Abstract
Many archaeal proteins undergo posttranslational modifications. S-layer proteins and flagellins have been used successfully to study a variety of these modifications, including N-linked glycosylation, signal peptide removal and lipid modification. Use of these well-characterized reporter proteins in the genetically tractable model organisms, Haloferax volcanii, Methanococcus voltae and Methanococcus maripaludis, has allowed dissection of the pathways and characterization of many of the enzymes responsible for these modifications. Such studies have identified archaeal-specific variations in signal peptidase activity not found in the other domains of life, as well as the enzymes responsible for assembly and biosynthesis of novel N-linked glycans. In vitro assays for some of these enzymes have already been developed. N-linked glycosylation is not essential for either Hfx. volcanii or the Methanococcus species, an observation that allowed researchers to analyze the role played by glycosylation in the function of both S-layers and flagellins, by generating mutants possessing these reporters with only partial attached glycans or lacking glycan altogether. In future studies, it will be possible to consider questions related to the heterogeneity associated with given modifications, such as differential or modulated glycosylation.
1. Introduction
Carl Woese initially defined the third form of life, the Archaea, on the basis of the novel oligonucleotide signatures of their small ribosomal subunit RNA [1–3]. Specifically, by generating phylogenetic trees based on 16S rRNA sequences, Woese clearly showed that Archaea formed a unique group, distinct from Bacteria or Eukarya. However, early analysis also revealed that this unusual group of microbes shared a variety of other characteristics, most notably ether-linked membrane lipids, a variety of unusual cell walls (none of which contained murein), atypical DNA-dependent RNA polymerases and later, their own variation of flagella [4, 5]. Indeed, cell wall composition was one of the very first phenotypical traits of the Archaea considered that allowed for differentiation from Bacteria [6] and was considered in the early days of archaeal research to be “the only useful phylogenetic criterion, other than direct molecular phylogenetic measurement” to distinguish between the two prokaryotic domains [7]. A common feature of many genera of Archaea, found in representatives of all the major lineages, is the presence of an outermost component of the cell envelope termed the surface (S)-layer, comprising protein or often glycoprotein subunits that form a regularly structured array.
In addition to their distinctive cell walls, cell surface structures of Archaea are also unusual [8]. While many, such as cannulae [9] and hami [10] are unique to Archaea, even the more commonly found flagella and pili are unlike their bacterial namesakes [11, 12]. Archaeal flagella are the best-studied of the archaeal appendages and are unusual in many aspects, including the initial biosynthesis of the component flagellins with N-terminal class III signal peptides that are cleaved by a prepilin peptidase-like enzyme. As considered below, these traits are all similar to those found in bacterial type IV pili systems but absent in bacterial flagella systems.
Both S-layer proteins and flagellins are among the most abundant proteins synthesized by the archaeal cell and both can be isolated with relative ease in substantial amounts for biochemical and structural studies. In Archaea, the majority of these proteins appear to be glycoproteins, mainly containing N-linked glycans, yet sometimes also containing glycans O-linked to threonine residues. In fact, archaeal S-layer proteins, especially those from extreme halophiles, served to identify a novel class of prokaryotic glycoproteins [13–16]. More recently, S-layer proteins and flagellins have been widely used as reporter proteins for the study of a variety of posttranslational modifications in Archaea [17–20], including both class I and class III signal peptide removal, N- and O-glycosylation and lipid modification. Despite these advances, it is only in a very small number of Archaea, including Haloferax volcanii and Methanococcus species, that genetic studies linking specific genes to a particular posttranslational modification have been performed.
2. The S-layer of Haloarchaea andMethanoarchaea
Although found in numerous archaeal species, the S-layers of haloarchaea remain the best studied. Indeed, the first description of a S-layer in Archaea was reported in 1956 when electron microscopic examination of Halobacterium halobium (salinarum) cells revealed a surface presenting morphological units organized in a hexagonal pattern [21]. Later examination of thin-sectioned haloarchaeal cells revealed the presence of a 17 nm thick cell wall beyond the plasma membrane [22–24]. Blaurock et al. [25] later relied on X-ray diffraction to demonstrate a protein layer laying beyond the haloarchaeal plasma membrane at a distance of 8 nm, with a periplasmic-like space being formed from morphological subunits assuming an “inverted-parabola shape”. Enzymatic iodination of Hbt. salinarum surface proteins, together with proteolytic treatment, revealed this surface (S)-layer to contain the S-layer glycoprotein [13]. Kessel et al. [26] next proposed a three-dimensional reconstruction of the Halobacterium (later renamed Haloferax) volcanii S-layer glycoprotein and cell envelope after considering the primary sequence of the Hbt. salinarum S-layer glycoprotein [14], as well as the earlier X-ray diffraction data and electron microscopic images of negatively stained cell envelopes. In this model, based on reconstruction to a 2 nm resolution, six S-layer glycoproteins form a 4.5 nm thick dome-shaped pore, with the open center expanding as one approaches the membrane. The deduced C-terminal transmembrane domain of each S-layer glycoprotein is thought to anchor the structure to the membrane, while an O-glycosylated domain of the S-layer glycoprotein lying upstream of the transmembrane domain is proposed to act as a spacer unit, propping up the domed structure. While the forces responsible for maintaining the integrity of such assemblies remain unknown, divalent cations have been shown to be important [13, 26]. Given the use of intact cells maintained in their growth medium and the high degree of sample preservation afforded by the rapid freezing, reconstruction of the Hbt. salinarum S-layer through the use of electron tomography offered a more realistic view of this structure [27]. It was thus shown that the Hbt. salinarum cell envelope assumes the same basic architecture as does the Hfx. volcanii S-layer. Despite similarities in their S-layer architecture, Hbt. salinarum and Hfx. volcanii cells assume very different shapes, with the former appearing as rods and the latter as indented disks, pointing to factors other than the S-layer as affecting cell shape.
Like their halophilic counterparts, numerous metha-noarchaeal species, such as members of the genus Methanococcus, are also surrounded by a glycoprotein-based S-layer [28]. For many methanogens, study of the S-layer has been relatively limited to identification of the major S-layer component, its response to glycoprotein staining procedures and determination of the lattice symmetry by electron microscopy [29–31]. In the case of M. voltae, the S-layer is formed from a 76 kDa protein arranged into an hexagonal lattice with a center to center spacing of 10 nm [32]. While the S-layer protein does not stain positively with the periodic acid Schiff reagent, suggesting the absence of protein glycosylation, subsequent mass spectrometry analysis has shown it to contain a N-linked glycan identical to that found on flagellins in this species [33]. Methodologies were developed to create protoplasts of M. voltae by removal and regeneration of the S-layer [34]. The protoplasts could be used for transformation of plasmid DNA either directly or by electroporation [35].
In some instances, such as Methanothermus fervidus [36], the S-layer surrounds a sacculus of pseudomurein, a peptidoglycan unique to certain methanoarchaeal species and distinct from bacterial murein. In some Methanosarcina species, the cell envelope is thought to consist of a protein-based S-layer surrounded by a rigid cell wall composed of methanochondroitin, a heteropolysaccharide reminiscent of eukaryotic chondroitin [37].
A model of the S-layer of Methanolobus limicola, formed from glycoprotein subunits, was determined using a variety of microscopy techniques. Using standard electron crystallographic techniques, it was determined that the S-layer had p6 symmetry with a lattice constant of 14.7 nm and a thickness of 4.5 nm [38]. Later examination by scanning tunneling microscopy resulted in a thickness determination of 6.5 nm [39]. As in Hbt. salinarum, the subunits are thought to assemble into a dome-shaped structure. Moreover, although no spacer elements have been identified linking the S-layer to the cytoplasmic membrane, negative staining does reveal a narrow space of about 5–10 nm between the two layers [38].
Methanospirillum hungatei cells present a very complicated envelope profile. Individual cells are surrounded by a cell wall consisting of an S-layer [40] and then a second unusual outer paracrystalline layer termed the sheath [41], consisting of individual and discrete hoop-like components [42]. The ensheathed cells are then separated from the surrounding environment or from neighboring cells by complicated end or spacer plugs [42].
Finally, in Methanocorpusculum sinense, examination of the role of the S-layer in cell-shape maintenance and cell division revealed that lattice faults in the normal p6 symmetry of the S-layer appear to be sites of incorporation of new subunits and initiation points for cell division [43].
3. Archaeal Flagella Assembly and Composition
Biochemical, genetic, and structural studies performed on flagella from several different archaeal species over the last 2 decades have demonstrated the unique nature of this motility apparatus [5, 44]. Flagella have been reported in all of the major subgroupings of cultivatable archaea, including species of extreme halophiles, haloalkaliphiles, methanogens, hyperthermophiles, and thermoacidophiles [45, 46]. Detailed biochemical, structural and/or genetic studies have been reported in a variety of archaeal genera, including Methanococcus [47–49], Methanospirillum [50–52], Halobacterium [53–56], Haloarcula [57], Haloferax [58], Sulfolobus [59], Natrialba [60, 61], Thermococcus [62], and Pyrococcus [63]. However, the bulk of published work on archaeal flagella is focused on Halobacterium and Methanococcus; that is, members of the Euryarchaeota, and it is not certain that findings in one organism or even within one of the archaeal domains are applicable to all Archaea or even to members of the other major archaeal domain (i.e., crenarchaeotes). There may be fundamental differences between the two domains; for instance, genes known to be essential for flagellation in Methanococcus are not found in crenarchaeotes [46, 64] and even something as fundamental as the presence of the hook may be variable, as hooks have not been observed in the crenarchaeote Sulfolobus [65].
Archaeal flagella are motility structures involved in swimming and, in the one example where this has been examined in any detail (i.e., Hbt. salinarum), the flagella can switch their direction of rotation [66–68]. Other than this superficial commonality, archaeal flagella do not bear other similarities to their bacterial counterparts. For example, there are no homologues of bacterial flagella structural or biosynthetic genes contained in any sequenced archaeal genome [67, 69]. Another fundamental difference between the two prokaryotic flagella organelles may be in the driving force for flagellar rotation. Proton or, more rarely, sodium gradients are used to power bacterial flagellar motion [70], while in the one instance where this has been examined in Archaea, again in Hbt. salinarum, flagellar motor rotation depends on ATP [71]. Structurally, archaeal flagella are similar to bacterial type IV pili, surface structures involved in a type of motility across solid surfaces called twitching [72], and, critically, lack a central channel that could allow the passage of subunits through the growing structure for assembly at the distal tip [73–75]. Indeed, the archaeal flagellum has been termed “a bacterial propeller with a pilus-like structure” [75]. Accordingly, archaeal flagella share several commonalities with bacterial type IV pili. Most strikingly, the major subunits of the archaeal flagellum, the flagellins, are made as preproteins with unusual, type IV pilin-like signal peptides (class III signal peptides) that are removed by a specific type IV prepilin signal peptidase homologue (FlaK/PibD; see below) [47, 48, 76]. In addition, both the archaeal flagella system and the type IV pili systems contain a homologous ATPase and a conserved membrane component that may serve as the platform for assembly of the structures [77, 78]. These similarities to type IV pili and the lack of a central channel indicate that assembly of the archaeal flagellum takes place by addition of subunits to the base of the structure, as is also the case in pili growth [45], and fundamentally different from the growth of bacterial flagella, where new subunits are added to the distal end after their passage through the central channel [79].
A single fla operon encompassing up to thirteen flagella-associated genes has been identified in various flagellated archaea, although the core composition of genes involved in flagellation and their arrangement in the genome can vary in different organisms [44]. Unlike most bacterial species, nearly all the flagellated archaeal species contain multiple (i.e., 2–6) flagellin genes, a rare exception being Sulfolobus spp., where only a single flagellin gene is found. Studies show that each flagellin has its own function as deletion of a single flagellin usually results in nonflagellated cells [64, 80, 81]. Interestingly, one of the flagellins forms the hook region in Methanococcus species [49, 64, 82] and Hbt. salinarum [82]. The fla operon typically begins with the multiple flagellin genes, followed by the conserved fla-associated genes, flaC-flaJ, or a subset thereof. The preflagellin peptidase gene is typically located outside this main locus. Deletion analysis has demonstrated that all of the successfully deleted fla-associated genes are essential for flagellation, even though some of these genes are not found in all flagellated archaea [64]. Of the fla-associated genes, flaHIJ are conserved in all flagellated archaea [5]. FlaI and FlaJ are homologous to ATPases (i.e., PilT/PilB) and conserved membrane protein (i.e., PilC/TadB) of type IV pili systems. FlaI has been shown to possess ATPase activity [83]. FlaH may also have ATPase activity, as it contains a conserved Walker box A, although a Walker box B has not been identified [84]. Thus, due to their universal presence in all flagellated archaea and their relationships to type IV pili-related proteins, FlaHIJ are most likely key components in the export and assembly of flagellin subunits. Deletions in any of these genes lead to nonflagellated cells [56, 59, 64, 84, 85]. The roles for the other fla associated genes are generally unknown, although recent reports indicate FlaCE and FlaD of Hbt. salinarum associate with various Che proteins of the chemotaxis system [86].
Finally, in addition to signal peptide removal, archaeal flagellins also undergo a second posttranslational modification, as most are glycoproteins. While Bacteria sometimes contain glycosylated flagellins, the flagellin glycan is always found in an O-linkage [87]. In Archaea, to date the glycan has always been found to be N-linked to the flagellins [33, 88, 89]. As considered below, the presence and completeness of the glycan has marked effects on the assembly and function of the archaeal flagella.
In bacteria, flagella do not function only as organelles for swimming but can also be involved in such diverse activities as swarming motility across surfaces, sensing wetness and playing a role in biofilm formation, for example [90–92]. Similarly, the flagella of Archaea have been recently shown to be involved in other important biological functions in addition to their presumably primary role in swimming. In Pyrococcus furiosus, flagella can form cable-like connections among cells and in adhesion to Methanopyrus kandleri cell surfaces [63]. Interactions of P. furiosus with Methanopyrus cells can occur through flagella, resulting in a two-component archaeal biofilm [93]. In Sulfolobus, in addition to mediating swimming and swarming, flagella, along with pili, have both been shown to be involved in surface adhesion [94]. On the other hand, in Hfx. volcanii, flagella were shown not to be involved significantly in surface adhesion [58]. Instead, attachment is mediated by other type IV pilin-like proteins processed by a type IV prepilin peptidase-like enzyme. Swimming without flagella can occur via different mechanisms in bacteria [12] but, to date, no such nonflagellar-driven swimming modes have been reported in Archaea.
4. Posttranslational Modification of S-Layer Glycoproteins and Flagellins
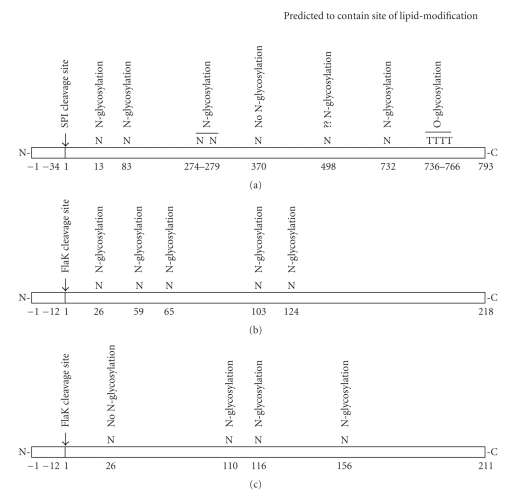
In addition to serving important structural and physiological roles, S-layer glycoproteins and flagellins, in particular those from halophilic and methanogen archaea, are important reporters of posttranslational modifications, including signal sequence cleavage, glycosylation and lipid attachment (Figure 1).
Figure 1.
Schematic depiction of the posttranslational modifications experienced by the Hfx. volcanii S-layer glycoprotein and Methanococcus flagellins. A. Hfx. volcanii S-layer glycoprotein; B. M. voltae FlaB1; C. M. maripaludis FlaB1. Below each sequence, depicted by the elongated rectangle, amino acid residue positions are provided. Above each sequence, the residue at that position is listed, as is the posttranslational modification experienced by that residue or region of the protein. For sequon Asn residues, N-glycosylation, no N-glycosylation or unverified (??) N-glycosylation is marked. Note that the three sequences are not drawn to scale.
4.1. Signal Peptide Cleavage
Bacteria can contain as many as three distinct signal peptidases, essential for removing the N-terminal signal peptides that target preproteins for export from the cytoplasm [95]. Signal peptidase I (SPI) is the housekeeping signal peptidase, responsible for cleaving the signal peptides from most preproteins secreted from the cell via either the Sec or TAT pathways. Signal peptidase II (SPII) removes signal peptides specifically from lipoproteins. Finally, type IV prepilin peptidases (TFPP, sometimes termed signal peptidase III, SPIII) are necessary for the cleavage of class III signal peptides from type IV pilins and related molecules. In Archaea, only SPI and TFPP have been identified [18].
Signal Peptidase I —
Archaeal signal peptidase I was first identified in Methanococcus voltae, where the gene was cloned and the protein expressed and studied biochemically in vitro, using a heterologously expressed, truncated S-layer protein as substrate [96]. Site-directed mutagenesis studies on the methanogen enzyme and that of Hfx. volcanii reached the same conclusion; namely, that the archaeal enzyme relies on a different grouping of essential amino acids than does either the typical prokaryotic (P-type) enzyme found in Bacteria or eukaryotic (ER-type) enzyme [97, 98]. Bacterial P-type SPIs, found as well in mitochondria and chloroplasts, utilize a Ser-Lys dyad for catalysis. Site-directed mutagenesis studies revealed that Ser90 and Lys145 of Escherichia coli SPI are critical for enzymatic activity [99], while subsequent mutagenesis studies, based on crystal structure analysis, lead to the identification of Ser278 as being necessary for optimal activity [100]. In the case of ER-type SPIs, no lysine residues are essential for activity, reflecting the use of a different catalytic mechanism. Indeed, the conserved lysine of the P-type SPI is replaced by a conserved histidine in the ER-type SPI [101]. Further mutagenesis studies identified a conserved serine, histidine and two aspartic acid residues as being important for activity of the eukaryal enzyme, suggesting a potential catalytic mechanism relying on a Ser-His-Asp catalytic triad or a Ser-His catalytic dyad [101, 102]. Archaeal SPI maintains the conserved amino acids of the ER-type enzyme, notably the replacement of the conserved lysine by a histidine. However, site-directed mutagenesis again revealed archaeal-specific features. While the essential natures of the conserved serine and histidine residues were demonstrated, only one of the conserved aspartic acid residues was shown to be essential, unlike the yeast SPI, where both are essential [97, 98]. Almost all archaeal SPIs contain these four conserved amino acids [18], such that the mechanism of catalysis likely involves the Ser-His-Asp triad, as in the ER-type enzyme.
In Hfx. volcanii, two functional SPIs, that is, Sec11a and Sec11b, were identified, and although both are expressed, only Sec11b was deemed essential [103]. Since the two enzymes cleaved substrates differentially in an in vitro assay, they may serve distinct physiological roles.
FlaK/PibD: Archaeal Type IV Prepilin-Like Peptidases —
The best studied of all archaeal signal peptidases are the TFPPs, represented mainly by FlaK/PibD [48, 76, 104], as well as by EppA [105]. These enzymes were first identified as TFPP homologues, able to cleave class III signal peptides from archaeal flagellins. Unlike bacterial flagellins, archaeal flagellins are synthesized as preproteins containing unusually short type IV pilin-like signal peptides. Such processing is an essential step in the assembly of archaeal flagella, as flaK mutants are nonflagellated [76].
Site-directed mutagenesis of both enzymes and substrate has greatly contributed to our knowledge of the mechanism of action and substrate range of archaeal TFPPs. Initial studies involving mutation of conserved amino acids in the signal peptide of model substrates, such as flagellins and sugar-binding proteins, revealed general similarities to the type IV pili system, where glycine at the −1 position of the signal peptide (i.e., the position immediately upstream of the cleavage site) was strongly preferred, with alanine shown to be an acceptable substitute. The −2 and −3 positions of the signal peptide are usually basic amino acids and, in the case of Methanococcus FlaK, the presence of lysine at the −2 position of the substrate is critical for cleavage [106]. Sulfolobus PibD is much less stringent in this regard, in keeping with the large number of potential substrates processed by this enzyme [104]. As revealed initially by signal peptide analysis of potential substrates [104] and later shown directly by in vitro assays [107], PibD is also able to accommodate extremely short signal peptides which are not processed by FlaK. Indeed, it has been suggested that PibD is a rarity among archaeal TFPPs in terms of its range of substrates [46], since, in Methanococcus for example, there are no sugar-binding proteins and type IV pilins are processed by a different dedicated TFPP, EppA [105]. Key to the activity of EppA seems to be the glutamine residue at position 1 found in all pilins but not in flagellins, since flagellins modified to include the pilin −2 to +2 amino acid region were cleaved [105]. In keeping with their processing of type IV prepilin-like molecules, site-directed mutagenesis of both FlaK [76] and PibD [108] revealed that the pair of conserved aspartic acid residues that align with two aspartic acid residues shown to be essential for the bacterial type IV prepilin peptidase activity are also essential in the archaeal enzymes. Other conserved aspartic acid residues are not essential. Thus, both the bacterial and archaeal enzymes rely on the same catalytic mechanism and belong to the same family of novel aspartic acid proteases.
In Methanococcus maripaludis, both flagella and type IV-like pili are composed of major structural proteins possessing class III signal peptides. Interestingly, this species expresses two TFPPs, with FlaK specifically processing flagellins and EppA specifically cleaving the signal peptides from the prepilins [105]. This is not the case in Sulfolobus solfataricus, where PibD has a much broader substrate range, cleaving flagellins and type IV pilins, as well as a number of sugar binding proteins which have been hypothesized to form a pilus-like extension from the cell surface, termed the bindosome [8, 109]. In addition, the Iho670 fibers of Ignicoccus hospitalis, representing a novel archaeal surface structure, are also composed of subunits that contain class III signal peptides cleaved by a TFPP [110]. It thus appears that assembly of surface structures throughout the archaeal domain may rely heavily on the type IV pilus-like model.
5. N-Glycosylation
While the glycoprotein-based composition of the Halobacterium cell envelope had been previously suggested [111, 112], it was Mescher and Strominger [13] who purified and characterized the Hbt. salinarum S-layer glycoprotein, at the same time presenting the first example of a noneukaryal N-glycosylated protein. Initial efforts at describing the process of archaeal N-glycosylation revealed similarities to the parallel eukaryal process. In both cases, oligosaccharides are assembled on dolichol lipid carriers and transferred to target proteins following their delivery across a membrane; namely, the ER membrane in Eukarya and the plasma membrane in Archaea (cf. [113]). However, it was only thirty years later, with the availability of complete genome sequences, that delineation of the biochemical pathways of archaeal N-glycosylation began in earnest. Almost simultaneously, agl (archaeal glycosylation) genes implicated in this posttranslational modification were identified in Hfx. volcanii [114] and M. voltae [115]. Subsequently, agl genes participating in the Methanococcus maripaludis N-glycosylation pathway were identified [116, 117].
Haloferax volcanii —
In Hfx. volcanii, agl genes implicated in the assembly and attachment of a pentasaccharide to select Asn residues of the S-layer glycoprotein were first identified on the basis of their homologies to known N-glycosylation components in Eukarya or Bacteria (i.e., Campylobacter jejuni), the only bacterium for which a complete N-glycosylation pathway has been defined (cf. [118]). Subsequently, additional agl genes were identified either based upon their proximity to previously identified agl sequences or upon reannotation of that region of the genome where all but one of the previously identified agl sequences clustered [119–123]. In this manner, AglJ, AglG, AglI, AglE, and AglD, glycosyltransferases responsible for assembly of the S-layer glycoprotein-modifying pentasaccharide, comprising a hexose, two hexuronic acids, a methylester of hexuronic acid and a final hexose, were identified [123, 124]. AglB was shown to be the oligosaccharyltransferase responsible for delivery of the pentasaccharide, and apparently its precursors, to at least two residues of the S-layer glycoprotein; namely, Asn-13 and Asn-83 [123]. In addition, AglF, AglM, and AglP have also been shown to participate in the assembly of the pentasaccharide [120, 122, 124].
While involvement of each of these gene products in S-layer glycoprotein N-glycosylation has been shown through a combination of gene deletion and mass spectrometry approaches, in several instances, biochemical characterization has been carried out. In the case of AglF and AglM, the proteins have been purified and assays compatible with hypersaline conditions have been developed. Accordingly, AglF was shown to be a glucose-1-phosphate uridyltransferase, able to generate UDP-glucose from glucose-1-phosphate and UTP in a NAD+-dependent manner, while AglM was revealed to function as a UDP-glucose dehydrogenase, generating UDP-glucuronic acid from UDP-glucose [122]. Indeed, a coupled reaction containing AglF, AglM, glucose-1-phosphate, UTP and NAD+ led to the appearance of UDP-glucuronic acid, thus representing the first step towards in vitro reconstitution of the Hfx. volcanii N-glycosylation process. In the case of AglP, purification and subsequent development of an in vitro assay to test the function of the protein confirmed AglP to be a S-adenosyl-L-methionine-dependent methyltransferase [124], as predicted by earlier bioinformatics analysis [121]. Specifically, AglP acts on the fourth subunit of the pentasaccharide decorating the Hfx. volcanii S-layer glycoprotein, adding a methyl moiety to a hexuronic acid to yield the 190 Da methyl ester of hexuronic acid found at this position [124]. These results are summarized in Table 1.
Table 1.
Effects of Hfx. volcanii agl deletions.
| Gene | Role | Effect1 of deletion on: | ||||||
|---|---|---|---|---|---|---|---|---|
| Hfx. volcanii Growth in high salt | S-layer Assembly | Shedding | Susceptibility to protease | S-layer glycoprotein SDS-PAGE migration | N-linked glycan structure | Reference | ||
| aglB | OTase2 | Decreased | No effect | Increased | No effect | Increased | No glycan | [114, 123] |
| aglD | GTase3 (sugar 5) | Decreased | Perturbed | Decreased | Decreased | Increased | Tetrasaccharide | [114, 123] |
| aglE | GTase (sugar 4) | No effect | n.d. | n.d. | No effect | No effect | Trisaccharide | [119] |
| aglF | Glucose-1-P uridyltransferase (sugar 3) | n.d.4 | n.d. | n.d. | Increased | Increased | Disaccharide | [120] |
| aglG | GTase (sugar 2) | n.d. | n.d. | n.d. | Increased | Increased | Monosaccharide | [120] |
| aglI | GTase (sugar 3) | n.d. | n.d. | n.d. | Increased | Increased | Disaccharide | [120] |
| aglM | UDP-glucose dehydrogenase (sugars 2, 3 (4?)) | n.d. | n.d. | n.d. | Increased | Increased | Monosaccharide | [122] |
| aglP | Methyltransferase (sugar 4) | n.d. | n.d. | n.d. | Increased | n.d. | Modified tetrasaccharide | [124] |
1Relative to level detected in parent strain; increased, decreased or no effect.
2OTase: oligosaccharyltransferase.
3GTase: glycosyltransferase.
4n.d.: not determined.
Methanococcus voltae —
The N-linked glycan described for M. voltae PS is a trisaccharide component with structure β-ManpNAcA6Thr-(1–4)-β-GlcpNAc3NAcA-(1–3)-β-GlcpNAc [33], although a strain harbouring a tetrasaccharide variant of this (the same trisaccharide as above with an extra 220 or 260 Da moiety attached) has been reported [125]. The glycan is linked to select asparagine residues present in flagellins and S-layer components via an N-acetylglucosamine, rather than the hexose observed in Hfx. volcanii. A combination of techniques, including insertional inactivation of targeted genes, immunoblot, heterologous expression studies and mass spectrometry analysis of purified flagella have identified the glycosyltransferases and the oligosaccharyltransferase required for the assembly and attachment of the glycan. AglH and AglA are responsible for the addition of the first and last sugar residues to the trisaccharide, respectively, [115, 126]. The role of AglH was elucidated by its ability to successfully complement a conditionally lethal mutation in alg7 (N-acetylglucosamine-1-phosphate transferase) in yeast. AglC and AglK have both been implicated in the transfer of the second sugar residue [125]. Mutants in the oligosaccharyltransferase (aglB) contain S-layer glycoproteins and flagellins presenting molecular masses smaller than observed in any of the other agl mutants, consistent with this enzyme being responsible for the transfer of the N-glycan. The viability of strains carrying a disruption of aglB indicates that the N-linked glycosylation pathway is not essential in M. voltae [115].
Methanococcus maripaludis —
With the development of advanced genetic tools [127], elucidation of the N-linked glycosylation pathway in methanogenic archaea has continued in M. maripaludis, where a tetrasaccharide glycan is N-linked to flagellin subunits [88]. The reported structure of the N-linked glycan was Sug-4-β-ManNAc3NAmA6Thr-4-β-GlcNAc3NAcA-3-β-GalNAc, where Sug was a previously unreported (5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose, representing the first example of a naturally occurring diglycoside of an aldulose [88]. Although the glycans of the two Methanococcus species are related, an obvious difference is that M. maripaludis uses N-acetylgalactosamine as the linking sugar, as compared to N-acetyglucosamine in M. voltae. In addition, the third sugar in both species is the same, except for a 3-acetamidino group addition in M. maripaludis that is carried out by the product of MMP1081 (K. F. J., unpublished results). Interestingly, a strong homologue of MMP1081 is also found in the sequenced genome of M. voltae A3. Should this gene also be present in M. voltae PS; namely, that strain used for glycan structural study, it is unclear why an acetamidino group would not also be added here.
The genes MMP1079, MMP1080 and MMP1088, designated aglO, aglA and aglL, respectively, have been implicated by deletion/complementation analysis and mass spectrometry as being the glycosyltransferases responsible for transfer of the second, third and fourth sugars to the glycan structure, respectively, [116]. As in M. voltae, but unlike the case in Hfx. volcanii, where all but one of the agl genes are found in one large cluster, aglB is located elsewhere on the M. maripaludis chromosome. Its deletion leads to the appearance of nonglycosylated flagellins [116]. The glycosyltransferase responsible for the transfer of the first sugar residue has yet to be identified. Interestingly, mutants harboring deletions in genes that lead to a nonflagellated phenotype (i.e., aglB and aglO) initially synthesize normal levels of the flagellins and other cotranscribed fla gene products. However, upon continued laboratory sub-culturing, these strains appear to stop transcription of the entire fla operon. Other genes identified as involved in the glycan synthesis include MMP0350, the product of which is likely responsible for addition of one of the two acetyl groups found on the second sugar [117] and MMP1085 which encodes a protein responsible for attachment of the methyl group to the terminal sugar (K. F. J. unpublished results). Available information on N-glycosylation in the two Methanococcus species is summarized in Table 2.
In addition to the genetic studies described, heterologous expression and in vitro biochemical and enzymatic studies of proteins predicted to be involved in the glycosylation pathway have helped described the biosynthesis of the acetamido sugar subunit precursors in methanococci [128].
Table 2.
Effects of M. maripaludis and M. voltae agl deletions.
| Gene | Role | Effect1 of deletion on: | ||||
|---|---|---|---|---|---|---|
| Cell flagellation | Motility | Flagellin SDS-PAGE migration | N-linked glycan structure | Reference | ||
| M. maripaludis | ||||||
| aglA | GTase2 (sugar 3) | Present | Decreased | Increased | Disaccharide | [116] |
| aglB | OTase3 | Absent | Non-motile | Increased | No glycan | [116] |
| aglL | GTase (sugar 4) | Present | Decreased | Increased | Modified trisaccharide | [116] |
| aglO | GTase (sugar 2) | Absent | Non-motile | Increased | Monosaccharide | [116] |
| MMP0350 | Acetyltransferase (sugar 2) | Absent | Non-motile | Increased | Monosaccharide | [117] |
| MMP1081 | Acetamidino transfer (sugar 3) | Absent | Decreased | Increased | Modified trisaccharide | unp |
| MMP1085 | Methyltransferase (sugar 4) | Present | n.d. | Increased | Modified tetrasaccharide | unp |
|
| ||||||
| M. voltae | ||||||
| aglA | GTase (sugar 3) | Present | n.d. | Increased | Disaccharide | [115] |
| aglB | OTase | Absent | Non-motile | Increased | No glycan | [115] |
| aglC | GTase (sugar 2) | Absent | n.d. | Increased | Monosaccharide | [125] |
| aglK | GTase (sugar 2) | Absent | n.d. | Increased | Monosaccharide | [125] |
N. B.: M. maripaludis wild type N-linked glycan is a tetrasaccharide, M. voltae wild type N-linked glycan is a trisaccharide.
1Relative to level detected in parent strain.
2GTase: glycosyltransferase.
3OTase: oligosaccharyltransferase.
4n.d.: not determined.
5unp: unpublished data.
6. O-Glycosylation
In addition to N-glycosylation, the Hbt. salinarum and Hfx. volcanii S-layer glycoproteins also undergo O-glycosylation. In each case, a Thr-rich region upstream of the predicted membrane-spanning domain of the protein is decorated at numerous positions by galactose-glucose disaccharides, linked through the galactose subunit [13, 129]. Essentially nothing is presently known of the archaeal O-glycosylation process.
7. Lipid Modification
In addition to signal peptide cleavage and glycosylation, haloarchaeal S-layer glycoproteins also experience covalent posttranslational attachment of lipids. This was first shown when Hbt. salinarum cells were incubated with [3H]-mevalonate and other tritiated lipid tracers, leading to selective incorporation of the radiolabel into the S-layer glycoprotein [130]. The linked radioactive moiety was subsequently revealed by mass spectrometry to be a novel diphytanylglycerol phosphate. Although the precise location of the attached lipid has yet to be defined, a 28 kDa trypsin-generated fragment derived from the C-terminal region of the protein (residues 731–816) was shown to contain the linked group. In terms of attachment of the lipid, it is thought that phosphodiester-based linkage to either a S-layer glycoprotein Ser or Thr residue is responsible. Hence, it would appear that in addition to the single membrane-spanning domain located close to the C-terminus of the haloarchaeal S-layer glycoprotein, deduced from primary sequence analysis, a lipid moiety also anchors the protein to the membrane. Moreover, given sequence similarities in the same C-terminal region of the Hbt. salinarum, Hfx. volcanii and Haloarcula japonica S-layer glycoproteins [14, 129, 131], it is likely that the latter two similarly experience lipid modification [130]. Indeed, such lipid modification has been demonstrated in the case of the Hfx. volcanii S-layer glycoprotein [132, 133].
8. Importance of Flagellin and S-Layer Glycoprotein Posttranslational Modifications
The ability to generate deletion mutants of Hfx. volcanii, M. voltae and M. maripaludis as well as the availability of other molecular tools have allowed for the importance of posttranslational modifications of reporter proteins in these species to be addressed.
Haloferax volcanii —
In Hfx. volcanii, the use of deletion strains has provided considerable insight into the importance of N-glycosylation to the cell. Strains lacking the ability to perform N-glycosylation, due to the absence of the oligosaccharyltransferase, AglB, or only able to partially recruit the N-glycosylation pathway, due to an absence of other Agl proteins, present various phenotypes, including an S-layer of modified architecture showing increased susceptibility to proteolytic digestion, enhanced S-layer glycoprotein release into the growth medium, and slower growth in medium of increasing salt [119, 120, 122, 123]. Indeed, differential transcription of the various Agl proteins in response to differing growth conditions, reflected by reverse transcription or real time PCR, points to N-glycosylation as being an adaptive process in Hfx. volcanii.
Studies addressing the biogenesis of the Hfx. volcanii S-layer glycoprotein have also provided insight into the importance of lipid modification. Metabolic [35S] pulse-chase radiolabelling, together with the use of the ribosome-targeted antibiotic, anisomycin, revealed the S-layer glycoprotein to undergo a posttranslational maturation step on the outer surface of the plasma membrane, reflected as an increase in the hydrophobicity and apparent molecular weight of the protein [133]. Support for lipid modification as being responsible for S-layer glycoprotein maturation came from experiments showing that growth in the presence of [3H] mevalonic acid led to radiolabel being incorporated into the S-layer glycoprotein and that mevinolin, an inhibitor of 3-HMG-CoA reductase (responsible for converting acetyl-CoA into mevalonic acid), prevented the maturation of the S-layer glycoprotein [132].
Moreover, such lipid modification-based maturation does not occur in the absence of Mg2+ [133], required for maintaining haloarchaeal S-layer integrity [13, 26]. As the Hbt. salinarum S-layer glycoprotein also undergoes a similar lipid-based maturation step [132], this posttranslational modification may be common to S-layer glycoprotein biogenesis in other haloarchaea.
Methanococcus voltae and Methanococcus maripaludis —
The flagellins expressed by both M. voltae and M. maripaludis undergo two major posttranslational modifications necessary for their correct assembly into flagella, namely signal peptide cleavage and N-linked glycosylation. Disruption or deletion of the signal peptidase gene (flaK) results in cells that are no longer able to assemble flagella, with the unprocessed flagellins remaining in the cytoplasmic membrane [76]. In the case of M. maripaludis, these cells are, however, still piliated, since the type IV pilin-like proteins are processed by a separate prepilin peptidase-like enzyme, EppA [105]. Deletion of eppA in a cell that is already deleted for flaK results in the appearance of nonflagellated and nonpiliated cells (K. F. J., unpublished results).
Early studies pointed to a critical role for glycosylation in flagella structure in Methanococcus. When incubated with bacitracin, a known inhibitor of glycosylation, Methanococcus deltae cells became nonflagellated, accompanied by a decrease in apparent molecular weight of the flagellins, suggestive of under-glycosylation [134]. More recently, in both M. voltae and M. maripaludis, it was shown that at least a two sugar-member glycan must be attached to flagellin subunits for proper assembly of the protein into flagella filaments [115, 116]. Deletion of genes involved in the N-glycosylation pathway of both M. voltae and M. maripaludis result in flagellin subunits that migrate faster on SDS-PAGE, with the enhanced migration corresponding incrementally to the degree of truncation of the glycan [115, 116]. Motility assays using semisolid agar demonstrated that strains harboring deletions of genes in this pathway that are still able to assemble flagella displayed impaired swimming capabilities, as compared to cells able to produce the native N-linked glycan [116]. Finally, deletion of a single gene, that is, MMP0350, assigned as an acetyltransferase necessary for the biosynthesis of the second sugar of the M. maripaludis N-linked glycan, resulted in defects in both flagellation and piliation. Since the glycan consisted of only a single sugar in this mutant, the fact that the cells were nonflagellated was not unexpected. However, further examination revealed that while these mutants were generally nonpiliated, apparently intact pili were found in the culture supernatants, indicating that a defect in pili anchoring had occurred [117].
9. Conclusions
As the research spotlight begins to shift from the genome to the proteome, it is becoming clear that numerous archaeal proteins experience posttranslational modifications [135–139]. As discussed here, the availability of well-characterized reporters of protein processing events, such as haloarchaeal and methanoarchaeal S-layer glycoproteins and flagellins, offer excellent models in studies attempting to dissect the pathways responsible for such modifications. Along with the identification of additional reporter proteins, it will be possible to consider questions related to the heterogeneity associated with a given modification, such as differential or modulated glycosylation. Moreover, with the development of appropriate in vitro assays for these novel reporters, future efforts can address the importance of posttranslational modifications to enzyme function, stability and other traits.
Acknowledgments
J. Eichler is supported by grants from the Israel Science Foundation (Grant 30/07) and the US Army Research Office (Grant W911NF-07-1-0260). K. F. Jarrell is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- 1.Woese CR. The archaeal concept and the world it lives in: a retrospective. Photosynthesis Research. 2004;80(1–3):361–372. doi: 10.1023/B:PRES.0000030445.04503.e6. [DOI] [PubMed] [Google Scholar]
- 2.Fox GE, Stackebrandt E, Hespell RB, et al. The phylogeny of prokaryotes. Science. 1980;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 3.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zillig W. Comparative biochemistry of Archaea and Bacteria. Current Opinion in Genetics and Development. 1991;1(4):544–551. doi: 10.1016/s0959-437x(05)80206-0. [DOI] [PubMed] [Google Scholar]
- 5.Jarrell KF, VanDyke DJ, Wu J. Archaeal flagella and pili. In: Jarrell KF, editor. Pili and Flagella: Current Research and Future Trends. Norfolk, UK: Caister Academic Press; 2009. pp. 215–234. [Google Scholar]
- 6.Kandler O, Konig H. The Bacteria. A Treatise on Structure and Function. Orlando, Fla, USA: Academic Press; 1985. Cell envelopes of archaebacteria. [Google Scholar]
- 7.Magrum LJ, Luehrsen KR, Woese CR. Are extreme halophiles actually “bacteria”? Journal of Molecular Evolution. 1978;11(1):1–8. doi: 10.1007/BF01768019. [DOI] [PubMed] [Google Scholar]
- 8.Ng SYM, Zolghadr B, Driessen AJM, Albers S-V, Jarrell KF. Cell surface structures of archaea. Journal of Bacteriology. 2008;190(18):6039–6047. doi: 10.1128/JB.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickell S, Hegerl R, Baumeister W, Rachel R. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. Journal of Structural Biology. 2003;141(1):34–42. doi: 10.1016/s1047-8477(02)00581-6. [DOI] [PubMed] [Google Scholar]
- 10.Moissl C, Rachel R, Briegel A, Engelhardt H, Huber R. The unique structure of archaeal ’hami’, highly complex cell appendages with nano-grappling hooks. Molecular Microbiology. 2005;56(2):361–370. doi: 10.1111/j.1365-2958.2005.04294.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang YA, Yu X, Ng SYM, Jarrell KF, Egelman EH. The structure of an archaeal pilus. Journal of Molecular Biology. 2008;381(2):456–466. doi: 10.1016/j.jmb.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nature Reviews Microbiology. 2008;6(6):466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 13.Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium . The Journal of Biological Chemistry. 1976;251(7):2005–2014. [PubMed] [Google Scholar]
- 14.Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. The Journal of Biological Chemistry. 1987;262(20):9724–9729. [PubMed] [Google Scholar]
- 15.Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. The Journal of Biological Chemistry. 1992;267(12):8182–8185. [PubMed] [Google Scholar]
- 16.Sumper M. Halobacterial glycoprotein biosynthesis. Biochimica et Biophysica Acta. 1987;906(1):69–79. doi: 10.1016/0304-4157(87)90005-0. [DOI] [PubMed] [Google Scholar]
- 17.Yurist-Doutsch S, Chaban B, VanDyke DJ, Jarrell KF, Eichler J. Sweet to the extreme: protein glycosylation in Archaea. Molecular Microbiology. 2008;68(5):1079–1084. doi: 10.1111/j.1365-2958.2008.06224.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng SYM, Chaban B, VanDyke DJ, Jarrell KF. Archaeal signal peptidases. Microbiology. 2007;153(2):305–314. doi: 10.1099/mic.0.2006/003087-0. [DOI] [PubMed] [Google Scholar]
- 19.Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Current Opinion in Structural Biology. 2008;18(5):544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Pohlschröder M, Giménez MI, Jarrell KF. Protein transport in Archaea: sec and twin arginine translocation pathways. Current Opinion in Microbiology. 2005;8(6):713–719. doi: 10.1016/j.mib.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Houwink AL. Flagella, gas vacuoles and cell-wall structure in Halobacterium halobium; an electron microscope study. Journal of General Microbiology. 1956;15:146–150. doi: 10.1099/00221287-15-1-146. [DOI] [PubMed] [Google Scholar]
- 22.Stoeckenius W, Rowen R. A morphological study of Halobacterium halobium and its lysis in media of low salt concentration. Journal of Cell Biology. 1967;34(1):365–393. doi: 10.1083/jcb.34.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steensland H, Larsen H. A study of the cell envelope of the halobacteria. Journal of General Microbiology. 1969;55(3):325–336. doi: 10.1099/00221287-55-3-325. [DOI] [PubMed] [Google Scholar]
- 24.Kirk RG, Ginzburg M. Ultrastructure of two species of Halobacterium . Journal of Ultrasructure Research. 1972;41(1-2):80–94. doi: 10.1016/s0022-5320(72)90040-8. [DOI] [PubMed] [Google Scholar]
- 25.Blaurock AE, Stoeckenius W, Oesterhelt D, Scherphof GL. Structure of the cell envelope of Halobacterium halobium . Journal of Cell Biology. 1976;71(1):1–22. doi: 10.1083/jcb.71.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessel M, Wildhaber I, Cohen S, Baumeister W. Three-dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead Sea. The EMBO Journal. 1988;7:1549–1554. doi: 10.1002/j.1460-2075.1988.tb02974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trachtenberg S, Pinnick B, Kessel M. The cell surface glycoprotein layer of the extreme halophile Halobacterium salinarum and its relation to Haloferax volcanii: cryo-electron tomography of freeze-substituted cells and projection studies of negatively stained envelopes. Journal of Structural Biology. 2000;130(1):10–26. doi: 10.1006/jsbi.2000.4215. [DOI] [PubMed] [Google Scholar]
- 28.Kandler O, Konig H. Cell envelopes of archaea: structure and chemistry. In: Kates M, Kusher DJ, Matheson AT, editors. The Biochemistry of Archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 223–259. [Google Scholar]
- 29.Konig H, Stetter KO, Postulka W, Klink F. Studies on archaebacterial S-layers. Systematic and Applied Microbiology. 1986;7:300–309. [Google Scholar]
- 30.Nusser E, Konig H. S layer studies on three species of Methanococcus living at different temperatures. Canadian Journal of Microbiology. 1987;33(3):256–261. [Google Scholar]
- 31.Akca E, Claus H, Schultz N, et al. Genes and derived amino acid sequences of S-layer proteins from mesophilic, thermophilic, and extremely thermophilic methanococci. Extremophiles. 2002;6(5):351–358. doi: 10.1007/s00792-001-0264-1. [DOI] [PubMed] [Google Scholar]
- 32.Koval SF, Jarrell KF. Ultrastructure and biochemistry of the cell wall of Methanococcus voltae . Journal of Bacteriology. 1987;169(3):1298–1306. doi: 10.1128/jb.169.3.1298-1306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voisin S, Houliston RS, Kelly J, et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae . The Journal of Biological Chemistry. 2005;280(17):16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- 34.Patel GB, Choquet CG, Nash JHE, Sprott GD. Formation and regeneration of Methanococcus voltae protoplasts. Applied and Environmental Microbiology. 1993;59(1):27–33. doi: 10.1128/aem.59.1.27-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel GB, Nash JHE, Agnew BJ, Sprott GD. Natural and electroporation-mediated transformation of Methanococcus voltae protoplasts. Applied and Environmental Microbiology. 1994;60(3):903–907. doi: 10.1128/aem.60.3.903-907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karcher U, Schroder H, Haslinger E, et al. Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus . The Journal of Biological Chemistry. 1993;268(36):26821–26826. [PubMed] [Google Scholar]
- 37.Sowers KR, Boone JE, Gunsalus RP. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Applied and Environmental Microbiology. 1993;59(11):3832–3839. doi: 10.1128/aem.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheong G-W, Cejka Z, Peters J, Stetter KO, Baumeister W. The surface protein layer of Methanoplanus limicola: three-dimensional structure and chemical characterization. Systematic and Applied Microbiology. 1991;14(3):209–217. [Google Scholar]
- 39.Cheong G-W, Guckenberger R, Fuchs K-H, Gross H, Baumeister W. The structure of the surface layer of Methanoplanus limicola obtained by a combined electron microscopy and scanning tunneling microscopy approach. Journal of Structural Biology. 1993;111(2):125–134. [Google Scholar]
- 40.Firtel M, Southam G, Harauz G, Beveridge TJ. Characterization of the cell wall of the sheathed methanogen Methanospirillum hungatei GP1 as an S layer. Journal of Bacteriology. 1993;175(23):7550–7560. doi: 10.1128/jb.175.23.7550-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart M, Beveridge TJ, Sprott GD. Crystalline order to high resolution in the sheath of Methanospirillum hungatei: a cross-beta structure. Journal of Molecular Biology. 1985;183(3):509–515. doi: 10.1016/0022-2836(85)90019-1. [DOI] [PubMed] [Google Scholar]
- 42.Beveridge TJ, Sprott GD, Whippey P. Ultrastructure, inferred porosity, and gram-staining character of Methanospirillum hungatei filament termini describe a unique cell permeability for this archaeobacterium. Journal of Bacteriology. 1991;173(1):130–140. doi: 10.1128/jb.173.1.130-140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pum D, Messner P, Sleytr UB. Role of the S layer in morphogenesis and cell division of the archaebacterium Methanocorpusculum sinense . Journal of Bacteriology. 1991;173(21):6865–6873. doi: 10.1128/jb.173.21.6865-6873.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarrell KF, Ng SY, Chaban B. Flagellation and chemotaxis. In: Cavicchioli R, editor. Archaea: Molecular and Cellular Biology. Washington, DC, USA: ASM Press; 2007. pp. 385–410. [Google Scholar]
- 45.Jarrell KF, Bayley DP, Kostyukova AS. The archaeal flagellum: a unique motility structure. Journal of Bacteriology. 1996;178(17):5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng SYM, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. Journal of Molecular Microbiology and Biotechnology. 2006;11(3-5):167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 47.Kalmokoff ML, Jarrell KF. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae . Journal of Bacteriology. 1991;173(22):7113–7125. doi: 10.1128/jb.173.22.7113-7125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bardy SL, Jarrell KF. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiology Letters. 2002;208(1):53–59. doi: 10.1111/j.1574-6968.2002.tb11060.x. [DOI] [PubMed] [Google Scholar]
- 49.Bardy SL, Mori T, Komoriya K, Aizawa S-I, Jarrell KF. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae . Journal of Bacteriology. 2002;184(19):5223–5233. doi: 10.1128/JB.184.19.5223-5233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faguy DM, Koval SF, Jarrell KF. Physical characterization of the flagella and flagellins from Methanospirillum hungatei . Journal of Bacteriology. 1994;176(24):7491–7498. doi: 10.1128/jb.176.24.7491-7498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southam G, Kalmokoff ML, Jarrell KF, Koval SF, Beveridge TJ. Isolation, characterization, and cellular insertion of the flagella from two strains of the archaebacterium Methanospirillum hungatei . Journal of Bacteriology. 1990;172(6):3221–3228. doi: 10.1128/jb.172.6.3221-3228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruden D, Sparling R, Markovetz AJ. Isolation and ultrastructure of the flagella of Methanococcus thermolithotrophicus and Methanospirillum hungatei . Applied and Environmental Microbiology. 1989;55(6):1414–1419. doi: 10.1128/aem.55.6.1414-1419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alam M, Oesterhelt D. Morphology, function and isolation of halobacterial flagella. Journal of Molecular Biology. 1984;176(4):459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- 54.Alam M, Oesterhelt D. Purification, reconstitution and polymorphic transition of halobacterial flagella. Journal of Molecular Biology. 1987;194(3):495–499. doi: 10.1016/0022-2836(87)90677-2. [DOI] [PubMed] [Google Scholar]
- 55.Gerl L, Deutzmann R, Sumper M. Halobacterial flagellins are encoded by a multigene family Identification of all five gene products. FEBS Letters. 1989;244(1):137–140. doi: 10.1016/0014-5793(89)81179-2. [DOI] [PubMed] [Google Scholar]
- 56.Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum . Molecular Microbiology. 2001;41(3):653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- 57.Pyatibratov MG, Beznosov SN, Rachel R, et al. Alternative flagellar filament types in the haloarchaeon Haloarcula marismortui . Canadian Journal of Microbiology. 2008;54(10):835–844. doi: 10.1139/w08-076. [DOI] [PubMed] [Google Scholar]
- 58.Tripepi M, Imam S, Pohlschroder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. Journal of Bacteriology. 2010;192(12):3093–3102. doi: 10.1128/JB.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szabó Z, Sani M, Groeneveld M, et al. Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus . Journal of Bacteriology. 2007;189(11):4305–4309. doi: 10.1128/JB.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pyatibratov MG, Leonard K, Tarasov VY, Fedorov OV. Two immunologically distinct types of protofilaments can be identified in Natrialba magadii flagella. FEMS Microbiology Letters. 2002;212(1):23–27. doi: 10.1111/j.1574-6968.2002.tb11239.x. [DOI] [PubMed] [Google Scholar]
- 61.Serganova I, Ksenzenko V, Serganov A, et al. Sequencing of flagellin genes from Natrialba magadii provides new insight into evolutionary aspects of archaeal flagellins. Journal of Bacteriology. 2002;184(1):318–322. doi: 10.1128/JB.184.1.318-322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagahisa K, Ezaki S, Fujiwara S, Imanaka T, Takagi M. Sequence and transcriptional studies of five clustered flagellin genes from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. FEMS Microbiology Letters. 1999;178(1):183–190. doi: 10.1111/j.1574-6968.1999.tb13776.x. [DOI] [PubMed] [Google Scholar]
- 63.Näther DJ, Rachel R, Wanner G, Wirth R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. Journal of Bacteriology. 2006;188(19):6915–6923. doi: 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaban B, Ng SYM, Kanbe M, et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis . Molecular Microbiology. 2007;66(3):596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- 65.Ellen AF, Zolghadr B, Driessen AJM, Albers SV. Shaping the Archaeal Cell Envelope. Archaea. 2010;2010 doi: 10.1155/2010/608243. Article ID 608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Rosario RCH, Diener F, Diener M, Oesterhelt D. The steady-state phase distribution of the motor switch complex model of Halobacterium salinarum . Mathematical Biosciences. 2009;222(2):117–126. doi: 10.1016/j.mbs.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 67.Nutsch T, Oesterhelt D, Gilles ED, Marwan W. A quantitative model of the switch cycle of an archaeal flagellar motor and its sensory control. Biophysical Journal. 2005;89(4):2307–2323. doi: 10.1529/biophysj.104.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marwan W, Alam M, Oesterhelt D. Rotation and switching of the flagellar motor assembly in Halobacterium halobium . Journal of Bacteriology. 1991;173(6):1971–1977. doi: 10.1128/jb.173.6.1971-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faguy DM, Jarrell KF. A twisted tale: the origin and evolution of motility and chemotaxis in prokaryotes. Microbiology. 1999;145(2):279–281. doi: 10.1099/13500872-145-2-279. [DOI] [PubMed] [Google Scholar]
- 70.Blair DF. Flagellar movement driven by proton translocation. FEBS Letters. 2003;545(1):86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- 71.Streif S, Staudinger WF, Marwan W, Oesterhelt D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. Journal of Molecular Biology. 2008;384(1):1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 72.Bradley DE. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Canadian Journal of Microbiology. 1980;26(2):146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 73.Cohen-Krausz S, Trachtenberg S. The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design. Journal of Molecular Biology. 2008;375(4):1113–1124. doi: 10.1016/j.jmb.2007.10.048. [DOI] [PubMed] [Google Scholar]
- 74.Cohen-Krausz S, Trachtenberg S. The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. Journal of Molecular Biology. 2002;321(3):383–395. doi: 10.1016/s0022-2836(02)00616-2. [DOI] [PubMed] [Google Scholar]
- 75.Trachtenberg S, Cohen-Krausz S. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. Journal of Molecular Microbiology and Biotechnology. 2006;11(3–5):208–220. doi: 10.1159/000094055. [DOI] [PubMed] [Google Scholar]
- 76.Bardy SL, Jarrell KF. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae . Molecular Microbiology. 2003;50(4):1339–1347. doi: 10.1046/j.1365-2958.2003.03758.x. [DOI] [PubMed] [Google Scholar]
- 77.Bayley DP, Jarrell KF. Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. Journal of Molecular Evolution. 1998;46(3):370–373. [PubMed] [Google Scholar]
- 78.Peabody CR, Chung YJ, Yen M-R, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149(11):3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 79.Macnab RM. How bacteria assemble flagella. Annual Review of Microbiology. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 80.Jarrell KF, Bayley DP, Florian V, Klein A. Isolation and characterization of insertional mutations in flagellin genes in the archaeon Methanococcus voltae . Molecular Microbiology. 1996;20(3):657–666. doi: 10.1046/j.1365-2958.1996.5371058.x. [DOI] [PubMed] [Google Scholar]
- 81.Tarasov VY, Pyatibratov MG, Tang S-L, Dyall-Smith M, Fedorov OV. Role of flagellins from A and B loci in flagella formation of Halobacterium salinarum . Molecular Microbiology. 2000;35(1):69–78. doi: 10.1046/j.1365-2958.2000.01677.x. [DOI] [PubMed] [Google Scholar]
- 82.Beznosov SN, Pyatibratov MG, Fedorov OV. On the multicomponent nature of Halobacterium salinarum flagella. Microbiology. 2007;76(4):435–441. [PubMed] [Google Scholar]
- 83.Albers S-V, Driessen AJM. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus . Microbiology. 2005;151(3):763–773. doi: 10.1099/mic.0.27699-0. [DOI] [PubMed] [Google Scholar]
- 84.Thomas NA, Jarrell KF. Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. Journal of Bacteriology. 2001;183(24):7154–7164. doi: 10.1128/JB.183.24.7154-7164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas NA, Mueller S, Klein A, Jarrell KF. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Molecular Microbiology. 2002;46(3):879–887. doi: 10.1046/j.1365-2958.2002.03220.x. [DOI] [PubMed] [Google Scholar]
- 86.Schlesner M, Miller A, Streif S, et al. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiology. 2009;9, article 56 doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Logan SM. Flagellar glycosylation—a new component of the motility repertoire? Microbiology. 2006;152(5):1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- 88.Kelly J, Logan SM, Jarrell KF, VanDyke DJ, Vinogradov E. A novel N-linked flagellar glycan from Methanococcus maripaludis . Carbohydrate Research. 2009;344(5):648–653. doi: 10.1016/j.carres.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 89.Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulfated glycoproteins. The Journal of Biological Chemistry. 1985;260(28):15180–15185. [PubMed] [Google Scholar]
- 90.Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annual Review of Microbiology. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 91.Anderson JK, Smith TG, Hoover TR. Sense and sensibility: flagellum-mediated gene regulation. Trends in Microbiology. 2010;18(1):30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. Sensing wetness: a new role for the bacterial flagellum. EMBO Journal. 2005;24(11):2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schopf S, Wanner G, Rachel R, Wirth R. An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri . Archives of Microbiology. 2008;190(3):371–377. doi: 10.1007/s00203-008-0371-9. [DOI] [PubMed] [Google Scholar]
- 94.Zolghadr B, Kling A, Koerdt A, Driessen AJM, Rachel R, Albers S-V. Appendage-mediated surface adherence of Sulfolobus solfataricus . Journal of Bacteriology. 2010;192(1):104–110. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal peptidases. Chemical Reviews. 2002;102(12):4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 96.Ng SYM, Jarrell KF. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae . Journal of Bacteriology. 2003;185(20):5936–5942. doi: 10.1128/JB.185.20.5936-5942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bardy SL, Ng SYM, Carnegie DS, Jarrell KF. Site-directed mutagenesis analysis of amino acids critical for activity of the type I signal peptidase of the archaeon Methanococcus voltae . Journal of Bacteriology. 2005;187(3):1188–1191. doi: 10.1128/JB.187.3.1188-1191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fink-Lavi E, Eichler J. Identification of residues essential for the catalytic activity of Sec11b, one of the two type I signal peptidases of Haloferax volcanii . FEMS Microbiology Letters. 2008;278(2):257–260. doi: 10.1111/j.1574-6968.2007.01000.x. [DOI] [PubMed] [Google Scholar]
- 99.Tschantz WR, Sung M, Delgado-Partin VM, Dalbey RE. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. The Journal of Biological Chemistry. 1993;268(36):27349–27354. [PubMed] [Google Scholar]
- 100.Paetzel M, Dalbey RE, Strynadka NCJ. Crystal structure of a bacterial signal peptidase apoenzyme. Implications for signal peptide binding and the Ser-Lys dyad mechanism. The Journal of Biological Chemistry. 2002;277(11):9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 101.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Science. 1997;6(6):1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.VanValkenburgh C, Chen X, Mullins C, Fang H, Green N. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. The Journal of Biological Chemistry. 1999;274(17):11519–11525. doi: 10.1074/jbc.274.17.11519. [DOI] [PubMed] [Google Scholar]
- 103.Fine A, Irihimovitch V, Dahan I, Konrad Z, Eichler J. Cloning, expression, and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii . Journal of Bacteriology. 2006;188(5):1911–1919. doi: 10.1128/JB.188.5.1911-1919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Albers S-V, Szabó Z, Driessen AJM. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. Journal of Bacteriology. 2003;185(13):3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Szabó Z, Stahl AO, Albers S-V, Kissinger JC, Driessen AJM, Pohlschröder M. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. Journal of Bacteriology. 2007;189(3):772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thomas NA, Chao ED, Jarrell KF. Identification of amino acids in the leader peptide of Methanococcus voltae preflagellin that are important in posttranslational processing. Archives of Microbiology. 2001;175(4):263–269. doi: 10.1007/s002030100254. [DOI] [PubMed] [Google Scholar]
- 107.Ng SYM, VanDyke DJ, Chaban B, et al. Different minimal signal peptide lengths recognized by the archaeal prepilin-like peptidases FlaK and PibD. Journal of Bacteriology. 2009;191(21):6732–6740. doi: 10.1128/JB.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szabó Z, Albers S-V, Driessen AJM. Active-site residues in the type IV prepilin peptidase homologue PibD from the archaeon Sulfolobus solfataricus . Journal of Bacteriology. 2006;188(4):1437–1443. doi: 10.1128/JB.188.4.1437-1443.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Albers S-V, Pohlschröder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13:403–410. doi: 10.1007/s00792-009-0241-7. [DOI] [PubMed] [Google Scholar]
- 110.Müller DW, Meyer C, Gürster S, et al. The Iho670 fibers of Ignicoccus hospitalis: a new type of archaeal cell surface appendage. Journal of Bacteriology. 2009;191(20):6465–6468. doi: 10.1128/JB.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koncewicz MA. Glycoproteins in the cell envelope of Halobacterium halobium . Biochemical Journal. 1972;128(4):p. 124. doi: 10.1042/bj1280124p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mescher MF, Strominger JL, Watson SW. Protein and carbohydrate composition of the cell envelope of Halobacterium salinarium . Journal of Bacteriology. 1974;120(2):945–954. doi: 10.1128/jb.120.2.945-954.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Eichler J, Adams MWW. Posttranslational protein modification in Archaea. Microbiology and Molecular Biology Reviews. 2005;69(3):393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abu-Qarn M, Eichler J. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Molecular Microbiology. 2006;61(2):511–525. doi: 10.1111/j.1365-2958.2006.05252.x. [DOI] [PubMed] [Google Scholar]
- 115.Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Molecular Microbiology. 2006;61(1):259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- 116.Vandyke DJ, Wu J, Logan SM, et al. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis . Molecular Microbiology. 2009;72(3):633–644. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- 117.VanDyke DJ, Wu J, Ng SYM, et al. Identification of a putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis . Journal of Bacteriology. 2008;190(15):5300–5307. doi: 10.1128/JB.00474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nature Reviews Microbiology. 2005;3(3):225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 119.Abu-Qarn M, Giordano A, Battaglia F, et al. Identification of AglE, a second glycosyltransferase involved in N glycosylation of the Haloferax volcanii S-layer glycoprotein. Journal of Bacteriology. 2008;190(9):3140–3146. doi: 10.1128/JB.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yurist-Doutsch S, Abu-Qarn M, Battaglia F, et al. AglF, aglG and aglI, novel members of a gene island involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Molecular Microbiology. 2008;69(5):1234–1245. doi: 10.1111/j.1365-2958.2008.06352.x. [DOI] [PubMed] [Google Scholar]
- 121.Yurist-Doutsch S, Eichler J. Manual annotation, transcriptional analysis, and protein expression studies reveal novel genes in the agl cluster responsible for N glycosylation in the halophilic archaeon Haloferax volcanii . Journal of Bacteriology. 2009;191(9):3068–3075. doi: 10.1128/JB.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yurist-Doutsch S, Magidovich H, Ventura VV, Hitchen PG, Dell A, Eichler J. N-glycosylation in Archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Molecular Microbiology. 2010;75(4):1047–1058. doi: 10.1111/j.1365-2958.2009.07045.x. [DOI] [PubMed] [Google Scholar]
- 123.Abu-Qarn M, Yurist-Doutsch S, Giordano A, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. Journal of Molecular Biology. 2007;374(5):1224–1236. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 124.Magidovich H, Yurist-Doutsch S, Konrad Z, et al. AglP is a S-adenosyl-L-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii . Molecular Microbiology. 2010;76(1):190–199. doi: 10.1111/j.1365-2958.2010.07090.x. [DOI] [PubMed] [Google Scholar]
- 125.Chaban B, Logan SM, Kelly JF, Jarrell KF. AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae . Journal of Bacteriology. 2009;91(1):187–195. doi: 10.1128/JB.00885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shams-Eldin H, Chaban B, Niehus S, Schwarz RT, Jarrell KF. Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementation in Saccharomyces cerevisiae . Journal of Bacteriology. 2008;190(6):2217–2220. doi: 10.1128/JB.01778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. Journal of Bacteriology. 2005;187(3):972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Namboori SC, Graham DE. Acetamido sugar biosynthesis in the Euryarchaea. Journal of Bacteriology. 2008;190(8):2987–2996. doi: 10.1128/JB.01970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii . Journal of Bacteriology. 1990;172(12):7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kikuchi A, Sagami H, Ogura K. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium . The Journal of Biological Chemistry. 1999;274(25):18011–18016. doi: 10.1074/jbc.274.25.18011. [DOI] [PubMed] [Google Scholar]
- 131.Wakai H, Nakamura S, Kawasaki H, et al. Cloning and sequencing of the gene encoding the cell surface glycoprotein of Haloarcula japonica strain TR-1. Extremophiles. 1997;1(1):29–35. doi: 10.1007/s007920050012. [DOI] [PubMed] [Google Scholar]
- 132.Konrad Z, Eichler J. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochemical Journal. 2002;366(3):959–964. doi: 10.1042/BJ20020757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eichler J. Post-translational modification of the S-layer glycoprotein occurs following translocation across the plasma membrane of the haloarchaeon Haloferax volcanii . European Journal of Biochemistry. 2001;268(15):4366–4373. doi: 10.1046/j.1432-1327.2001.02361.x. [DOI] [PubMed] [Google Scholar]
- 134.Bayley DP, Kalmokoff ML, Jarrell KF. Effect of bacitracin on flagellar assembly and presumed glycosylation of the flagellins of Methanococcus deltae . Archives of Microbiology. 1993;160(3):179–185. [Google Scholar]
- 135.Falb M, Aivaliotis M, Garcia-Rizo C, et al. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. Journal of Molecular Biology. 2006;362(5):915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 136.Aivaliotis M, Gevaert K, Falb M, et al. Large-scale identification of N-terminal peptides in the halophilic Archaea Halobacterium salinarum and Natronomonas pharaonis . Journal of Proteome Research. 2007;6(6):2195–2204. doi: 10.1021/pr0700347. [DOI] [PubMed] [Google Scholar]
- 137.Saunders NFW, Ng C, Raftery M, Guilhaus M, Goodchild A, Cavicchioli R. Proteomic and computational analysis of secreted proteins with type I signal peptides from the antarctic archaeon Methanococcoides burtonii . Journal of Proteome Research. 2006;5(9):2457–2464. doi: 10.1021/pr060220x. [DOI] [PubMed] [Google Scholar]
- 138.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase a mutant of the haloarchaeon Haloferax volcanii . Journal of Bacteriology. 2008;190(1):193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Francoleon DR, Boontheung P, Yang Y, et al. S-layer, surface-accessible, and concanavalin a binding proteins of Methanosarcina acetivorans and Methanosarcina mazei . Journal of Proteome Research. 2009;8(4):1972–1982. doi: 10.1021/pr800923e. [DOI] [PMC free article] [PubMed] [Google Scholar]