Abstract
Telomerase maintains the simple sequence repeats at chromosome ends, protecting cells from genomic rearrangement, proliferative senescence and death. The telomerase reverse transcriptase (TERT) and telomerase RNA (TER) alone can assemble into active enzyme in a heterologous cell extract, but the physiological process of telomerase biogenesis is more complex. The endogenous accumulation of Tetrahymena thermophila TERT and TER requires an additional telomerase holoenzyme protein, p65. Here, we reconstitute this cellular pathway for telomerase ribonucleoprotein biogenesis in vitro. We demonstrate that tandem RNA interaction domains in p65 recognize the sequence of the TER 3′ stem. Notably, the p65–TER complex recruits TERT much more efficiently than does TER alone. Using bacterially expressed p65 and TERT polypeptides, we show that p65 enhances TERT-TER interaction by a mechanism involving a conserved bulge in the protein-bridging TER molecule. These findings reveal a pathway for telomerase holoenzyme biogenesis that preassembles TER for TERT recruitment.
Cellular ribonucleoprotein (RNP) biogenesis has gained attention as a dynamic, regulated cascade of events that can be affected in human disease1–3. RNPs assemble in vivo aided both by assembly chaperones, which contribute transient interactions, and by architectural RNA-binding proteins, which remain integral components of an assembled RNP4. Progress in the elucidation of physiological RNP biogenesis mechanisms has been impeded by the limited success in reconstitution of ordered protein and RNA assembly pathways in vitro.
TERT and TER can assemble in rabbit reticulocyte lysate (RRL) to form a minimal recombinant telomerase RNP5. The physiological process of telomerase enzyme biogenesis, however, seems more complex. In vivo, human TER accumulation and telomere length maintenance are compromised by disease-associated single-residue substitutions in the RNA-binding protein dyskerin6,7. In budding yeast, TER accumulation requires RNA-binding proteins from the Sm family8. The potentially pleiotropic impact of vertebrate dyskerin or yeast Sm protein alterations complicates the interpretation of their direct roles in telomerase RNP assembly. Additionally, studies of these RNP assembly pathways in vitro are hampered by the inability to reconstitute a dyskerin or Sm RNP efficiently in the absence of other, typically concerted cellular events. In the biological context, it was thus unresolved whether TERT assembles with TER unassisted or only transiently chaperoned, or whether TERT-TER interaction is modulated by other steps of the physiological telomerase holoenzyme assembly process.
Endogenous TER accumulation in the ciliate T. thermophila requires the telomerase holoenzyme protein p65 (ref. 9). Tetrahymena thermophila p65 and its probable ortholog Euplotes aediculatus p43 each copurify with active telomerase and reciprocally copurify the vast majority of telomerase RNP9–12. Genetic depletion of T. thermophila p65 reduces TER accumulation without impact on other small nuclear RNAs, suggesting that in contrast to vertebrate dyskerin or yeast Sm proteins, p65 has a telomerase-specific function in RNP biogenesis9. Based on these and additional findings, we proposed a p65-dependent pathway for the physiological biogenesis of T. thermophila telomerase RNP. Here, we test the proposed telomerase RNP assembly pathway by reconstituting the interactions of recombinant p65, TERT and TER in vitro. We show that p65 binds TER directly and with a novel specificity. We also find that p65 markedly improves TER association with TERT.
RESULTS
Specific interaction of recombinant p65 and TER
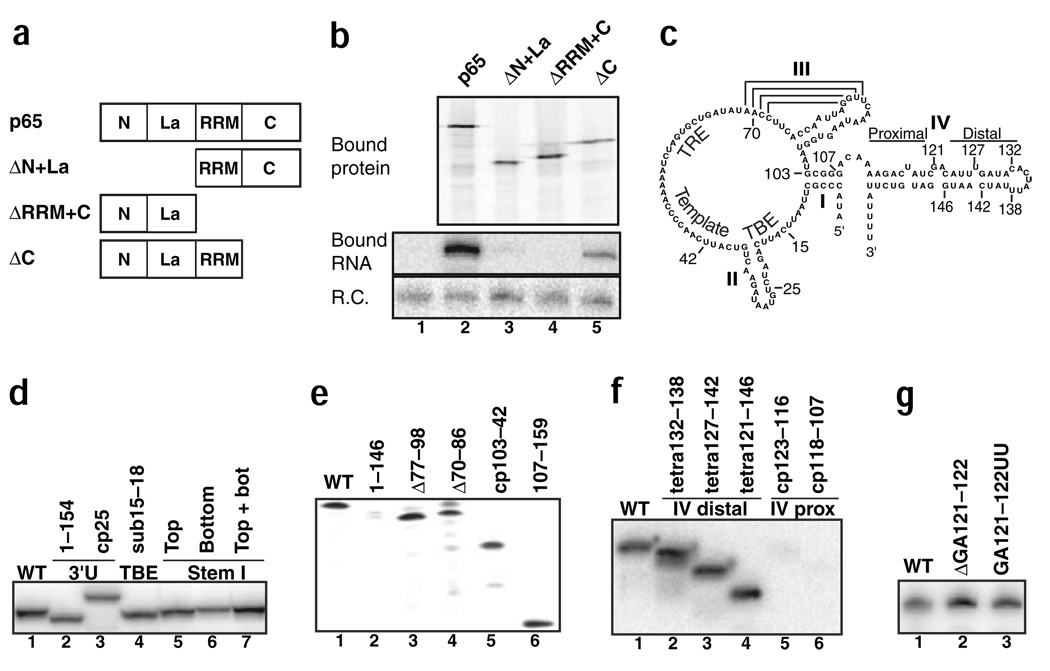
The overall domain structure of p65 (Fig. 1a) parallels that of eukaryotic La, an RNA-binding protein best characterized by its ability to recognize the 3′ poly(U) tract of RNA polymerase III transcripts13. The p65 La motif and putative RNA recognition motif (RRM) show relatively high divergence from consensus but still represent the most probable sites of TER interaction9. Because T. thermophila TERT expressed in RRL can assemble with TER to generate an accurately template-guided polymerase14, we used a similar strategy of expression and RNP assembly to investigate the interaction of T. thermophila p65 and TER. We expressed p65 with an N-terminal, tandem protein A tag (ZZ tag) to allow affinity purification from RRL with IgG agarose. Protein expression lysates were supplemented with TER after protein synthesis and then subjected to immunopurification at approximately physiological ionic strength in the presence of nonspecific protein and RNA competitors (see Methods). Aliquots of washed purification resin were analyzed by SDS-PAGE to detect radiolabeled protein or by northern blot hybridization to detect TER. Immunopurification from a mock expression reaction did not recover TER, whereas readily detectable TER was recovered in association with p65 (Fig. 1b, lanes 1 and 2).
Figure 1.
Specific p65-TER interaction. (a) Schematic of full-length p65 and domain truncations. (b) Equal amounts of RRL expression reactions containing no expressed protein (lane 1) or the tagged protein indicated (lanes 2–5) were supplemented with TER. Immunopurified samples were split for analysis by SDS-PAGE and phosphorimaging (top) or RNA extraction and blot hybridization for TER (bottom). R.C. is a truncated TER fragment added to immunopurified samples before extraction as a recovery control. (c) Schematic of T. thermophila TER with labeled position numbers and secondary structure elements (I–IV). Stem IV proximal and distal regions are separated by a GA bulge. The template is also labeled as are three regions of TER that make interactions with TERT: the TBE (the high-affinity TERT-binding site) and the TRE and NPE (lower-affinity contact sites; see text for description). (d–g) Equal aliquots of a single ZZ-p65 expression reaction were combined with the TER variants indicated and analyzed as in b. WT, wild type. Blot hybridization for TER is shown; SDS-PAGE analysis revealed an equal recovery of protein after immunopurification (data not shown). Each experiment also included controls to verify negligible TER background binding to resin and equal sample recovery from RNA extraction by addition of a truncated TER fragment as the recovery control (data not shown).
To investigate which regions of p65 are required for TER binding, we expressed a series of truncated p65 polypeptides, each with an N-terminal ZZ tag (Fig. 1a). Truncation from the p65 N terminus, including the La motif, or truncation from the C terminus, including the putative RRM, markedly reduced or eliminated TER interaction (Fig. 1b, lanes 3 and 4). A less severe reduction in TER binding was observed upon removal of only the p65 C-terminal extension, leaving the La motif and putative RRM intact (Fig. 1b, lane 5). These results suggest that p65 relies on cooperation of its La motif domain and adjacent RRM-like domain to establish high-affinity RNA interaction, in notable parallel to La13.
We next assessed the sequence specificity of p65-TER interaction. The 159-nucleotide T. thermophila TER (Fig. 1c) accumulates as an unprocessed, primary transcript of RNA polymerase III predicted to have substantial secondary structure15,16. We compared the ability of p65 to copurify wild-type and variant TERs using equal aliquots of a single protein expression reaction supplemented with a fixed concentration of each in vitro–transcribed, purified TER. We first tested p65 interaction with TER variants predicted to show decreased binding to a eukaryotic La (note that T. thermophila La has not been characterized). A TER variant with a five-nucleotide 3′ truncation and a TER circular permutation (cp) variant with joined wild-type 5′ and 3′ ends could be recovered in association with p65 (Fig. 1d, lanes 1–3) despite the absence of the 3′ poly(U) or of the immediately following 3′ OH typically recognized by La13. We also tested a TER variant bearing a four-nucleotide sequence substitution of the high-affinity TERT-binding site17,18 (the TERT-binding/template boundary element or TBE; Fig. 1c). This TER variant also copurified with p65 (Fig. 1d, lane 4), indicating that p65 and TERT have distinct RNA-binding specificity.
Recombinant E. aediculatus p43 has recently been suggested to interact with stem I of its cognate TER12. Substitutions of T. thermophila TER stem I top strand, bottom strand or both strands to their sequence complement did not prevent p65 binding (Fig. 1d, lanes 5–7). Internal deletions of either pseudoknot stem-loop or circular permutation to collectively remove the entire pseudoknot, an adjacent TERT-interaction site at the template recognition element19,20 (TRE; Fig. 1c) and the template, did not reduce p65 interaction (Fig. 1e, lanes 3–5). In contrast, a truncation removing only 13 nucleotides from the 3′ end severely reduced TER copurification with p65 (Fig. 1e, lane 2). Ultimately, we found that a TER fragment containing only stem IV and the terminal poly(U) tract could be recovered in association with p65 (positions 107–159, Fig. 1e, lane 6).
Residues within the stem IV loop (the nucleotide addition processivity element or NPE; Fig. 1c) can be crosslinked to TERT20. Internal deletions that replaced this distal stem IV region with a tetraloop (Fig. 1f, lanes 1–4) or that altered the conserved central stem IV GA bulge (Fig. 1g) did not prevent p65 binding. In contrast, internal deletions in the region of stem IV adjacent to stem I (proximal stem IV; Fig. 1c) strongly compromised p65 interaction. Variant cp TERs lacking segments of the top strand of proximal stem IV (Fig. 1f, lanes 5 and 6), like the TER variant lacking the bottom strand of proximal stem IV (Fig. 1e, lane 2), were not recovered in stable association with p65. These findings demonstrate that p65-TER interaction requires the base of the 3′ terminal stem, a region not involved in any known TERT-TER interaction.
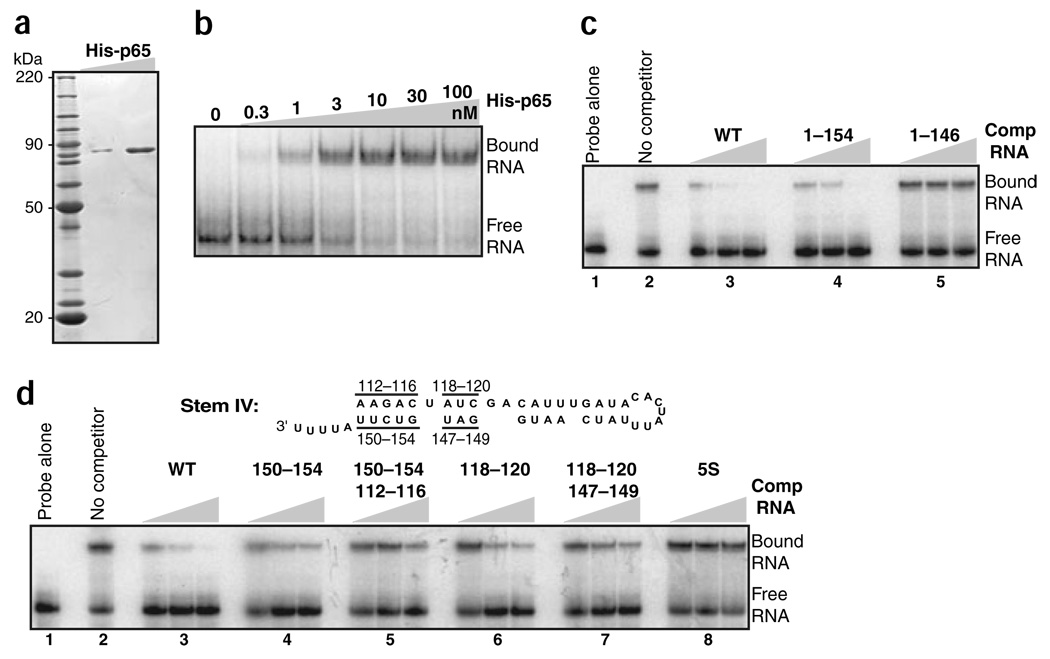
The in vitro assembly of T. thermophila or vertebrate TERT with TER is strongly stimulated by the presence of a eukaryotic cell lysate21,22. In contrast, we found that the assembly of p65 with TER was equally efficient before or after p65 immunopurification from RRL (data not shown). We bacterially expressed and purified recombinant p65 bearing an N-terminal His6-tag (Fig. 2a) in readily soluble, apparently monomeric form (data not shown). By electrophoretic gel mobility shift assay (EMSA) using a substoichiometric concentration of radiolabeled TER, we determined that p65 bound TER with an affinity of ~2–5 nM under a range of conditions including physiological ionic strength (Fig. 2b; additional data not shown).
Figure 2.
Direct interaction of p65 and TER. (a) SDS-PAGE and Coomassie blue staining of purified recombinant p65. Migration of molecular mass markers is indicated. (b) EMSA analysis of radiolabeled TER (~0.1 nM) in the presence of the indicated concentrations of p65 (0.3–100 nM). Similar results were observed using dilutions of the radiolabeled TER (data not shown). (c) EMSA competition for p65 (2 nM) binding to radiolabeled wild-type (WT) TER using the indicated TER variants (3, 10 or 30 nM). (d) EMSA competition for p65 (4 nM) binding to radiolabeled wild-type TER using E. coli 5S RNA (lane set 8) and the indicated TER variants (all at 3, 10 or 30 nM). Each TER variant is described by the positions altered to the wild-type sequence complement. Note that TER variants in lanes 4 and 6 unpair predicted duplex whereas TER variants in lanes 5 and 7 have compensatory changes in the second strand to restore base-pairing potential.
The specificity of p65-TER interaction established above using coimmunoprecipitation was verified by mobility shift competition. A titration of wild-type, nonradiolabeled TER in approximately three-fold steps could compete the p65 mobility shift of radiolabeled TER as expected (Fig. 2c, lane sets 1–3), whereas the same titration of a TER variant lacking the 13 nucleotides at the 3′ end could not (lane set 5). Notably, a subtle but reproducible, approximately three-fold loss of competition was evident for a TER variant lacking just the poly(U) tract (Fig. 2c, lane set 4). Substitution of the entire stem I top strand, bottom strand or both strands to restore base-pairing potential also resulted in approximately three-fold reduction in mobility shift competition relative to wild-type TER (Supplementary Fig. 1 online). Notably, substitutions that altered proximal stem IV sequence decreased the ability of TER variants to compete for p65 binding by at least ten-fold (Fig. 2d, lane sets 1–7). Defects resulting from changes in one strand were not rescued by compensatory substitutions in the alternate strand to restore base-pairing potential. As an additional control for the specificity of p65-TER interaction, we determined that Escherichia coli 5S RNA was incapable of p65 mobility shift competition despite its similar size to TER and substantial duplex structure (Fig. 2d, lane set 8). These results demonstrate that p65 recognizes the sequence of proximal stem IV.
The experiments described above suggest that proximal stem IV is the most critical of the regions disrupted independently for p65 binding. Other regions of TER, including the 3′ poly(U) tract, stem I and the central stem IV GA bulge (see below), when altered individually, impose a limited, approximately three-fold decrease in p65 interaction. Together, however, these regions beyond proximal stem IV are likely to contribute substantially to p65-TER interaction.
Assembly of a p65–TER–TERT complex
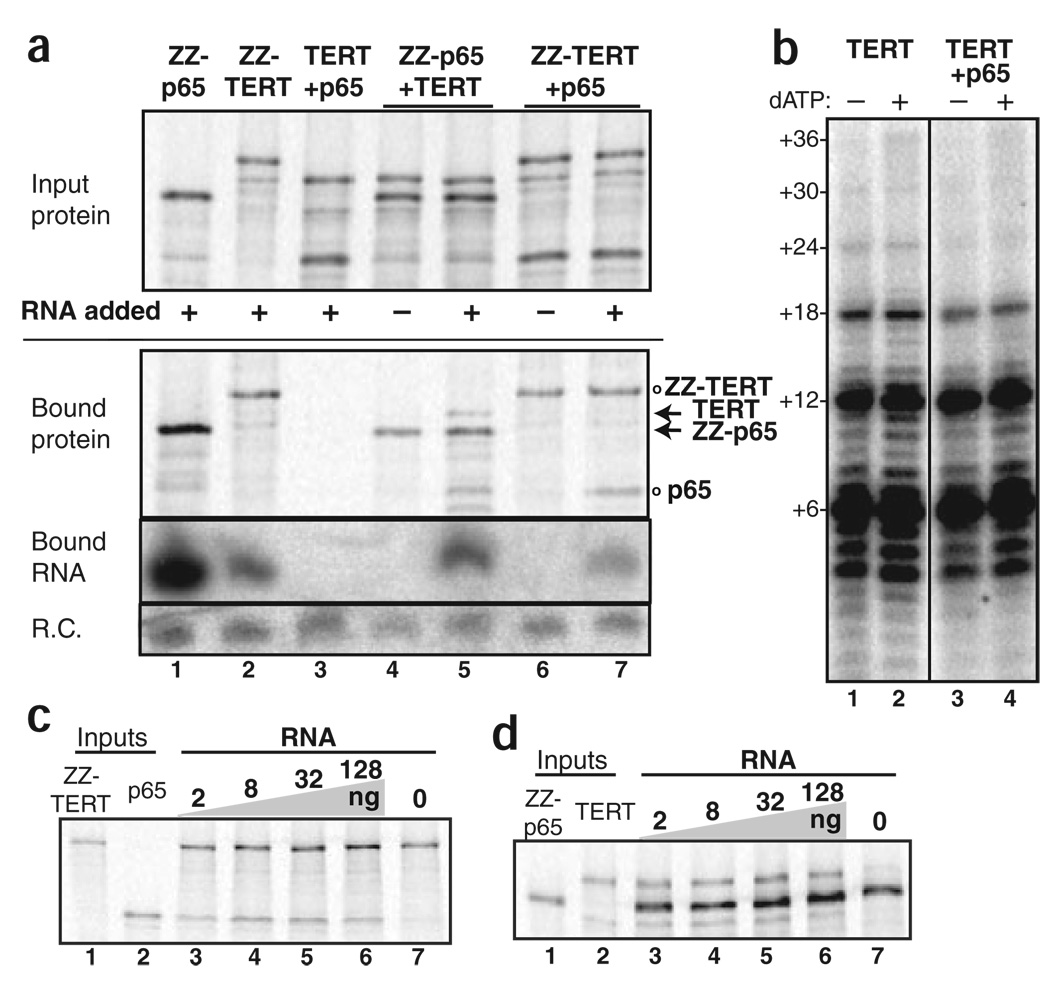
In vivo, p65 depletion severely reduces the accumulation of both TER and TERT9. To investigate whether p65 interacts directly with TERT, we used RRL to express epitope-tagged and untagged versions of both proteins. N-terminally tagged TERT (ZZ-TERT) or p65 (ZZ-p65) copurified TER whereas TER was not recovered in a mock immunopurification of the untagged proteins (Fig. 3a, lanes 1–3). From mixtures of one tagged and one untagged protein without TER, only the tagged protein was recovered above background (Fig. 3a, lanes 4 and 6). From mixtures of one tagged and one untagged protein supplemented with TER, however, ZZ-p65 copurified untagged TERT and ZZ-TERT copurified untagged p65 (Fig. 3a, lanes 5 and 7). These findings demonstrate that in the presence of TER, p65 and TERT associate to form a ternary complex.
Figure 3.
Formation of ternary complex with p65, TER and TERT. (a) Input proteins were combined, supplemented with TER, and then immunopurified to recover complexes that included the tagged protein (ZZ-TERT or ZZ-p65). Immunopurified samples were split for analysis by SDS-PAGE and phosphorimaging (top) or RNA extraction and blot hybridization for TER (bottom). Input SDS-PAGE samples correspond to a 12% loading of the bound SDS-PAGE samples. R.C. is a truncated TER fragment added to immunopurified samples before extraction as a recovery control. (b) Equal aliquots of a TERT RRL expression reaction were combined with a fixed volume of either mock or p65 RRL expression reaction and then TER. Both TER and p65 were in excess of TERT. Telomerase primer-extension products were analyzed by denaturing gel electrophoresis. Products representing sequential rounds of synthesis to the template 5′ end are indicated. (c,d) Input proteins were combined, supplemented with TER, and then immunopurified to recover complexes that included the tagged protein (ZZ-TERT or ZZ-p65). Immunopurified samples were analyzed by SDS-PAGE and phosphorimaging.
Adjusting for the relative content of radiolabeled methionine in the tagged and untagged p65 and TERT polypeptides, the copurification of untagged protein could match but not exceed a 1:1 molar ratio with the tagged protein. We did not detect substantial multimerization of TERT or p65 even when assayed in the physiological combination of both proteins and TER (Supplementary Fig. 2 online). This is consistent with the monomeric state of T. thermophila TERT9 and TER (K.C. and D. Cunningham, University of California Berkeley, unpublished data) in the endogenously assembled holoenzyme and in RRL-reconstituted complexes of TERT and TER alone23. Coassembly of p65 did not affect the primer-extension activity of T. thermophila TERT reconstituted with TER in RRL (Fig. 3b). A similar profile of products was observed in the absence (Fig. 3b, lanes 1 and 2) or presence (lanes 3 and 4) of p65 in reactions with template-cognate TTP and radiolabeled dGTP alone (lanes 1 and 3) or reactions with added dATP to allow loss of fidelity at the template 5′ boundary (lanes 2 and 4).
We next examined TERT preference for association with the p65–TER complex relative to TER alone. We titrated the amount of TER added to mixtures of p65 and TERT from approximately equimolar to ~50-fold excess (Fig. 3c,d). If TERT prefers the p65–TER complex to TER alone, excess TER will not reduce formation of ternary complex. In the absence of TER, as expected, ZZ-TERT did not associate with p65 and ZZ-p65 did not associate with TERT (Fig. 3c,d, lane 7). In the presence of approximately stoichiometric TER (Fig. 3c,d, lane 3) or up to ~50-fold excess TER (lanes 4–6), each tagged protein copurified the untagged protein. These results reveal that the p65–TER complex recruits TERT in strong preference to TERT association with TER alone. After these experiments were completed, a ten-fold excess of T. thermophila TER was shown to reduce the association of E. aediculatus p43 with T. thermophila TERT24. Tetrahymena thermophila p65 and E. aediculatus p43 have limited sequence conservation beyond their shared homology with La proteins, differing even in the predicted consensus residues of the p65 RRM9; their cognate TER stem IV regions share no primary sequence conservation and have major differences in stem lengths, bulges and potential branches25. Because T. thermophila and E. aediculatus are highly divergent ciliates, heterologous combinations of telomerase subunits would not reconstitute a physiological ternary complex.
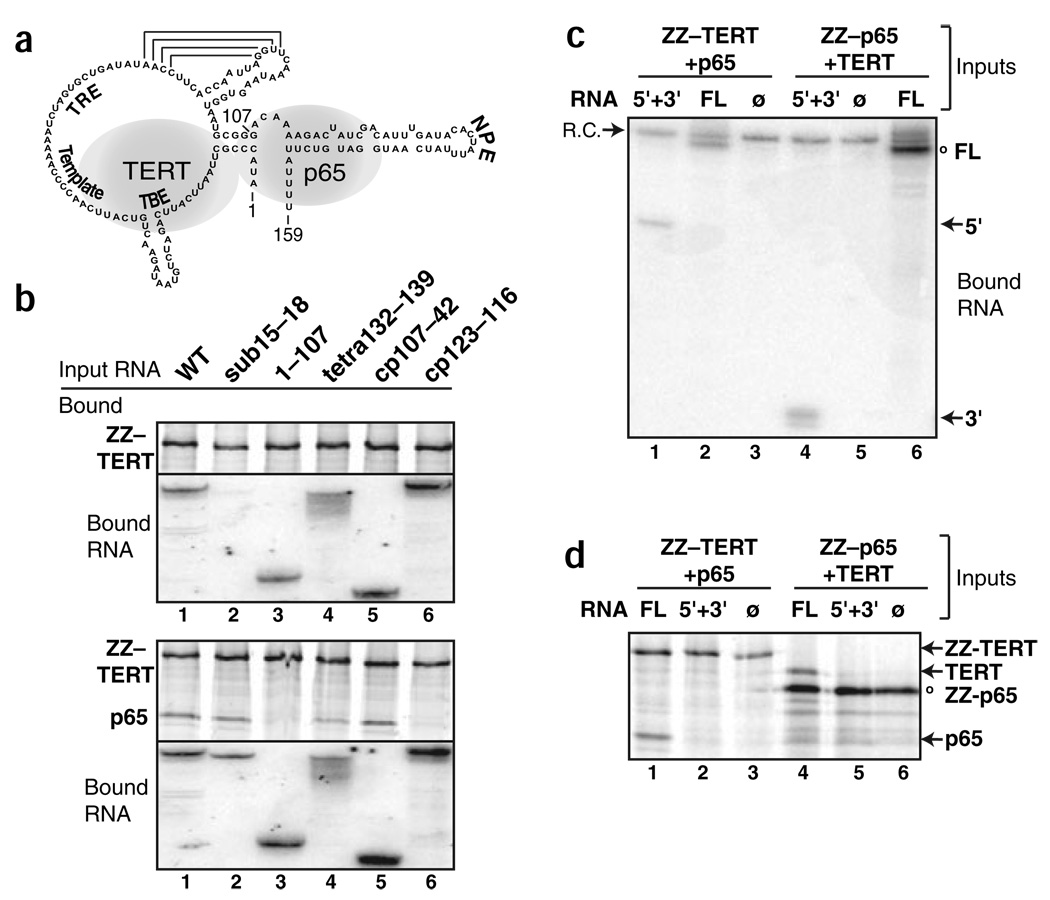
To test more directly whether TERT has greater affinity for TER bound to p65 than TER alone, we compared ZZ-TERT copurification of TER variants in the presence or absence of p65. We used a panel of TER variants altered at sites of TERT-TER and p65-TER interaction (described above and summarized in Fig. 4a). A four-nucleotide substitution in the TER TBE markedly reduced TERT interaction (Fig. 4b, lane 2, top), but in the presence of p65 even this TER variant was recovered with TERT (lane 2, bottom). TER variants lacking lower-affinity TERT contact sites in the NPE of distal stem loop IV (Fig. 4b, lane 4) or in the TRE-template region (lane 5) could also form ternary complexes. TER variants compromised for p65 binding by deletion of the entire stem IV region (Fig. 4b, lane 3) or some of the top strand of stem IV (lane 6) could bind ZZ-TERT but not form the ternary complex.
Figure 4.
Protein association through a TER bridge. (a) Schematic of TER with the regions most critical for each protein interaction shaded. Numbering indicates TER fragment boundaries. (b–d) Equal aliquots of mixed protein expression reactions were combined with the indicated TER variants and then immunopurified to recover complexes that included the tagged protein. Immunopurified samples were split for analysis by SDS-PAGE and phosphorimaging (b,d) or RNA extraction and blot hybridization for TER (b,c). Note that the relative levels of TER and TER fragments cannot be compared in c owing to the mixture of complementary oligonucleotide probes required for detection of nonoverlapping TER fragments. R.C. is a truncated TER fragment added to immunopurified samples before extraction as a recovery control. All experiments included controls to verify negligible TER background binding to resin (data not shown). WT, wild type; FL, full-length TER; 5′+3′, mixed 5′ and 3′ TER fragments; ø, no TER.
We also tested whether a ternary complex could form if the full-length sequence of TER were partitioned into 5′ and 3′ fragments, physically separating the high-affinity interaction sites for TERT and p65 (positions 1–107 and 107–159, respectively). We assayed protein association in the presence of full-length TER, mixed 5′ and 3′ TER fragments, or no TER using ZZ-TERT and p65 or ZZ-p65 and TERT. Tagged TERT copurified full-length TER or the 5′ TER fragment, whereas tagged p65 copurified full-length TER or 3′ TER fragment (Fig. 4c). Formation of a ternary complex was detected only in the presence of full-length TER (Fig. 4d). These experiments demonstrate that p65 and TERT copurify together only if there is a protein-bridging molecule of TER.
Mechanism of p65-enhanced TERT RNP assembly
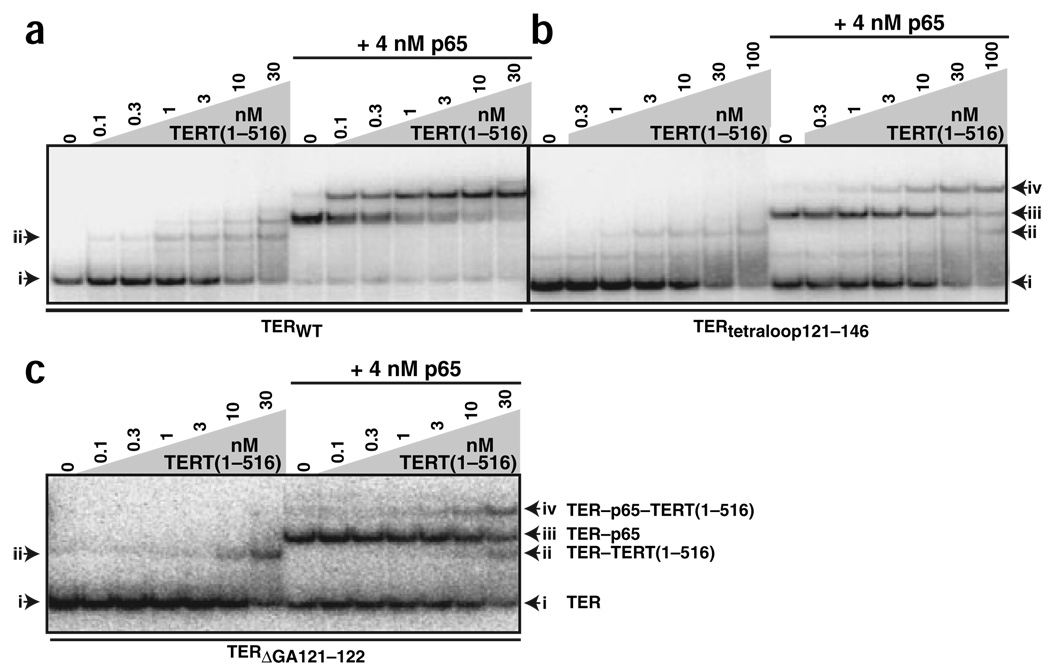
RRL provides factors that are critical for the in vitro assembly of full-length T. thermophila TERT and TER even after protein expression is complete21. In contrast, N-terminal domains of T. thermophila TERT harboring the sites of TER interaction can associate directly with TER after protein purification (C.M.O. and K.C., unpublished data). We bacterially expressed and purified a polypeptide including the N-terminal 516 residues of T. thermophila TERT (termed TERT (1–516)) containing all of the sites of TERT-TER interaction characterized for full-length TERT in RRL (C.M.O. and K.C., unpublished data). TERT(1–516) bound to radiolabeled wild-type TER with ~10–20 nM affinity (Fig. 5a, left). When high concentrations of TERT(1–516) were present, a single TER molecule could bind multiple protein molecules owing to the multiple sites of TER interaction in each TERT(1–516) polypeptide. We compared TERT(1–516)-TER interaction to TERT(1–516) interaction with the p65–TER complex. Notably, TERT(1–516) bound to the p65–TER complex with ~30-fold greater affinity and more homogeneity than it bound to TER alone (Fig. 5a). This unprecedented, entirely recombinant telomerase RNP reconstitution assay confirms and extends the conclusion that p65 enhances TERT assembly into RNP.
Figure 5.
Stimulation of TERT RNP assembly by p65. (a–c) Radiolabeled wild-type TER (a) and the TER variants indicated (b,c) were mixed with buffer (left) or a final concentration of 4 nM p65 (right) before addition of the indicated concentrations of TERT(1–516) and EMSA.
We exploited this assay to determine features of TER required for the p65 stimulation of TERT-TER interaction. When TER distal stem IV was replaced with a tetraloop (TERtetraloop121–146), the ~30-fold enhancement of TERT RNP assembly by p65 was lost (Fig. 5b). This TER variant had about a two- to three-fold reduced affinity for p65 or TERT(1–516) alone, which does not affect the direct comparison between the mobility shift of free TER (left) and p65-bound TER (right) by TERT(1–516). Notably, a much smaller deletion of only the central stem IV dinucleotide bulge (TERΔGA121–122) also strongly compromised p65 enhancement of TERT RNP assembly (Fig. 5c). These assays reveal that p65 promotes TERT RNP assembly by a mechanism requiring the TER central stem IV dinucleotide bulge. The central stem IV bulge and its immediately flanking duplex region are universally conserved in sequence among Tetrahymena species’ TERs26, supporting the biological importance of p65 function in the assembly of active telomerase RNP.
DISCUSSION
Tetrahymena thermophila p65 was identified as part of a telomerase holoenzyme complex containing TER and five proteins9. The domain structure of p65 largely parallels that of eukaryotic La and some La motif–containing proteins13. Similarly to La, p65 relies on a cooperative effort of the La motif and an adjacent RNA-binding domain to accomplish high-affinity RNA interaction. Also similarly to La, the p65 La motif could contact the TER poly(U) 3′ end. Distinctly from La, however, full-length p65 recognizes a region of TER predicted to contain base-paired duplex26–29. The specificity of p65-TER interaction also differs from the specificity of vertebrate dyskerin or yeast Sm protein interaction with the cognate TER. Despite this difference in sequence-specificity of TER binding, the known proteins required for cellular production of a stable telomerase RNP in vertebrates, budding yeast and T. thermophila share the property of binding near the TER 3′ end. These proteins may additionally share common functions in blocking exonuclease access and favoring TER assembly with TERT. Distinguishing the relative importance of these two roles for p65 will require the discovery of p65 or TER variants that retain wild-type p65-TER and TERT-TER interaction yet eliminate p65 enhancement of TERT RNP assembly.
Chaperone activities in a heterologous cell lysate such as RRL can promote the assembly of an active T. thermophila telomerase enzyme from TERT and TER alone, overcoming the essential requirement for p65 observed in vivo. RRL reconstitution conditions for T. thermophila TERT and TER have been optimized to maximally assemble TERT into RNP; we are not surprised that under these conditions RNP reconstitution efficiency and total catalytic activity are not increased by p65. The percentage of RRL-expressed TERT that does not reconstitute RNP seems grossly misfolded because it is incapable even of binding to affinity purification resin21. The findings above imply that RRL reconstitution differs in mechanism from the physiological RNP assembly process: chaperones and general RNP assembly factors would lack the ability of p65 to form sequence-specific interactions with TER. Active telomerase enzymes of many species have been refractory to reconstitution by expression of TERT and TER in a heterologous system. In these cases, general chaperone activities may not effectively substitute for holoenzyme protein roles in telomerase RNP assembly.
Numerous proteins assemble with TER and TERT to form a stable, endogenous cellular telomerase holoenzyme9,30. The rationale for evolution of multisubunit telomerase holoenzymes has not been clear. We suggest that this cellular commitment developed in concert with an expansion of TER structural and functional complexity. TER motifs establish specific features of the telomerase catalytic cycle and can regulate telomerase interaction with chromosome ends31,32. These nontemplate functions of TER distinguish telomerase from a viral protein reverse transcriptase. TER functional specialization could have been facilitated by the appropriation of RNA-binding proteins that served to stabilize favorable TER conformations. Telomerase may thus have gained a stepwise, hierarchical biogenesis strategy in its evolution from functional protein to cofunctional RNP, a strategy previously implicated in the evolution of ancestral RNAs to modern-day RNP machines such as the ribosome33–35.
METHODS
Protein expression and purification
Synthetic genes were used to express T. thermophila TERT14 and p65 (from the first in-frame TAP65 AUG codon9). We note that for unclear reasons, under some SDS-PAGE conditions only, recombinant p65 migrates slower than expected. RRL expression vectors were constructed using pCITE4a. Constructs for expression of untagged proteins were designed such that an NdeI site encodes the initiating methionine and a BamHI site follows the stop codon. Constructs for the expression of ZZ-tagged proteins were created using a DNA fragment encoding a His6 leader, two protein A domains and a cleavage site for TEV protease inserted at the NdeI site. Escherichia coli expression of p65 was directed by pET15 using the His6-tag of the vector.
Protein expression in RRL was done by standard methods as described20. Protein yields from RRL expression reactions were estimated using the known amount of untagged TERT produced per microliter of RRL expression reaction (~2 ng) by comparison of relative protein radiolabeling normalized for methionine content. TERT expression level was established by immunoblotting of RRL reactions versus purified recombinant protein standards under conditions in which radiolabel and immunoblot signals were distinguishable. Bacterial expression of p65 was carried out using BL21(DE3). Cells were grown at 37 °C and then induced with 1 mM IPTG at 16 °C. Extract of cells lysed in T2MG (20 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10% (v/v) glycerol) was supplemented with 0.3 M NaCl and 20 mM imidazole, cleared by centrifugation, then bound to Ni-NTA agarose in batch. Elution was accomplished with 0.5 M imidazole in the same buffer. Bacterial expression of TERT(1–516) was carried out as described (C.M.O. and K.C., unpublished data).
RNA expression and variants
TERs were synthesized in vitro with T7 RNA polymerase using PCR-generated or plasmid templates and purified as described20. TERs were additionally analyzed by SYBR Gold (Molecular Probes) staining after denaturing gel electrophoresis to verify RNA integrity and relative concentrations. Unless indicated otherwise, sequence substitutions were to the complement of wild-type sequence. Stem I substitutions were of the entire top strand (positions 103–107) or bottom strand (positions 4–8) or both; substitution of both strands restores base-pairing potential. Tetraloop substitutions of distal stem IV replaced the indicated wild-type TER positions with the tetraloop UUCG. Circular permutations added a ten-nucleotide linker to bridge an extended U tract with the wild-type TER 5′ end as described19. Internal deletion cpTERs are designated using both new ends (for example, cp123–116 begins at position 123 and ends at position 116). The control cp25 TER disrupts the distal end of stem II, a region dispensable for function19. Any cpTER lacking guanosine at the 5′ end was expressed with this added.
Assays
For RNP assembly in RRL, protein expression reactions were supplemented with TER and then immediately incubated at 30 °C for 15 min. Unless indicated otherwise, TER was typically added in approximately ten-fold excess of protein. Primer-extension activity assays were carried out for 30 min at 30 °C using 0.6 µM [32P]dGTP (800 Ci mmol−1), 5 µM unlabeled dGTP, 200 µM TTP, with or without 200 µM dATP and with 1 µM (TG)8T2G3 in reaction buffer with 50 mM Tris-acetate, pH 8.0, 10 mM spermidine, 5 mM β-mercaptoethanol and 2 mM MgCl2. RNP assembly reactions were diluted at least ten-fold into the final activity assay volume. Products were precipitated and resolved by denaturing gel electrophoresis. For immunopurification, ZZ-tagged p65 or TERT was recovered using IgG agarose at 4 °C for 1 h in T2MG with 0.1 M NaCl, 0.1% (v/v) NP-40 and 10 µg ml−1 each of tRNA and BSA. Northern blot hybridizations were carried out as described20 using a single, end-labeled oligonucleotide probe fully complementary to all TER variants within the panel tested unless indicated otherwise. Control experiments verified that TER was not degraded during the RNP assembly reaction or immunopurification, in the presence or absence of p65 and/or TERT.
Radiolabeled TER was synthesized by T7 RNA polymerase using [32P]UTP. Full-length RNA was gel-purified and passively eluted. EMSA reactions for p65 alone contained T2MG, 0.1 M NaCl, 5 mM DTT, 500 ng BSA, 1 µg tRNA, 0.25 µl RNasin, bromophenol blue, xylene cyanol and an ~0.1 nM final concentration of TER. Samples were mixed on ice in a 10 µl final volume then incubated at room temperature for 10 min before electrophoresis in a 5% (v/v) acrylamide native gel (37.5:1 acrylamide/bisacrylamide, 4% (v/v) glycerol, 0.5× TBE) run with 0.5× TBE at 200 V for 3 h at 4 °C. EMSA reactions including TERT(1–516) contained T2MG, 0.1 M NaCl, 5 mM DTT, 5 µg BSA, 0.25 µl RNasin, 250 ng tRNA and an ~0.1 nM final concentration of TER. Samples were mixed on ice in a 10 µl final volume then incubated at 30 °C for 20 min before electrophoresis as described above. Time courses of incubation before electrophoresis revealed that binding reactions were at equilibrium (data not shown).
Supplementary Material
ACKNOWLEDGMENTS
We thank Collins laboratory members and the RNA community for experimental discussion and comments on the manuscript. Funding was provided by a predoctoral fellowship from the US National Science Foundation (C.M.O.) and by US National Institutes of Health grant GM54198 (K.C.).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 2.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 3.Wong JMY, Collins K. Telomere maintenance and disease. Lancet. 2003;362:983–988. doi: 10.1016/S0140-6736(03)14369-3. [DOI] [PubMed] [Google Scholar]
- 4.Weeks KM. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 5.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;7:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PJ. Stem cells, telomerase and dyskeratosis congenita. Bioessays. 2003;25:126–133. doi: 10.1002/bies.10229. [DOI] [PubMed] [Google Scholar]
- 8.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 9.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aigner S, et al. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aigner S, Postberg J, Lipps HJ, Cech TR. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. doi: 10.1021/bi034121y. [DOI] [PubMed] [Google Scholar]
- 13.Wolin SL, Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 14.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 16.ten Dam E, van Belkum A, Pleij K. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. USA. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt SE, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol. Biol. Cell. 2003;14:4794–4804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aigner S, Cech TR. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA. 2004;10:1108–1118. doi: 10.1261/rna.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 26.McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharyya A, Blackburn EH. Architecture of telomerase RNA. EMBO J. 1994;13:5721–5731. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 30.Harrington L. Biochemical aspects of telomerase function. Cancer Lett. 2003;194:139–154. doi: 10.1016/s0304-3835(02)00701-2. [DOI] [PubMed] [Google Scholar]
- 31.Blackburn EH. The end of the (DNA) line. Nat. Struct. Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- 32.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Williamson JR. After the ribosome structures: how are the subunits assembled? RNA. 2003;9:165–167. doi: 10.1261/rna.2164903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nottrott S, Urlaub H, Luhrmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagai K, et al. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.