Abstract
About 15 years ago, several groups including ours had used matched pairs of cell lines carrying wild type or mutant p53 genes to ascertain a role for p53 in cell survival. These were isogenic cell lines differing only by p53 status. The trend at that time was to support p53-mediated apoptosis. Accordingly, p53-wildtype cells were sensitive to DNA damage compared to p53-mutant cells which were thought to evade apoptosis. However, this finding was not universal. In particular, after UV-radiation, p53-mutant cells were more sensitive than their wild type p53 counterparts in several studies. The finding that p53 controlled a major DNA repair pathway, nucleotide excision repair (NER) which repairs UV-damage, provided a mechanism for the observations. We coined the term “the two faces of tumor suppressor p53” to illustrate that p53 can on one hand induce apoptosis leading to cell sensitivity, but p53 can also enhance the rate of DNA repair thereby protecting cells from DNA damage. This concept has gained acceptance and has been expanded to other DNA-damaging agents. New insights into how p53 is “switched” from a protective function to an apoptotic function are reviewed.
Keywords: DNA-repair, DNA-damage protection
A simple way to view p53 is to view it through the lens of genomic DNA damage. A cell never carries “zero” DNA damage. Cells endure a baseline level damage arising from endogenous sources. At the baseline level, cellular DNA repair pathways can repair the damage at a rate that allows cell cycle progression to occur/continue/resume. Typically, damage persists only a few hours. In this “baseline” setting, p53 acts as “Guardian of the Genome” by 1) halting the cell cycle for sufficient time to repair; and 2) transcriptionally regulating genes whose products promote DNA repair, especially the Xpc gene and its encoded protein XPC that is required for nucleotide excision DNA repair (NER). The baseline activity is a “constitutive” function of p53 in that DNA damage is continually being induced, repaired, and induced again elsewhere in the genome. The baseline is never ”zero”.
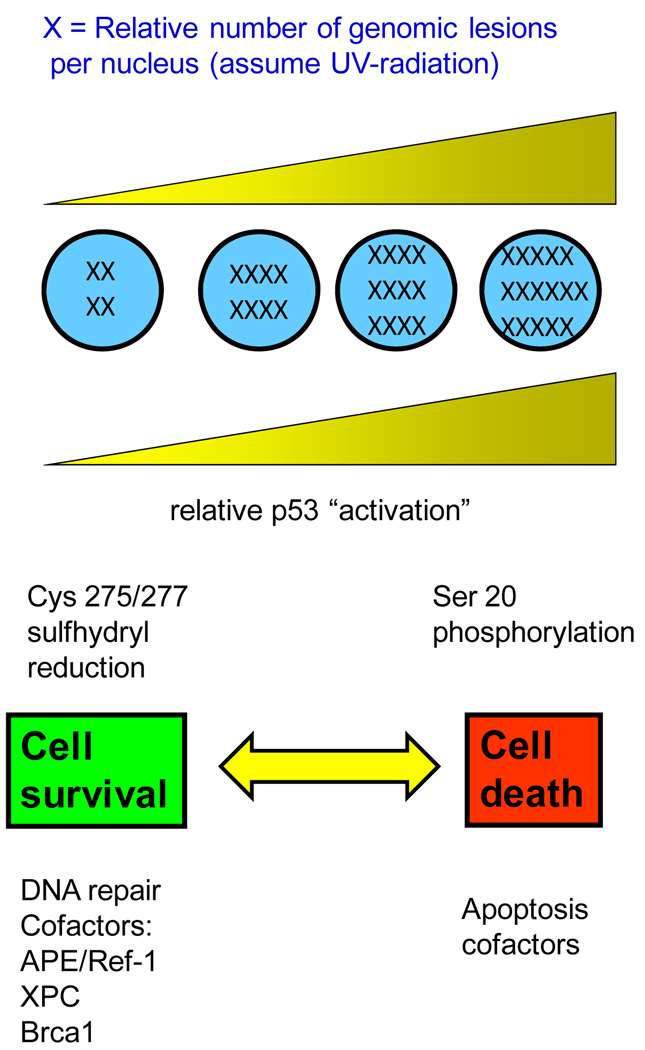
Exogenous sources of DNA damage have the effect of raising the level of DNA damage, perhaps even to irreparable levels. What happens then? Cells die, perhaps by apoptosis. By “irreparable” levels of DNA damage, we mean that the rate of lesion removal is not fast enough to keep up with lesion formation or that the lesion frequency exceeds the capacity for removal (Figure 1).
Figure 1. “The two faces of p53” adapted from Smith and Fornace, 1996, 1997 (3, 4).
The baseline level of DNA damage is never “zero”. Endogenous sources of DNA damage provide a baseline for p53 “activation”. “Activation” is defined as transcriptional induction of p53-regulated effector genes. As DNA damage levels increase, p53 becomes phosphorylated which may promote apoptosis. A number of DNA repair proteins are involved in the p53-mediated protective mechanism, including APE/Ref-1, XPC, gadd45, and Brca1.
Post-translational modifications to p53 protein probably signal the transition from “cell protector” to “cell killer”. In particular, the reduction of p53 cysteine sulfhydryl residues at codons 275 and/or 277 can promote DNA repair and cell survival without promoting apoptosis (1). After UV-radiation, an exogenous source of DNA damage, p53 serine residues become phosphorylated. It is likely that p53-mediated apoptosis is triggered by p53 phosphorylation (2). If key p53 residues are not phosphorylated, the result is to promote cell survival. Probably, the exact balance in a particular cell type depends on UV-wavelength known to affect the utilization of divergent DNA repair pathways, and of course the dose of UV-radiation (3, 4).
If one thinks about it, the first response of the cell is to attempt to repair the damage. Only secondly, having failed that, would apoptosis be invoked (Figure 1). Thus, the lesion frequency (DNA damage) promotes p53 “activation” (defined as transcriptional induction of p53-regulated genes). But, p53 accelerates the rate of lesion removal largely through elevating XPC, the rate-limiting protein for NER (5). Thus is attained a steady-state level of DNA damage that cells can tolerate.
We first discussed the protective role of p53 in the context of UV-radiation some 15 years ago (3,4,6,7). At that time, only a handful of genes were known to be p53-regulated. Certainly XPC, because it is required and rate-limiting for NER, is probably a key player in cellular protection (5). XPC plays an important role in bone marrow protection because Xpc−/− mice died from myelosuppression (8). Other p53-regulated genes such as Gadd45, may also contribute (7). Recombinant Gadd45 protein was found to bind selectively to UV-damaged chromatin (9). And Gadd45−/− mice were defective in NER and exhibited increased mutagenesis (10).
We and others showed that p53-mutant or p53-null cells were preferentially sensitive to UV-radiation (3, 4, 6, 7). An implication that followed was that UV-mimetic cancer chemotherapy drugs would likewise be effective in killing p53-mutant or p53-null cancer cells. Sure enough, UV-mimetics cisplatin and carboplatin yielded results similar to UV-radiation, namely that p53-mutants were more sensitive than p53-wildtype cells in the same cellular background (11, 12). Some investigators are revisiting this idea (13). One possibility is that p53-mutant cells are initially more sensitive to DNA damage, owing to defective NER DNA repair. But, after many passages in culture or multiple DNA-damaging treatments p53-mutant cells become resistant. A p53-mutant background may be mutagenic and therefore additional mutations throughout the genome would accumulate in p53-mutant cells.
A second major DNA repair pathway, base excision DNA repair (BER) is also influenced by p53 status. Cells with wildtype p53 carry out normal BER while cells with mutant p53 or null for p53 are defective in BER (14). BER is responsible for removal of base damage such as 8-oxoguanine or N7-methyladenine. The exact mechanism by which p53 regulates BER is not clear. The APE endonuclease a required enzyme in BER interacts directly with p53 suggesting that the mechanism may not require p53-downstream effector genes. However recombinant p53 did not affect APE endonuclease activity in vitro.
Acknowledgements
Supported by NIH 5R01 HL086978 to M.L.S.
Footnotes
Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a Ref 1 dependent redox mechanism. Proc Natl Acad Sci USA. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.She QB, Ma WY, Dong Z. Role of MAP kinases in UVB-induced phosphorylation of p53 at serine 20. Oncogene. 2002;21:1580–1589. doi: 10.1038/sj.onc.1205239. [DOI] [PubMed] [Google Scholar]
- 3.Smith ML, Fornace AJ., Jr The two faces of tumor suppressor p53. Am J Path. 1996;148:1019–1022. [PMC free article] [PubMed] [Google Scholar]
- 4.Smith ML, Fornace AJ., Jr p53–mediated protective responses to UV irradiation. Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci USA. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ML, Chen IT, Zhan Q, O'Connor PM, Fornace AJ., Jr Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 7.Smith ML, Ford JM, Hollander MC, et al. p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer JL, Kumar MA, Day TW, et al. The Xpc gene markedly affects cell survival in mouse bone marrow. Mutagenesis. 2009;24:309–316. doi: 10.1093/mutage/gep011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrier F, Georgel PT, Pourquier P, et al. Gadd 45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollander MC, Kovalsky O, Salvador JM, et al. Dimethylbenzanthracene carcinogenesis in Gadd 45a-null mice is associated with decreased DNA repair and increased mutation frequency. Cancer Res. 2001;61:2487–2491. [PubMed] [Google Scholar]
- 11.Fan S, Smith ML, Rivet DJ, et al. Disruption of p53 sensitizes breast cancer MCF7 cells to cisplatin and pentoxifylline. Cancer Res. 1995;55:1649–1654. [PubMed] [Google Scholar]
- 12.Seo YR, Chen EI, Smith ML. Sensitivity of p53 – deficient cells to oxaliplatin and thio-TEPA (N,N’,N” triethylenethiophosphoramide) Breast Cancer Res Treat. 2002;72:255–263. doi: 10.1023/a:1014913708916. [DOI] [PubMed] [Google Scholar]
- 13.Gudkov A, Komarova EA. Dangerous habits of a security guard: the two faces of p53 as a drug target. Human Mol Genet. 2007;16:67–72. doi: 10.1093/hmg/ddm052. [DOI] [PubMed] [Google Scholar]
- 14.Seo YR, Fishel ML, Amundson SA, Kelley MR, Smith ML. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21:731–737. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]