Abstract
High-throughput sequencing promises to accelerate the discovery of sequence variants, but distinguishing oncogenic mutations from irrelevant "passenger" mutations remains a major challenge. Here we present an analysis of two sequence variants of the MET receptor (hepatocyte growth factor receptor) R970C and T992I (also designated R988C and T1010I). Previous reports indicated these sequence variants are transforming and contribute to oncogenesis. We screened patients with chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), colorectal cancer, endometrial cancer, thyroid cancer, or melanoma as well as individuals without cancer and found these variants at low frequencies in most cohorts, including normal individuals. No evidence of increased phosphorylation or transformative capacity by either sequence variant was found. Since small-molecule inhibitors for MET are currently in development, it will be important to distinguish between oncogenic sequence variants and rare single-nucleotide polymorphisms to avoid the use of unnecessary and potentially toxic cancer therapy agents.
Keywords: cancer genetics, cancer genomics, targeted therapy
Introduction
Tyrosine kinases play a critical role in numerous cellular processes(1). Therapies targeted to dysreglated kinases have proven more successful than conventional approaches(2–5). One emerging kinase target is the hepatocyte growth factor receptor, MET, identified as part of the fusion oncogene, TPR-MET(6, 7). Subsequently, point mutations in MET were identified in renal papillary carcinoma(8–10). More recently, MET variants, R970C and T992I (also designated R988C and T1010I), have been found in lung cancer cell lines as well as individuals with lung, thyroid, renal, breast cancer, CLL, and lymphoma(11–18). These variants have been characterized by Ba/F3 transformation and phospho-tyrosine immunoblots among other assay systems and were concluded to be transforming(13, 14).
To better understand the cancer genetics of these MET variants we determined R970 and T992 genotypes for patients with CLL, AML, CMML, colorectal cancer, endometrial cancer, thyroid cancer, or melanoma as well as normal individuals. This process revealed low frequency METR970C and METT992I in most of the cohorts, including the normal individuals. Since these sequence variants were identified in a wide variety of different malignancies including individuals without clinically detectable cancer, we hypothesized that these were rare single nucleotide polymorphisms not relevant to oncogenesis. In support of this, we examined the human and murine versions of each of these sequence variants in Ba/F3 transformation assays as well as for tyrosine phosphorylation status and found no evidence of a transforming phenotype.
Material and Methods
Plasmid Construction
Human MET was purchased from Origene (Rockville, MD). Murine Met and TPR-MET were generously provided by Dr. George F. Vande Woude (Van Andel Research Institute, Grand Rapids, MI). All three genes were cloned into MSCV-IRES-GFP (MIG) using the Gateway Cloning system (Invitrogen Corporation, Carlsbad, CA). All point mutations were created using the Quikchange kit (Agilent Technologies, Cedar Creek, TX) according to the manufacturer’s instructions.
Cell culture
Ba/F3 cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in RPMI 1640 medium (Invitrogen) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA), L-glutamine (Invitrogen), penicillin/streptomycin (Invitrogen), fungizone (Invitrogen), and WEHI-conditioned media. Ba/F3 cells were infected with retrovirus expressing human or murine MET WT, R970C (R968C in murine version), T992I (T990I in murine version), or TPR-MET. Stable cell lines were sorted for GFP expression on a FACSAria flow cytometer (BD Biosciences, San Jose, CA). Cells were counted daily using Guava ViaCount reagent and Guava Personal Cell Analysis flow cytometer (Guava Technologies, Hayward, CA).
WEHI-independence Assays
Parental Ba/F3 cells or those expressing human or murine MET WT, R970C (R968C in murine version), T992I (T990I in murine version), or TPR-MET were washed three times and resuspended in RPMI media with 10% FBS, penicillin/streptomycin, and fungizone and counted daily.
Immunoblotting
Human MET WT, R970C, T992I, or TPR-MET were transfected into 293 T17 cells using Fugene6 transfection reagent (Roche, Indianapolis, IN). After 48 hours, 293 T17 cells were lysed in 1 × lysis buffer (Cell Signaling, Danvers, MA), and diluted in sample buffer (75 mM Tris pH 6.8, 3% SDS, 15% glycerol, 8% β-mercaptoethanol, 0.1 % bromophenol blue). Whole cell lysates were separated by SDS-PAGE. Proteins were transferred to PVDF membranes (Millipore, Billerica, MA) and subjected to immunoblot analysis with antibodies specific for phospho-tyrosine (4G10, Millipore), total or phospho-MET (Cell Signaling), or β-actin (Millipore).
Genotypic Analysis
All patient material was de-identified and obtained with informed consent with approval from the institutional review board of Oregon Health & Science University. MET juxtamembrane and activation loop domains were sequenced in CLL, AML, CMML, and normal individuals as described(19). For colorectal, endometrial, thyroid, and melanoma patient samples, mass spectrometric genotyping of MET R970 and T992 was performed with the Sequenom MassARRAY system and the OncoCarta assay panel (Sequenom, San Deigo, CA) as described(20).
Statistical Analysis
The incidence of METR970C and METT992I in each cohort was estimated by the proportion of individuals with mutations (i.e., the number of individuals with mutations divided by the total number of individuals in a cohort) and is expressed as a percentage. The 95% confidence intervals were calculated by using the exact binomial distribution. For Ba/F3 transformation experiments and densitometry of immunoblots, a Student’s t-test was performed for each MET variant compared with METWT and samples with p value less than 0.05 were deemed significant.
Results and Discussion
To determine the frequency of the sequence variants METR970C and METT992I in patients with a variety of malignancies, we screened DNA from 96 CLL, 191 AML, 32 CMML, 109 colorectal cancer, 73 endometrial cancer, 168 thyroid cancer, and 115 melanoma patients as well as 96 normal individuals. Screening of these cohorts revealed that all groups contained low frequency METR970C or METT992I sequence variants, and most cohorts contained both variants. Matched buccal cell samples were available for 3 cases exhibiting each variant (T992I – 2 melanoma and 1 endometrial cancer; R970C – 2 thyroid and 1 endometrial cancer) and all 6 cases showed presence of the variant allele in the germline material. Interestingly, we also identified one normal individual with METR970C and one normal individual with METT992I (Table 1). These alleles have not been detected previously in normal samples. Thus, our findings as well as those by others suggest these sequence variants occur at similar frequencies among many different types of malignancy as well as in individuals without cancer (Table 1)(11–18, 21). In addition, 95% confidence intervals of the incidence of each sequence variant are overlapping for each diagnosis compared with all other diagnoses and compared with the individuals without cancer (Table 1). This indicates a lack of evidence for statistically significant differences in the incidence of either sequence variant in individuals with or without cancer. Since we (and others) have also found that these variants are generally germline, we functionally characterized these variants for transforming capacity.
| Cohort (n) | Incidence of R970C | 95% Exact CI | Reference |
|---|---|---|---|
| Normal (96) | 1 (1%) | 0–6% | Reported Here |
| CLL (96) | 0 | 0–4% | Reported Here |
| AML (191) | 0 | 0–2% | Reported Here |
| CMML (32) | 2 (6%) | 0–21% | Reported Here |
| Colorectal Cancer (109) | 1 (1%) | 0–5% | Reported Here |
| Endometrial Cancer (73) | 1 (1%) | 0–7% | Reported Here |
| Thyroid Cancer (168) | 2 (1%) | 0–4% | Reported Here |
| Melanoma (115) | 1 (1%) | 0–5% | Reported Here |
| Total (880) | 8 (0.9%) | 0–2% | |
| Papillary renal carcinoma (129) | 0 | 0–3% | Schmidt et al. 1999 |
| Other solid tumors (201) | 0 | 0–2% | Schmidt et al. 1999 |
| Breast Cancer (30) | 0 | 0–12% | Lee et al. 2000 |
| CLL (95) | 0 | 0–4% | Brown et al. 2008 |
| CLL (8) | 1 (13%) | 0–53% | Tjin et al. 2006 |
| Follicular Lymphoma (15) | 1 (7%) | 0–32% | Tjin et al. 2006 |
| Burkitt's Lymphoma (12) | 1 (8%) | 0–38% | Tjin et al. 2006 |
| Diffuse Large B-cell Lymphoma (39) | 1 (3%) | 0–13% | Tjin et al. 2006 |
| Small Cell Lung Cancer (32) | 0 | 0–11% | Ma et al. 2003 |
| Non-small cell lung carcinoma (20) | 0 | 0–17% | Teng et al. 2006 |
| Lung carcinoma (361) | 0 | 0–1% | Teng et al. 2006 |
| Fillicular Carcinoma (21) | 0 | 0–16% | Wasinius et al. 2005 |
| Papillary Carcinoma (53) | 0 | 0–7% | Wasinius et al. 2005 |
| Medullary Carcinoma (13) | 0 | 0–25% | Wasinius et al. 2005 |
| Mesothelioma (43) | 0 | 0–8% | Jagadeeswaran et al. 2006 |
| Gastric Carcinoma (85) | 0 | 0–4% | Lee et al. 2000 |
| Total (1157) | 4 (0.35%) | 0–1% | |
| Cohort (n) | Incidence of T992I | 95% Exact CI | Reference |
| Normal (96) | 1 (1%) | 0–6% | Reported Here |
| CLL (96) | 2 (2%) | 0–7% | Reported Here |
| AML (191) | 2 (1%) | 0–4% | Reported Here |
| CMML (32) | 0 | 0–11% | Reported Here |
| Colorectal Cancer (109) | 0 | 0–3% | Reported Here |
| Endometrial Cancer (73) | 2 (3%) | 0–10% | Reported Here |
| Thyroid Cancer (168) | 0 | 0–2% | Reported Here |
| Melanoma (115) | 2 (2%) | 0–6% | Reported Here |
| Total (880) | 9 (1.0%) | 0–2% | |
| Papillary renal carcinoma (129) | 1 (1%) | 0–4% | Schmidt et al. 1999 |
| Other solid tumors (201) | 0 | 0–2% | Schmidt et al. 1999 |
| Breast Cancer (30) | 1 (3%) | 0–17% | Lee et al. 2000 |
| CLL (95) | 1 (1%) | 0–6% | Brown et al. 2008 |
| Small Cell Lung Cancer (32) | 1 (3%) | 0–16% | Ma et al. 2003 |
| Non-small cell lung carcinoma (20) | 1 (5%) | 0–25% | Teng et al. 2006 |
| Lung carcinoma (361) | 5 (1%) | 0–3% | Teng et al. 2006 |
| Follicular Carcinoma (21) | 2 (10%) | 1–30% | Wasinius et al. 2005 |
| Papillary Carcinoma (53) | 3 (6%) | 1–16% | Wasinius et al. 2005 |
| Medullary Carcinoma (13) | 1 (8%) | 0–36% | Wasinius et al. 2005 |
| Mesothelioma (43) | 0 | 0–8% | Jagadeeswaran et al. 2006 |
| Gastric Carcinoma (85) | 0 | 0–5% | Lee et al. 2000 |
| Total (1083) | 16 (1.5%) | 0–2% |
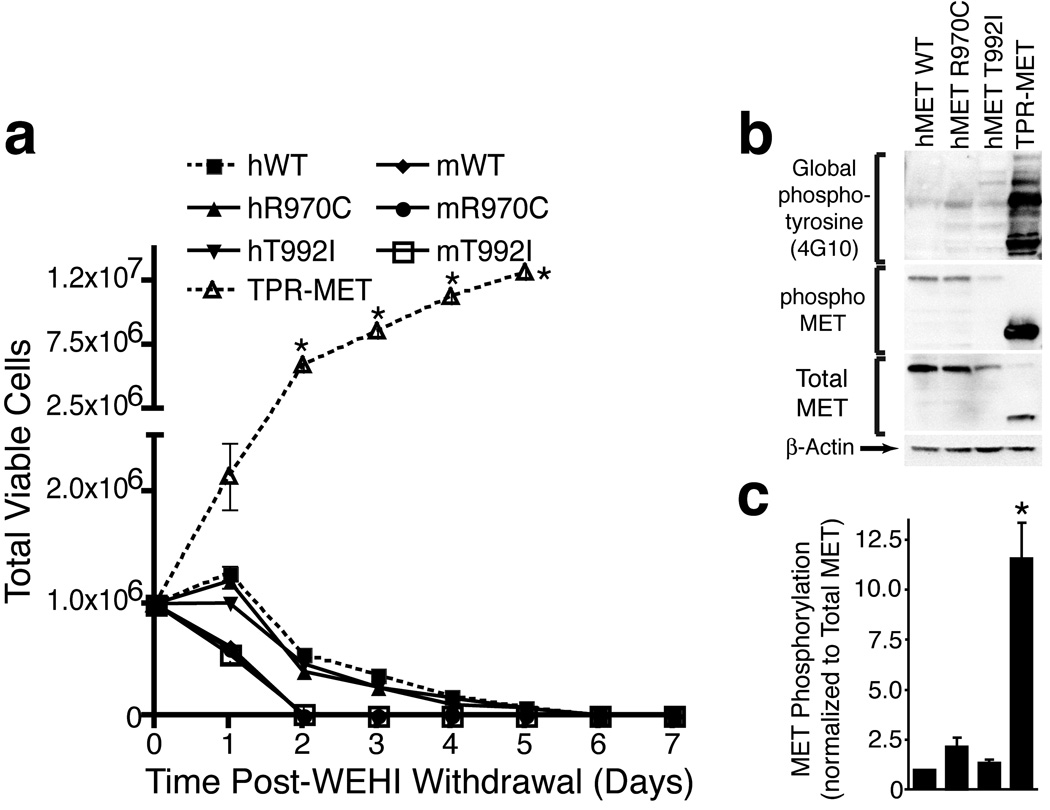
We created Ba/F3 cells stably expressing human or murine METWT, human METR970C, human METT992I, murine MetR968C, murine MetT990I, or TPR-MET and withdrew IL-3 from culture media. Ba/F3 cells that have been transformed by an oncogene such as TPR-MET are capable of proliferating without IL-3 as shown in Figure 1A; however, we did not observe transformation by any of the full-length MET transgenes, including the sequence variants (Figure 1A). Importantly, Ba/F3 cells are not a universally effective system for evaluation of tyrosine kinase oncogenecity, depending on the particular kinase and the physical location of the mutation in the protein. However, previous reports of the METR970C and METT992I variants suggested that these variants were capable of transforming Ba/F3 cells, hence this system was employed to directly test this prior claim(14). In addition to the Ba/F3 system, we next assessed the phosphorylation status of wild-type MET compared with each variant—a technique that was also previously employed to demonstrate increased phosphorylation of these sequence variants(14). We expressed human METWT, METR970C, METT992I, or TPR-MET in 293 T17 cells and immunoblotted with antibodies specific for global phospho-tyrosine (4G10), phospho-MET, total MET, or β-actin. While cells expressing TPR-MET exhibited markedly increased levels of global phosphotyrosine as well as phospho-MET, we did not observe any differences in phosphorylation levels with the MET R970C or T992I variants, again arguing against a direct transforming or dysregulated role for METR970C or METT992I (Figure 1B, C).
Figure 1. Characterization of MET Sequence Variants in Ba/F3 Cells.
(A) Ba/F3 cells overexpressing human or murine METWT, human METR970C, human METT992I, murine MetR968C, murine MetT990I, or TPR-MET were plated in media without IL-3. Cells were counted daily for one week. Values represent mean ± s.e.m. (n=3). * indicates p < 0.002 with t-test in comparison to human METWT.
(B) Whole cell lysates from 293 T17 cells overexpressing human METWT, METR970C, METT992I, or TPR-MET were subjected to immunoblot analysis with antibodies specific for phospho-tyrosine (4G10), phoshpo-MET, total MET, or β-actin.
(C) Densitometric analysis of immunoblots shown in panel (B). Phospho-MET is normalized to total MET and values are expressed as fold of WT. Values represent mean ± s.e.m. (n=3). * indicates p < 0.05 with t-test in comparison to human METWT.
Several forms of MET have been implicated in oncogenesis including fusion with TPR and point mutation at residues D1228, Y1230, and M1250(6, 7, 9, 10). The capacity for METR970C and METT992I to contribute to oncogenesis has been a topic of debate. First identified by Schmidt et al.(15), METT992I was thought to represent a rare polymorphism due to lack of disease segregation and failure to induce focus formation or phosphorylation in NIH3T3 cells. Lee et al. also observed no functional consequence of these variants using similar assays but did observe slightly faster tumor growth in nude mice(12). Consequences of these variants were identified in assays for cytoskeletal function(13, 14, 21). Of note, neither variant was previously observed in samples from healthy individuals(13, 14). We find no difference in transformative capacity or phosphorylation status of either variant compared with wild-type MET and suggest these alleles are not transforming. This conclusion is also based on the presence of these variants in a wide variety of malignancies as well as individuals without cancer. We cannot rule out the possibility that these alleles may play a role in disease pathogenesis in some other manner such as promoting changes in migration or differentiation. In this way, these alleles may predispose an individual towards cancer when combined with an oncogene that drives cellular proliferation. However, we find no evidence that these alleles directly promote transformation. Given the recent emergence of MET inhibitors as clinical cancer therapeutics(22), it will be important to carefully evaluate these alleles for their role in oncogenesis.
Acknowledgements
Supported in part by The Leukemia and Lymphoma Society, the TJ Martell Foundation, and the Doris Duke Charitable Foundation. J.W.T. is supported by grants from the William Lawrence and Blanche Hughes Fund and the Oregon Clinical and Translational Research Institute (OCTRI) grant number UL1 RR024140 from the National Center for Research Resources (NRCC), a component of the NIH, and NIH Roadmap for Medical Research. M.C.H. is supported in part by a VA Merit Review Grant. B.J.D. is an investigator of the Howard Hughes Medical Institute and principal investigator of the OHSU cancer center support grant 5 p30 CA069533. M.M.L is supported by the Leukemia and Lymphoma Society and the TJ Martell Foundation. Biostatistics support (MM) is in part provided by the Biostatistics Shared Resource of the Knight Cancer Institute (5P50CA069533) and the Biostatistics & Design Program of the Oregon Clinical & Translational Research Institute (UL1 RR024140).
References
- 1.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Smith I, Procter M, Gelber RD, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369:29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 6.Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 7.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci U S A. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffers M, Schmidt L, Nakaigawa N, et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci U S A. 1997;94:11445–11450. doi: 10.1073/pnas.94.21.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt L, Junker K, Weirich G, et al. Two North American families with hereditary papillary renal carcinoma and identical novel mutations in the MET proto-oncogene. Cancer Res. 1998;58:1719–1722. [PubMed] [Google Scholar]
- 11.Brown JR, Levine RL, Thompson C, Basile G, Gilliland DG, Freedman AS. Systematic genomic screen for tyrosine kinase mutations in CLL. Leukemia. 2008;22:1966–1969. doi: 10.1038/leu.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Han SU, Cho H, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–4953. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- 13.Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 14.Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 15.Schmidt L, Junker K, Nakaigawa N, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene. 1999;18:2343–2350. doi: 10.1038/sj.onc.1202547. [DOI] [PubMed] [Google Scholar]
- 16.Tengs T, Lee JC, Paez JG, et al. A transforming MET mutation discovered in non-small cell lung cancer using microarray-based resequencing. Cancer Lett. 2006;239:227–233. doi: 10.1016/j.canlet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Tjin EP, Groen RW, Vogelzang I, et al. Functional analysis of HGF/MET signaling and aberrant HGF-activator expression in diffuse large B-cell lymphoma. Blood. 2006;107:760–768. doi: 10.1182/blood-2005-05-1929. [DOI] [PubMed] [Google Scholar]
- 18.Wasenius VM, Hemmer S, Karjalainen-Lindsberg ML, Nupponen NN, Franssila K, Joensuu H. MET receptor tyrosine kinase sequence alterations in differentiated thyroid carcinoma. Am J Surg Pathol. 2005;29:544–549. doi: 10.1097/01.pas.0000156103.37756.e2. [DOI] [PubMed] [Google Scholar]
- 19.Loriaux MM, Levine RL, Tyner JW, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troxell ML, Levine J, Beadling C, et al. High prevalence of PIK3CA/AKT pathway mutations in papillary neoplasms of the breast. Mod Pathol. 23:27–37. doi: 10.1038/modpathol.2009.142. [DOI] [PubMed] [Google Scholar]
- 21.Jagadeeswaran R, Ma PC, Seiwert TY, et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006;66:352–361. doi: 10.1158/0008-5472.CAN-04-4567. [DOI] [PubMed] [Google Scholar]
- 22.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]