depletion causes telomere shortening without loss of the RNP catalytic core (Lingner et al., 1997; Witkin and Collins, 2004; Witkin et al., 2007). At a biochemical level, it has not been possible to assign activities or even protein-protein interaction specificities to p45 and p75, despite substantial effort.
To gain additional insight about telomerase holoenzyme proteins and their roles beyond assembly of the catalytic core, we sought to identify additional T. thermophila telomerase-associated proteins. Previous identification of p65, p45, and p75 involved large-scale purification of TERT-TAP (TERT tagged with a C-terminal calmodulin-binding peptide and tandem protein A domains) overexpressed from an ectopically integrated transgene (Witkin and Collins, 2004). Although steady-state TERT accumulation was not increased in cells overexpressing TERT-TAP, continuous degradation of overexpressed protein led to identification of a TERT-associated, but not holoenzyme-associated, p20 protein, the Skp1 subunit of an abundant ubiquitin ligase. Genetic depletion of Skp1 increased rather than decreased telomere length, suggesting that this protein has a role in turnover of factor(s) that promote telomere elongation (Witkin et al., 2007). Here, we exploit an improved affinity purification strategy and recent T. thermophila macronuclear genome annotation to discover three additional telomerase holoenzyme proteins designated p19, p50, and p82. The p19 subunit is part of a three-subunit “telomere adaptor” subcomplex (p75-p45-p19). The DNA-binding p82 subunit joins the p75-p45-p19 subcomplex to confer the high repeat addition processivity of endogenous holoenzyme, while the substoichiometric p50 subunit modulates association of the p75-p45-p19 subcomplex with the catalytic core. Overall, this work reveals an intricacy of holoenzyme protein interactions that bridge the telomerase active site with a subunit capable of sequence-specific binding to telomeric ssDNA.
RESULTS
Affinity Purification and Mass Spectrometry Identify Additional Telomerase Proteins
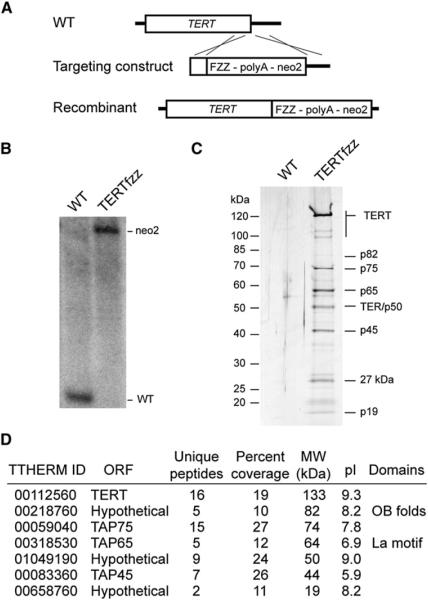
Based on previous studies, we designed a new strain for telomerase holoenzyme affinity purification. To avoid tagged TERT overexpression, we integrated a C-terminal tag cassette at the endogenous TERT locus such that tagged protein accumulation was dependent on expression from the endogenous promoter and translation from the endogenous open reading frame. In addition, we replaced the calmodulin-binding peptide of the previous TAP tag with 3 × FLAG peptide because the calmodulin-binding peptide reduces fusion protein expression level in T. thermophila and does not have high purification yield (Witkin et al., 2007). The new FZZ expression cassette (3 × FLAG epitope/ tobacco etch virus [TEV] protease cleavage site/ tandem protein A domains) was integrated at TERT by selection for a linked neomycin resistance cassette (Figure 1A). Complete replacement of recombinant for endogenous chromosomes was obtained (Figure 1B), indicating that tagged TERT retains biological function.
Figure 1. Affinity Purification and Mass Spectrometry Identify Additional TERT-Associated Proteins.
(A) Diagram of FZZ tagging strategy at the TERT locus.
(B) FZZ cassette integration. Replacement of the wild-type (WT) TERT locus by the FZZ-tagged recombinant (neo2) locus was detected by Southern blot.
(C) Two-step affinity purification of TERTfzz and associated proteins.
(D) TERTfzz-associated proteins identified by mass spectrometry.
Tandem steps of affinity purification were performed from whole-cell extract using two resins. The protein A modules of the tagged protein were first bound to IgG agarose, followed by elution with TEV protease. The protease-cleaved fusion protein retained a 3 × FLAG epitope and was next bound to FLAG antibody resin, followed by elution with 3 × FLAG peptide or denaturant. To examine the protein profile of two-step purified samples, we subjected an aliquot of the FLAG antibody elution to SDS-PAGE and silver staining. Affinity purification of TERTfzz copurified several polypeptides not recovered in the parallel mock purification from extract without tagged protein (Figure 1C). Proteins with the mobilities of p75, p65, and p45 were readily detectable. In addition, we observed a yellow-orange band around 50 kDa that results from silver staining of TER. Additional polypeptides were also evident, only some of which could be assigned by immunoblotting to be degradation products of known holoenzyme proteins (for example, the indicated doublet of TERT N-terminal truncation products at ~100–110 kDa in Figure 1C).
The polypeptides specifically and reproducibly coenriched with TERT were identified by mass spectrometry comparison of TERTfzz and mock affinity purifications. To be comprehensive, we used the entire mixture of TERT-associated proteins eluted by denaturation for peptide sequencing, as described previously for human telomerase (Fu and Collins, 2007). T. thermophila gene identification was performed by searching the predicted proteome of the macronuclear genome (Eisen et al., 2006). In addition to the known telomerase holoenzyme proteins TERT, p75, p65, and p45, the complete list of polypeptides specifically enriched with TERTfzz (Figure 1D) includes three proteins annotated as hypothetical in the Tetrahymena Genome Database. Each of the TERT-associated proteins was identified by at least two unique peptides (Figure 1D). Beyond these peptides, the open reading frames of the additional hypothetical telomerase proteins could have been incorrectly predicted by automated genome annotation. Therefore, mRNAs encoding these proteins were amplified by RT-PCR, were cloned, and were sequenced. The cloned cDNAs encoded proteins with predicted molecular weights of 19, 50, and 82 kDa (Figure 1D). In accordance with nomenclature rules, the genes were designated TAP19, TAP50, and TAP82 for telomerase- or TERT-associated proteins; the proteins are described as p19, p50, and p82.
The p19 and p50 do not share obvious homology with any nonciliate protein. In contrast, the domain architecture and primary sequence of p82 suggest that it has an evolutionary relationship with the replication protein A (RPA) largest subunit, RPA1. Domain structure modeling of p82 compiled by 3D-Jury suggests tandem OB folds most similar to those in human RPA1 (Figure S1A available online). RPA1 proteins are top BLAST hits of p82 as well, with homology extending over the three RPA1 OB-fold DNA-binding domains (Figure S1B). However, reciprocal BLAST of the T. thermophila genome with human RPA1 identifies a protein other than p82 as the top BLAST hit. The top BLAST hit is likely to be the actual T. thermophila ortholog of RPA1 based on sequence conservation and high expression level relative to p82. This putative T. thermophila RPA1 shares greater primary sequence identity with human RPA1 than either RPA1 shares with p82 (Figure S1C), indicating that TAP82 has substantially diverged from the putative ancestral RPA1 (as also supported by biochemical data described below).
The predicted SDS-PAGE mobilities of the three additional telomerase proteins match polypeptides detected by silver staining, although at low silver-staining intensity (Figure 1C). In addition to the full-length proteins, small polypeptides are enriched that are likely degradation products of larger subunits (for example, the 27 kDa polypeptide described below). The p19 polypeptide is likely the same protein observed in TERT-TAP purifications (Witkin and Collins, 2004) that appeared stoichiometric with TERT, p75, p65, and p45. On the other hand, p50 and p82 appear substoichiometric in their holoenzyme association. Their identification suggests that we have deeply surveyed holoenzyme protein composition. However, because protein detection by mass spectrometry is not quantitative with regard to protein abundance, it remains possible that additional holoenzyme subunits remain to be discovered.
Additional Telomerase Proteins Have Essential Functions in Telomere Maintenance
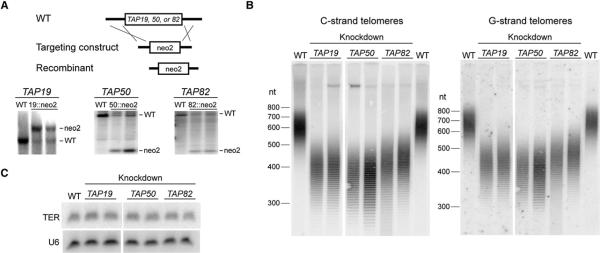
To compare the biological function(s) of the additional telomerase proteins with known telomerase holoenzyme proteins, we first examined whether TAP19, TAP50, and TAP82 are essential genes. For this purpose, we targeted each locus for replacement with a neomycin resistance cassette (Figure 2A). Because the expressed macronuclear genome is polyploid, initial transformants will be drug resistant but retain copies of the wild-type chromosome as well. Chromosome segregation is amitotic, resulting in some daughter cells with an increased ratio of recombinant to wild-type chromosomes. If the targeted gene is not essential for growth, continued selective pressure yields cells with only the recombinant chromosome. If the gene is essential, cells will retain a mixture of wild-type and recombinant chromosomes. In multiple independent selections, endogenous wild-type TAP19, TAP50,or TAP82 chromosomes could not be fully replaced (Figure 2A). Thus, like the previously characterized T. thermophila telomerase holoenzyme components, p19, p50, and p82 are essential.
Figure 2. Additional Telomerase Proteins Are Essential for Telomere Maintenance.
(A) Partial gene disruption for TAP19, TAP50, and TAP82. A diagram of the gene disruption strategy is shown.
(B) Telomere length in TAP19, TAP50, and TAP82 knockdown cultures.
(C) Northern blot for accumulation of TER and U6 RNA.
Cells depleted for p19, p50, or p82 by gene knockdown were examined for a telomere length phenotype. Limiting the dose of TER, TERT, or any other holoenzyme subunit reduces the length of macronuclear telomeric repeats from the usual ~300 bp (Miller and Collins, 2000; Witkin and Collins, 2004; Witkin et al., 2007). Half of the T. thermophila macronuclear telomeres share the same subtelomeric sequence due to the amplification of a palindromic chromosome encoding the large rRNA precursor (the rDNA). If genomic DNA is digested with a specific restriction enzyme and then denatured, resolved by PAGE, and hybridized with a subtelomeric-sequence oligonucleotide probe complementary to the G-rich or C-rich repeat strand, it is possible to detect change in length of both strands independently (Figure 2B). The denatured DNA strands resolve into a ladder that reflects the presence of a preferred telomeric repeat permutation at chromosome 3′ and 5′ termini (Jacob et al., 2001). As observed previously in telomerase holoenzyme component knockdown strains, knockdown strains of p19, p50, and p82 had shorter G-strand and C-strand telomere lengths without loss of a preferential end permutation (Figure 2B). This telomere-shortening phenotype differs from the telomere elongation phenotype observed upon shutoff of the major T. thermophila ssDNA end-binding protein, Pot1a (Jacob et al., 2007). Because p19, p50, and p82 knockdown strains retain normal TER accumulation relative to a small nuclear RNA-loading control (Figure 2C), as do p45 and p75 knockdown strains (Witkin and Collins, 2004; Witkin et al., 2007), these proteins influence a telomere maintenance step subsequent to stable assembly of the telomerase catalytic core.
Physical Associations Suggest an Overall Holoenzyme Subunit Architecture
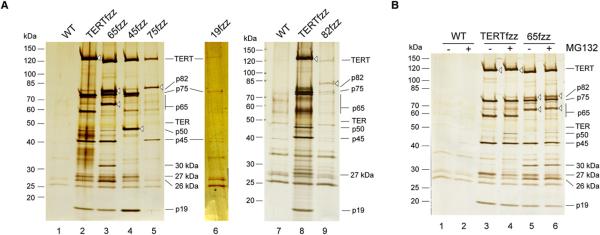
To reciprocally confirm the association of the additional telomerase proteins with telomerase and to compare the physical interaction partners of all telomerase holoenzyme proteins in parallel, we took advantage of the two-step affinity purification developed for TERTfzz. Although mass spectrometry yields of a relative scarce enzyme such as telomerase require many liters of culture input, ~1 liter of culture input is sufficient to detect TERTfzz-copurified proteins by SDS-PAGE and optimized silver staining. All proteins from the comprehensive list identified in association with TERTfzz (Figure 1D) were individually tagged at their endogenous locus using the C-terminal FZZ cassette. C-terminal TAP tagging of p65 and p45 was previously shown to produce biologically functional protein (Witkin and Collins, 2004). For these two subunits and the others discussed above (p75, p19, p50, and p82), C-terminal FZZ-cassette tagging also produced biologically functional protein, as demonstrated by complete substitution of recombinant for endogenous chromosomes (Figures S2A and S2B and data not shown). Telomeres remained near wild-type length in cells expressing 65fzz, 45fzz, or 50fzz and were only slightly shorter than wild-type length in cells expressing 75fzz or 82fzz (Figure S2C and data not shown). Telomeres had stable but notably shortened length in cells expressing 19fzz (Figure S2C), suggesting that tagging this small protein partially compromised its biological stability or function.
Affinity purifications of each tagged protein were carried out under parallel conditions to compare the profile of copurified proteins. After two-step affinity purification, each tagged subunit migrates with an ~3 kDa difference from its endogenous counterpart due to the 3 × FLAG tag fusion (Figure 3A, tagged subunits are indicated with open arrowheads). TERTfzz, 65fzz, 45fzz, 75fzz, 19fzz, and 82fzz copurified polypeptides with the migration of TERT and other holoenzyme proteins (Figure 3A; see Figure S3A for immunoblots), as well as TER and telomerase activity (see below), reciprocally confirming their association. Compared to the other purifications, tagged subunit yields from 19fzz or 82fzz purification were reproducibly lower. The low yield of 19fzz holoenzyme is likely a technical artifact of protein tagging, judging from the reduced telomere length of the 19fzz strain (described above) and the higher level of untagged p19 copurified with TERTfzz, 65fzz, and 45fzz (Figure 3A, compare lane 6 to lanes 2–4). The low yield of 82fzz holoenzyme instead appears to derive from ongoing subunit degradation and/or dissociation in extract: single-step affinity purifications recovered a level of holoenzyme comparable to TERTfzz (see below), but more extensive purification disproportionally reduced tagged p82 recovery. A scaled-up two-step affinity purification of 82fzz allowed holoenzyme copurification to be detected against high nonspecific background (Figure 3A, lanes 7–9). In contrast, even scaled-up 50fzz purifications failed to enrich a protein detectable against the nonspecific background, likely due to the extremely low expression level of p50 (50fzz was undetectable by immunoblot against the tag in whole-cell lysates; data not shown).
Figure 3. FZZ-Tagged Proteins Purify Other Telomerase Holoenzyme Subunits.
Two-step affinity purifications were performed to enrich fzz-tagged protein complexes. Open arrowheads indicate the tagged protein.
(A) Copurification of telomerase holoenzyme proteins. Purifications in lanes 7–9 used more input extract and thus have greater nonspecific background.
(B) Effect of MG132 on purified holoenzyme subunit composition.
Across a range of extract preparation and purification conditions, the polypeptides that copurified with tagged holoenzyme subunits had the SDS-PAGE mobilities of other holoenzyme subunits, not of unknown proteins. These findings suggest that the T. thermophila telomerase proteins are likely telomerase specific in assembly and function, unlike the telomerase-associated proteins of mammalian cells. Several additional observations emerged from the course of these purification assays. Addition of the proteasome inhibitor MG132 to extracts before purification improved the recovery of p82 and p50 with other holoenzyme components (Figure 3B, compare lanes 3 and 4). In addition, we observed variable mobility of p65, including at least two forms enriched by the C-terminal tag (Figure 3B, lanes 5 and 6). Finally, small polypeptides not corresponding to full-length holoenzyme proteins varied in their extent of copurification with the tagged subunits. A 27 kDa polypeptide showed decreased recovery in purifications with MG132 (Figure 3B, compare lanes 3 and 5 with lanes 4 and 6) and generally showed a reciprocal relationship with full-length p82, suggesting that it could be a p82 proteolytic fragment. In addition, 30 and 26 kDa polypeptides were enriched preferentially by 65fzz (Figure 3B, compare lanes 5 and 6 with 3 and 4), which could reflect p65 proteolysis or the participation of additional proteins in an early stage of telomerase RNP assembly.
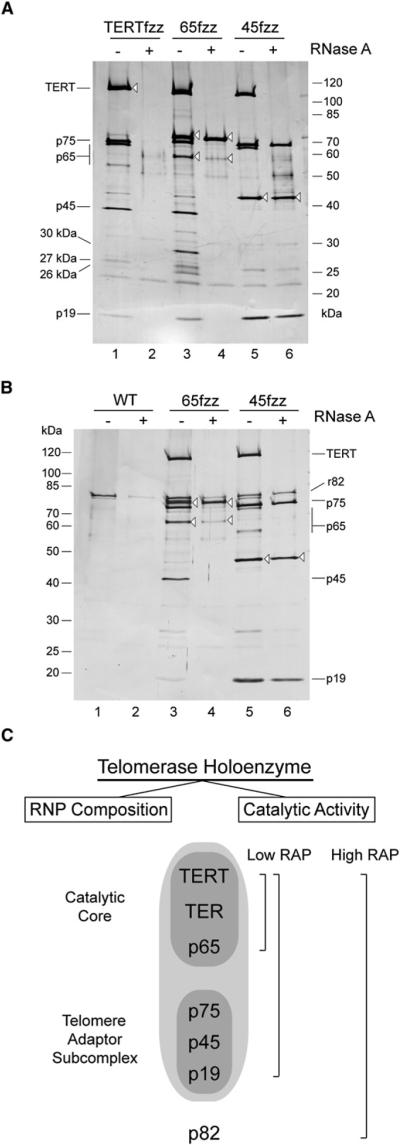
We exploited the relatively robust yield of TERTfzz, 65fzz, and 45fzz purifications to assay protein-protein interactions within the holoenzyme. Leaving the second-step FLAG-tagged complexes immobilized on antibody resin, we performed an incubation with RNase A or buffer control, washed to remove dissociated subunits, and eluted with FLAG peptide prior to SDS-PAGE and silver staining. RNase treatment of TERTfzz holoenzyme induced an apparent misfolding of TERT that affected protein recovery, likely by interfering with tag accessibility (Figure 4A, lanes 1 and 2). RNase treatment of p65fzz holoenzyme depleted all holoenzyme subunits except p65 (Figure 4A, lanes 3 and 4). RNase treatment of 45fzz holoenzyme depleted TERT and p65, and p45 and p75 remained associated with each other as an RNase-resistant subcomplex (Figure 4A, lanes 5 and 6; see Figure S3B for p65, p45, and p75 immunoblots). This subcomplex also contained p19 (Figure 4A, lanes 5 and 6; see Figure S3B for p19 immunoblot). This physical interaction of p75, p45, and p19 subunits is consistent with their shared in vivo depletion phenotype of telomere shortening without loss of TER RNP.
Figure 4. Protein Interactions Establish a Second Holoenzyme Subcomplex.
(A) Identification of a p75-p45-p19 subcomplex. Immobilized holoenzyme complexes were treated with RNase A.
(B) Association of p82 with the p75-p45-p19 subcomplex. Recombinant p82 (r82) was added to immobilized holoenzyme followed by treatment with RNase A.
(C) Summary of telomerase subcomplexes.
To bypass the limitation of low endogenous p82 or p50 recovery with two-step purified holoenzyme, we attempted to bacterially express and purify recombinant p82 and p50 for holoenzyme supplementation in vitro. This was successful only for recombinant p82 (r82), which could be purified as a soluble 6 × His-tagged protein (see below). After supplementing purified 65fzz or 45fzz holoenzyme with r82 and washing to remove excess protein, we examined the RNase sensitivity of protein-protein interactions. Some nonspecific association of r82 with antibody resin was detected in the absence of RNase A (Figure 4B, lanes 1 and 2). RNase treatment of r82-supplemented 65fzz holoenzyme released all proteins other than p65 itself, with only background levels of r82 association (Figure 4B, lanes 3 and 4). On the other hand, RNase treatment of r82-supplemented 45fzz holoenzyme reproducibly retained an elevated level of r82 in association with the p75-p45-p19 subcomplex (Figure 4B, lanes 5 and 6). These results suggest that the p75-p45-p19 subcomplex recruits p82 through protein-protein interactions. Overall, the findings described above suggest that a biologically stable assembly of the RNP catalytic core (p65-TER-TERT) associates directly with a second holoenzyme protein subcomplex (p75-p45-p19) that is required as an adaptor for p82 binding and telomerase function at telomeres in vivo (Figure 4C).
Holoenzyme Proteins Differentially Enrich High Repeat Addition Processivity
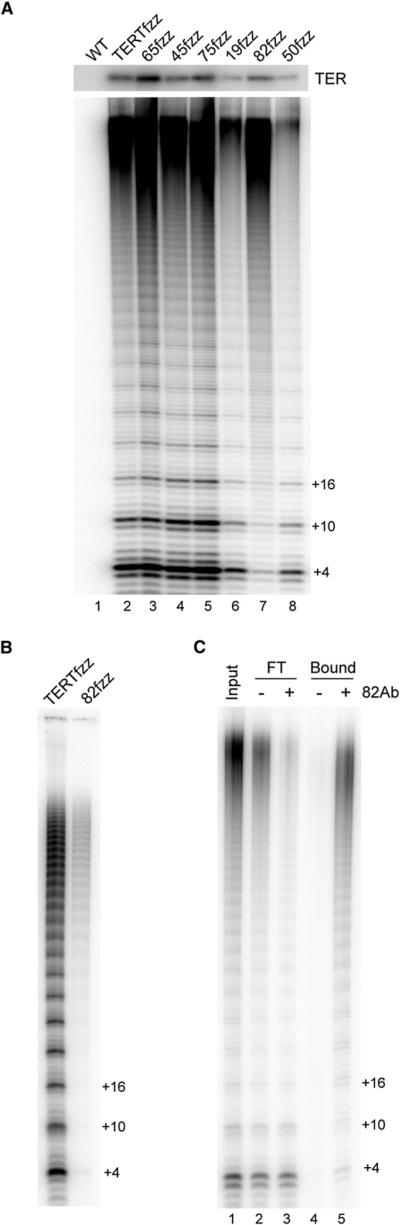
Tagged TERT, TER, p75, p65, and p45 copurify telomerase catalytic activity from cell extracts (Witkin and Collins, 2004; Cunningham and Collins, 2005; Witkin et al., 2007). Using the panel of FZZ strains created above, we compared the ability of known and additional telomerase proteins to associate with telomerase catalytic activity. Complexes recovered from extract by single-step binding to FLAG antibody resin were divided for parallel analysis of TER (Figure 5A, top) and telomerase activity (Figure 5A, main panel). Activity was assayed by direct primer extension of the telomeric repeat oligonucleotide (GT2G3)3 in reactions with dTTP and radiolabeled dGTP. All of the tagged proteins copurified telomerase activity, with amounts of activity proportional to the relative enrichment of TER (Figure 5A).
Figure 5. Tagged Holoenzyme Proteins Copurify TER and Telomerase Activity.
(A) TER and telomerase activity enriched by fzz-tagged proteins using single-step purification on FLAG antibody resin.
(B) High-RAP telomerase activity enriched by 82fzz. TERTfzz and 82fzz cell extracts were used for two-step affinity purification followed by activity assay. The same samples were analyzed by SDS-PAGE in Figure 3A, lanes 7–9 (the mock purification had no activity).
(C) Immunodepletion of p82. Unbound flowthrough (FT) and bound fractions from immunodepletion with preimmune or p82 antibody serum were assayed for telomerase activity.
Telomerase holoenzyme purified by TERTfzz, 65fzz, 45fzz, 75fzz, 19fzz, or 50fzz catalyzed the synthesis of a similar product profile, with an accumulation of both short and long products (Figure 5A). In distinction, telomerase holoenzyme purified by 82fzz catalyzed predominantly long-product synthesis (Figure 5A, lane 7). This selective enrichment of high-RAP activity was also observed with two-step affinity purification of 82fzz (Figure 5B). To confirm this TERTfzz versus 82fzz enzyme population difference, we performed a reaction time course (Figure S4). The TERTfzz-enriched holoenzyme population had high-RAP activity, generating products that gained length over time, and also had low-RAP activity, producing short products that increased in abundance, but not length, due to enzyme turnover. The 82fzz-enriched holoenzyme had high-RAP activity but much less low-RAP product synthesis; a small amount of low-RAP activity could derive from p82 dissociation from holoenzyme in the activity assay reaction.
To confirm that untagged p82 is also preferentially associated with high-RAP activity, we used TERTfzz-purified holoenzyme for an immunoenrichment assay with preimmune or p82 antiserum (Figure 5C). Incubation with p82 antibody, but not preimmune serum, depleted high-RAP activity, whereas low-RAP activity remained in the unbound fraction (Figure 5C, lanes 1–3). The p82 antibody selectively retained high-RAP activity (Figure 5C, lanes 4 and 5), similar to the enrichment observed by affinity purification of 82fzz (Figures 5A and 5B). These experiments reveal that the telomerase holoenzyme population assayed in cell extract includes at least two catalytically distinct forms, differentiated in subunit composition by the presence or absence of p82 (Figure 4C). This finding provides a rationale for the variability in RAP observed across T. thermophila extracts or fractionations: conditions that allow proteolysis diminish the recovery of high-RAP activity (Figure S5A).
Recombinant p82 Stimulates RAP and Binds Telomeric ssDNA
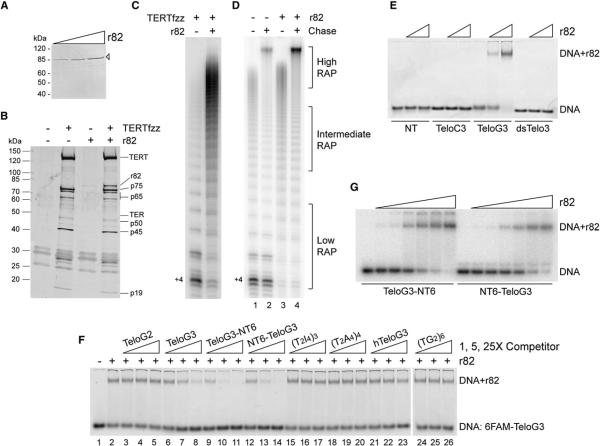
Among the seven tagged holoenzyme proteins that individually copurified active telomerase, only 82fzz did not enrich low-RAP activity (Figure 5A). This observation and the immunodepletion of high-RAP, but not low-RAP, activity with p82 antibody (Figure 5C) suggested that p82 could directly affect holoenzyme RAP. We therefore tested whether a TERTfzz-purified holoenzyme population with predominant low-RAP activity could be rescued to high RAP by addition of p82 alone. We added bacterially expressed and purified r82 (Figure 6A) or buffer control to two-step purified TERTfzz holoenzyme or a mock affinity purification immobilized on FLAG antibody resin, and then we washed and eluted with FLAG peptide. Holoenzyme incorporation of r82 (Figure 6B) induced a loss of low-RAP activity and a gain of high-RAP activity (Figure 6C). Similar results were observed if r82 was added directly to the activity assay itself (Figure S5B). On the other hand, r82 addition to the T. thermophila telomerase catalytic core reconstituted in RRL did not stimulate RAP (data not shown), consistent with holoenzyme association of p82 via the p75-p45-p19 subcomplex (Figure 4B).
Figure 6. Recombinant p82 Confers Processivity to Telomerase Holoenzyme and Binds Telomeric ssDNA.
(A) Purification of r82. N terminally 6 × His-tagged r82 was stained with Coomassie blue R-250. An open arrowhead marks the recombinant protein.
(B) Assembly of r82 with purified holoenzyme. Extra staining in the 25–32 kDa region derives from blocking proteins in the wash buffer.
(C) Activity assay of holoenzyme with or without added r82.
(D) Chase assay to establish RAP. Telomerase was preincubated with r82 (25 nM) and primer (50 nM) before the addition of nucleotides. After 10 min, samples without chase were stopped, while samples with chase were supplemented with 100-fold excess unlabeled dGTP followed by another 10 min extension.
(E) DNA-binding activity of r82. Fluorescein-labeled substrates NT (6FAM-(A2T)6), TeloC3 (6FAM-(C4A2)3), TeloG3 (6FAM-(T2G4)3), and dsTelo3 (TeloC3 annealed with unlabeled TeloG3) were incubated at 10 nM final concentration with 0, 5, or 100 nM r82.
(F) Competition assay for DNA-binding specificity. The 6FAM-TeloG3 probe (10 nM) and r82 (5 nM) were bound with or without competitor oligonucleotides (1-, 5-, 25-fold excess over probe).
(G) Binding affinity of r82. Radiolabeled TeloG3-NT6 or NT6-TeloG3 (1 pM) was incubated with r82 (4-fold concentrations steps from 2 to 2000 pM).
The high-RAP products of T. thermophila telomerase holoenzyme continue to elongate over an extended reaction time course (Greider, 1991; Cunningham and Collins, 2005). Using an initial pulse of radiolabeled dGTP and a chase of unlabeled dGTP, the longest products do not reach a steady-state distribution, but products with short and intermediate lengths can be resolved that are resistant to chase and thus can be used as elongation endpoints for a RAP calculation (Figure 6D). Quantification of the chased low-RAP products (Figure 6D, lane 2; bracketed low-RAP products) normalized for dGTP incorporation revealed that the efficiency of next repeat synthesis approaches only 30%, similar to the maximal efficiency obtained by T. thermophila TERT+TER reconstituted in RRL (Hardy et al., 2001). Addition of r82 eliminated the synthesis of low-RAP products, shifting the holoenzyme population to intermediate- and high-RAP product synthesis (Figure 6D, compares lanes 1 and 2 with lanes 3 and 4). Minimal estimates of RAP, based on the chased intermediate length products (Figure 6D, lanes 2 and 4; bracketed intermediate-RAP products), indicate a 50%–80% efficiency of next repeat synthesis, much greater than that supported by the TERT+TER RNP alone.
One obvious mechanism by which p82 could act as a processivity factor would be to contact ssDNA, thereby stabilizing holoenzyme association with its product. Sequence analysis of p82 predicted the presence of tandem OB folds similar to the three OB-fold DNA-binding domains of RPA1 (Figure S1). RPA1 provides the primary contact surface for DNA within the RPA heterotrimer, which recognizes an extended 20–30 nt length of ssDNA largely independent of sequence (Iftode et al., 1999; Bochkarev and Bochkareva, 2004). We therefore assayed the DNA-binding activity of r82 by electrophoretic mobility shift assay using 5′ fluorescein labeled (6FAM) oligonucleotides with either nontelomeric sequence, three G-rich telomeric repeats, three C-rich telomeric repeats, or the G-rich and C-rich repeats annealed to form a duplex (Table S1). Remarkably, despite its similarity to RPA1, r82 shifted only the G-rich telomeric repeat ssDNA (Figure 6E). The nontelomeric and C-rich telomeric repeat ssDNAs and the telomeric repeat duplex were not recognized by p82. Thus, although p82 shows biochemical similarity to RPA1 in its ability to bind ssDNA, it differs from RPA1 in the requirement for a specific ssDNA sequence. This distinction in DNA interaction specificity is consistent with the sequence divergence of p82 from both T. thermophila and human RPA1 (Figure S1C).
Binding of r82 to the 6FAM-labeled three-repeat G-rich telomeric strand was competed by addition of unlabeled three-repeat, but not two-repeat, G-rich telomeric DNA (Figure 6F, lanes 1–8). Four-repeat ssDNA forms folded structures under assay conditions, but extending the three-repeat sequence with six additional nontelomeric 3′ or 5′ nucleotides increased competition activity (Figure 6F, lanes 9–14), suggesting that r82 binds better to the longer ssDNAs. In addition, these results suggest that p82 does not specifically recognize a telomeric repeat at the ssDNA 3′ terminus, unlike the telomere end-binding protein complex from Oxytricha nova (Horvath et al., 1998). Competition was ineffective if the telomeric repeat guanosines were substituted by other purines or if human telomeric repeats or scrambled T. thermophila telomeric repeats were used (Figure 6F, lanes 15–26). Some of these modified sequences showed limited competition at much higher concentration (Figure S6). We used 5′-radiolabeled rather than 6FAM-labeled versions of the optimal competitor oligonucleotides to calculate r82 binding affinities for TeloG3-NT6 and NT6-TeloG3, which were 80 and 200 pM, respectively (Figure 6G).
Overexpression of p82 or p50 Alters Holoenzyme Composition and Telomere Maintenance
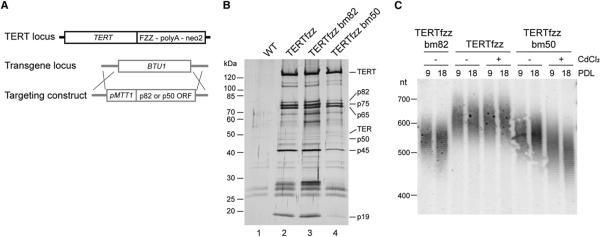
The substoichiometry of p50 and p82 in TERTfzz-purified telomerase holoenzyme suggested that cells could regulate telomerase function by limiting the expression of peripheral holoenzyme subunits. To investigate whether increasing the level of p50 or p82 expression would affect their association with holoenzyme and/or affect their telomerase function at telomeres, we introduced transgenes directing cadmium-inducible expression of untagged p50 or p82 into the TERTfzz strain background (Figure 7A). The MTT1 promoter used for cadmium-inducible protein overexpression to high cellular level (Shang et al., 2002) also has a basal level of transcription that represents modest mRNA overexpression for scarce telomerase subunits (Lee and Collins, 2007; Miao et al., 2009).
Figure 7. Overexpression of p82 or p50 Affects Holoenzyme Composition and Telomere Length.
(A) Strategy for construction of cell lines overexpressing p82 or p50 in a TERTfzz background.
(B) Holoenzyme purification from cells overexpressing p82 or p50. Two-step affinity purification of TERTfzz was performed from extracts of cells harvested 3 hr after addition of 1 μg/ml CdCl2.
(C) Telomere length in bm82 and bm50 cell lines. Cells were cultured for the indicated number of population doublings (PDL) at 30°C with or without 0.1 μg/ml CdCl2. Southern blots were performed using an rDNA subtelomeric probe against the G-rich strand.
Curiously, even vast overexpression of p50 with cadmium induction did not increase p50 association with TERTfzz-purified holoenzyme (Figure 7B). Instead, although TERTfzz still copurified p65, association of the RNP catalytic core with p75, p45, p19, and p82 was reduced (Figure 7B, compare lanes 2 and 4). As might be expected from this change in holoenzyme composition, p50 overexpression also induced telomere shortening (Figure 7C). Cadmium-induced overexpression of p82 increased its copurification with TERTfzz holoenzyme and also enhanced the copurification of smaller polypeptides that may be p82 degradation products (Figure 7B, compare lanes 2 and 3). Vast p82 overexpression caused rapid cell-cycle arrest (data not shown), whereas modest overexpression of p82 without cadmium allowed culture growth but induced telomere shortening (Figure 7C). These experiments suggest that the expression level of the holoenzyme subunits p50 and p82 must be regulated to achieve normal telomere length homeostasis.
DISCUSSION
New Telomerase Proteins Add Subunits and Subcomplexes to Holoenzyme Architecture
The aim of this work was to identify and biologically characterize all subunits of a telomerase holoenzyme, enabled by the use of T. thermophila as a model system. By fusing an optimized affinity purification tag to the C terminus of TERT at its endogenous locus, we were able to coenrich known and unknown telomerase holoenzyme proteins. The first insights about telomerase holoenzyme architecture that emerge from characterization of an eight-factor set of components include several surprises. At a first level, it is striking that telomerase holoenzymes from a single-celled eukaryote harbor so many subunits, each of which is required for telomere maintenance. Also, in T. thermophila, all of the subunits appear to be telomerase specific in their associations, rather than shared among multiple classes of nuclear RNP. Furthermore, six T. thermophila telomerase holoenzyme subunits are approximately stoichiometric when purified from asynchronously dividing cells (TER, TERT, p65, p75, p45, p19), but at least one subunit appears biologically substoichiometric in association and function (p50). The p82 subunit is also under-represented in purified holoenzyme. The Saccharomyces cerevisiae telomerase proteins Est1 and Est3 may be substoichiometric with other holoenyzme components as well because they are depleted from the catalytic core in G1 phase of the cell cycle (Osterhage et al., 2006). Thus, even in organisms with constitutive telomere maintenance and relatively many telomeres per genome content of DNA, telomerase regulation gives rise to holoenzyme complexes with differential subunit composition.
Subunit interaction studies uncovered the presence of holoenzyme subcomplexes. Previous genetic and biochemical assays demonstrate codependent assembly of p65, TER, and TERT to form a stable catalytic core (Witkin and Collins, 2004; Prathapam et al., 2005; O'Connor and Collins, 2006; Stone et al., 2007). Studies of the expanded inventory of TERTfzz-copurified proteins revealed another holoenzyme subcomplex, the p75-p45-p19 telomere adaptor complex, built by protein interactions among subunits required for telomere maintenance but dispensable for formation of the RNP catalytic core. Two additional subunits, p82 and p50, join the holoenzyme in a manner that is peripheral to stability of the RNP catalytic core or telomere-adaptor subcomplex. The extremely low endogenous expression level of p50 and the refractory behavior of recombinant protein expressed in E. coli limited the scope of biochemical analysis. However, we note that the profile of telomerase product synthesis associated with 50fzz (Figure 5A) indicates that this subunit associates with both low-RAP and high-RAP activity. Thus, p50 can join holoenzyme complexes with or without functional p82.
Sequence-Specific ssDNA Binding by a Telomerase Holoenzyme Protein Processivity Factor
Sequence analysis of p82 suggests that its structural similarity with RPA1 encompasses the RPA1 DNA-binding domains. Indeed, although their specificity for sequence differs, p82 and RPA1 share a binding preference for a relatively extended, ~20 nt length of ssDNA (Iftode et al., 1999; Bochkarev and Bochkareva, 2004). The 3–4 telomeric repeat length of ssDNA required for recognition by p82 is much longer than the ssDNA length required for sequence-specific binding by telomere proteins, which are sensitized to recognition of a few critical residues within 1–2 repeats (Croy and Wuttke, 2006). Considering the different biological roles of p82 and a telomere protein, this fundamental difference in recognition specificity makes sense. Telomerase holoenzyme should engage its substrate during genome replication, when long tracts of G-rich ssDNA are exposed, but not during interphase, when only shorter tracts of G-rich repeats are accessible. Also, considering p82 function within a telomerase holoenyzme, recognition of a long surface of ssDNA by p82 could allow dynamic individual contacts while maintaining stable protein-DNA association overall.
The telomerase holoenzyme context of p82 function obliges a major distinction from RPA1 in protein interaction specificity: instead of the RPA heterotrimer, p82 associates with the telomere adaptor subcomplex of telomerase holoenzyme. How the specificity of p82 interaction with p75, p45, and/or p19 is achieved remains to be explored after the development of a reconstitution system for the telomere adaptor subcomplex. In S. cerevisiae, the telomeric repeat binding protein Cdc13 alternates between a partnership with telomerase holoenzyme, which stimulates telomere elongation, or a partnership with Stn1 and Ten1, which provides telomere end-protective function (Pennock et al., 2001; Li et al., 2009). An RPA-like architecture of complex formation has been proposed to underlie interactions in the Cdc13-Stn1-Ten1 complex (Gao et al., 2007). It is possible that p75, p45, and/or p19 provide an interface for RPA-like multi-merization with p82. It is also possible that p82 contacts telomere proteins, potentially in a transient fashion. Approaches other than affinity purification under native conditions will be necessary for exploring the full scope of p82 interactions in vivo.
Endogenous p82 differentiates the purified holoenzyme population with high-RAP activity from the population with low-RAP activity, and supplementation of a mixed holoenzyme population with recombinant p82 converted the low-RAP enzyme to higher RAP. Like p82, the telomeric repeat ssDNA-binding complex POT1-TPP1 can stimulate human telomerase RAP in vitro, but unlike p82, association between these telomere proteins and telomerase holoenzyme is transient and potentially indirect (Xin et al., 2007; Wang et al., 2007). The existence of a holoenzyme subunit beyond the RNP catalytic core with independent DNA-binding activity provides a mechanism to improve RAP but presents an interesting challenge for telomerase negotiation with its product: as each newly synthesized repeat is displaced from the active site, does new product associate with p82, or does p82 remain bound to the initial site of interaction? These alternatives have biological implications in the coordination of telomere maintenance and will be fascinating to explore as a novel mechanism of processivity. The extremely high RAP that p82-bound holoenzyme can accomplish in vitro over an extended time course is unlikely to occur in the context of a replicating telomere in vivo (Yu and Blackburn, 1991). Instead, high RAP may be the signature of telomerase holoenyzme ability to associate tightly with telomeric repeat ssDNA. The insights about telomerase holoenyzme protein architecture and DNA-binding specificity uncovered here will enable direct assays of how telomerase subunit interactions with the telomere are regulated and coordinated with other chromosome replication machineries.
EXPERIMENTAL PROCEDURES
Affinity Purification and Mass Spectrometry
Two-step purification was carried out by batch binding for 2–4 hr at 4°C with 200 μl rabbit IgG-agarose (Sigma) for each 10 ml of extract, representing 500 ml of initial cell culture. Subsequent steps were done at room temperature in T2EG50 (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10% glycerol, 50 mM NaCl, and 5 mM β-mercaptoethanol or 5 mM DTT) supplemented with 0.1% Igepal CA-630. Washed IgG agarose was incubated with 30 μg/ml TEV protease for 0.5–1 hr. Eluted samples were batch bound to 10–15 μl of EZView Red anti-FLAG M2 resin (Sigma) in low-retention tubes for 1–2 hr. Washed resin was incubated with 150 ng/μl of 3 × 3 FLAG peptide (Sigma) for 30–60 min. For mass spectrometry, samples were eluted from M2 resin with 100 mM glycine (pH 2.5), neutralized with 2 M Tris base, precipitated with trichloracetic acid, and then processed as described previously (Fu and Collins, 2007). Single-step purification was performed by batch binding extract to 20 μl FLAG antibody resin for 2–4 hr at 4°C.
Additional methods are provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ramadevi Prathapam for creating 65fzz and 45fzz expression strains and Lori Kohlstaedt and Daniela Mavrici at the QB3 Vincent J. Coates Proteomics/Mass Spectrometry Laboratory for mass spectrometry analyses. This work was supported by National Institutes for Health grants GM54198 (K.C.) and the University of California Berkeley Cancer Research Laboratory training grant (B.M.).
Footnotes
ACCESSION NUMBERS The GenBank accession numbers for T. thermophila TAP19, TAP50, TAP82, and RPA1 are EU873082, EU873083, EU873081, and GQ274003, respectively.
SUPPLEMENTAL DATA Supplemental Data include Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://www.cell.com/molecular-cell/supplemental/S1097-2765(09)00701-1.
REFERENCES
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Chan A, Boulé JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Forms and functions of telomerase RNA. In: Walter NG, Woodson SA, Batey RT, editors. Non-Protein Coding RNAs. Springer-Verlag; Berlin: 2009. pp. 285–301. [Google Scholar]
- Croy JE, Wuttke DS. Themes in ssDNA recognition by telomereend protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Collins K. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 2005;25:4442–4454. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington TM, Fu D, Wong JM, Collins K. Disease-associated human telomerase RNA variants show loss of function for telomere synthesis without dominant-negative interference. Mol. Cell. Biol. 2008;28:6510–6520. doi: 10.1128/MCB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Chartrand P. Telomerase biogenesis: The long road before getting to the end. RNA Biol. 2008;5:212–215. doi: 10.4161/rna.7115. [DOI] [PubMed] [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CD, Schultz CS, Collins K. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 2001;276:4863–4871. doi: 10.1074/jbc.M005158200. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 2001;20:4299–4308. doi: 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol. Cell. Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol. Cell. 2000;6:827–837. doi: 10.1016/s1097-2765(05)00078-x. [DOI] [PubMed] [Google Scholar]
- O'Connor CM, Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell. Biol. 2006;26:2029–2036. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O'Connor CM, Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2005;12:252–257. doi: 10.1038/nsmb900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E. La sets the tone for telomerase assembly. Nat. Struct. Mol. Biol. 2007;14:261–262. doi: 10.1038/nsmb0407-261. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Prathapam R, Collins K. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 2007;27:2074–2083. doi: 10.1128/MCB.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Yu GL, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.