Summary
Neuronal migration is a fundamental component of brain development whose failure is associated with various neurological and psychiatric disorders. Reelin is essential for the stereotypical inside-out sequential lamination of the neocortex, but the molecular mechanisms of its action still remain unclear. Here we show that regulation of Notch activity plays an important part in Reelin signal-dependent neuronal migration. We found that Reelin-deficient mice have reduced levels of the cleaved form of Notch intracellular domain (Notch ICD) and that loss of Notch signaling in migrating neurons results in migration and morphology defects. Further, overexpression of Notch ICD mitigates the laminar and morphological abnormalities of migrating neurons in Reeler. Finally, our in vitro biochemical studies show that Reelin signaling inhibits Notch ICD degradation via Dab1. Together, our results indicate that neuronal migration in the developing cerebral cortex requires a Reelin-Notch interaction.
Keywords: migration, cerebral cortex, Reelin, Notch
Introduction
Cerebral cortical development is comprised of multiple processes, including neuronal production from neuroepithelium, migration of neurons to their proper positions and neuronal maturation (Rakic, 1988; Kriegstein and Noctor, 2004). These steps are tightly controlled by various molecular pathways (Caviness and Rakic 1978; Walsh and Goffinet, 2000; Lambert and Goffinet, 2001; Olson and Walsh, 2002; Bielas and Gleeson, 2004; LoTurco and Bai, 2006; Ayala et al., 2007; Kawauchi and Hoshino, 2008). For example, the precise positioning of radially migrating neurons is critically controlled by the Reelin signaling pathway and is indispensable for forming a stereotypical inside-out 6-layered pattern (Bar et al., 2000; Magdaleno and Curran, 2001; Rice and Curran, 2001; Tissir and Goffinet, 2003; Soriano and Del Rio, 2005; Kanatani et al., 2005; Forster et al., 2006; D’Arcangelo, 2006).
Reelin deficiency (Reeler) is characterized by an inverted lamination of the neocortex, and the human Reelin (RELN) mutation has been linked to lissencephaly, autism and other disorders (Hong et al., 2000; Zaki et al., 2007). Reelin encodes an extracellular matrix-associated glycoprotein that is secreted by Cajal-Retzius cells in the developing cerebral cortex. Very low density lipoprotein receptor (Vldlr) and apolipoprotein E receptor type2 (ApoER2) are canonical Reelin-binding receptors that subsequently activate intracellular Dab1 (Sheldon et al., 1997; Howell et al., 1997; Ware et al., 1997; Rice et al., 1998; Trommsdorff et al., 1999; Hiesberger et al., 1999; Howell et al., 1999; D’Arcangelo et al., 1999; Howell et al., 2000) and mediate divergent roles in neuronal migration (Hack et al., 2007). Dab1 interacts with multiple molecules, but most of these interactions have yet to be examined formally in migrating neurons (Bock et al., 2003; Ballif et al., 2004, Suetsugu et al., 2004; Chen et al., 2004; Pramatarova et al., 2003, 2008; Jossin and Goffinet, 2007). Thus the underlying molecular mechanisms of Reelin signaling that contribute to Reeler pathogenesis remain elusive.
Notch signaling represents another molecular pathway that is integral to cortical development. Delta and Serrate (known as Jagged in mammals) ligand binding to Notch receptors causes proteolytic release of the Notch ICD, the active form of Notch, which translocates to the nucleus and induces transcription of multiple target genes by forming a transcriptional complex with Rbpj (also known as CBF-1), a transcriptional factor which mediates canonical Notch signaling. This pathway has well-characterized roles during neurogenesis including cell elimination by controlling apoptosis and dendrite morphogenesis (reviewed by Yoon and Gaiano, 2005; Louvi and Artavanis-Tsakonas, 2006).
Previous studies have shown that Notch1 protein strongly localizes in the nuclei of cortical neurons as they accumulate beneath the marginal zone (MZ) (Sestan et al., 1999; Redmond et al., 2000), which consists of Reelin-expressing Cajal-Retzius cells. Although a Reelin homologue has not been identified in invertebrates including Drosophila, it also has been shown that Disabled (a Drosophila homologue of mammalian Dab1) binds Notch in vitro (Giniger, 1998; Le Gall and Giniger, 2004; Le Gall et al., 2008). Furthermore, reduction of a Notch downstream gene has been reported in Reeler mutant mice (Baba et al., 2006). These led us to hypothesize that Notch may play a role in Reelin-regulated lamination of the mammalian neocortex. In the present study, we provide evidence for a Reelin and Notch signaling pathway interaction that regulates neuronal migration during cerebral cortical development.
Results
Notch activity in migrating neurons is reduced in Reeler cerebral cortex
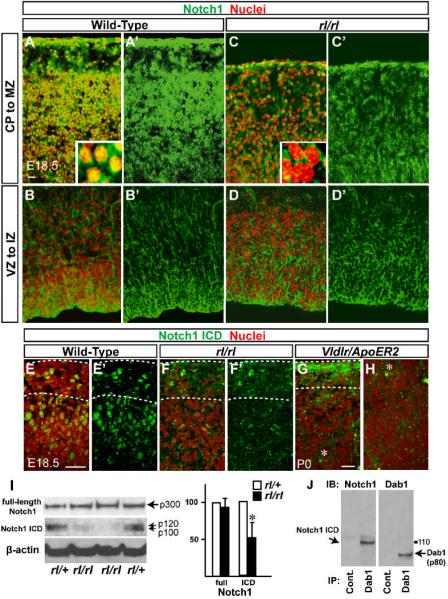
To test our hypothesis, we first examined whether the expression pattern of Notch1 is altered in Reelin-deficient cortex during the peak period of radial migration to layers II and III of the developing cortical plate (CP) by immunostaining. Using an antibody targeted against the Notch1 intracellular domain, we observed strong expression of Notch1 in the nuclei of wild-type cortical neurons beneath the MZ as previously reported (Sestan et al., 1999; Redmond et al., 2000), which gradually became weaker toward lower layers and the intermediate zone (IZ) (Figures 1A and 1A’). Strikingly, nuclear Notch1 expression is severely reduced in Reeler (rl/rl, Figures 1C and 1C’) Upon stimulation by its ligands, the Notch receptor undergoes several cleavages to become its activated form which translocates to the nucleus. Since the Notch1 antibody recognizes the intracellular domain of both cleaved (active) and uncleaved (non-active) forms of Notch1 gene products, nuclear Notch1 label presumably represents cleaved Notch1 ICD. In contrast, cytoplasmic expression of Notch1 in Reeler appeared to be comparable to that in wild-type (Figures 1A-1D’). To confirm the reduction of Notch1 ICD in Reeler cortex, we performed immunostaining using a different antibody that specifically recognizes the Notch1 ICD form (Val 1744). The presence of nuclear Notch1 ICD was strong in the cortex of wild-type, but dramatically reduced in Reeler mice (Figures 1E-1F’). Reduced Notch1 ICD was also observed in the Vldlr/ApoER2 Reelin signaling receptors double knockout (dKO) (Figures 1G and 1H) and signaling mediator Dab1 knockout (Scrambler)(data not shown).
Figure 1. Nuclear Notch ICD is reduced in cortical neurons in Reeler.
(A-H) Nuclei (red) and Notch1 (green in A-D’, antigen is Notch1 intracellular domain) and Notch1 ICD cleaved form (green in E-H) immunostaining in the cortex of wild-type and mutants as indicated. Higher magnifications are shown in the insets. Dashed lines in (E-G) indicate the pial surface and the border between CP and MZ in wild-type or superplate (SPP) in the mutants. (G) and (H) show upper CP and lower CP/IZ, respectively (the asterisk indicate the same cell). Note that Notch1 ICD expression at the lower CP (inverted layer II/III) is significantly decreased in Vldlr/ApoER2 mutant (H) which is similar to Reeler (not shown). Green staining at the pial surface in (G) is nonspecific. Bars = 25 μm. (I) Immunoblots of full-length Notch1 (p300), Notch1 ICD and β-actin from the same cortical lysates of 2 rl/+ and 2 rl/rl. Notch1 ICD bands include p110 non-phosphorylated and p120 phosphorylated forms. Right panel: Relative values of the band intensity of indicated proteins from rl/rl against those from rl/+, which were set as 100. Band intensities for each mutant were normalized to β-actin. The data represent the mean ± SD of 5 brains from independent experiments. *p<0.01 by paired t-test. (J) Immunoprecipitation (IP) of brain lysates with a non-specific goat IgG as negative control (left) or Dab1-specific antibody (E-19; right) followed by Notch1 and Dab1 immunoblotting (IB).
To confirm our immunohistochemistry, we probed protein extracts from Reeler neocortex with each of the Notch1 antibodies. We observed an ~50% decrease in Notch1 ICD, but no significant difference in the full-length (Figure 1I) and membrane-anchored transient intermediate forms of Notch1 (Figure S1). Similar results were obtained from the Vldlr/ApoER2 dKO and Dab1 knockout neocortex (data not shown). Previous studies have shown that inhibition of S1 or S2-3 cleavage of Notch results in accumulation of full-length or transient intermediate Notch proteins respectively. The evidence that no significant increases in full length or transient Notch1 precursors were observed in Reeler (Figure 1I, S1) suggests that Reelin signaling does not affect the proteolytic processing of the Notch receptor during cortical development. Therefore, the loss of Notch1 ICD in Reeler is likely due to other mechanisms, such as enhanced degradation that would prevent Notch1 ICD from accumulating in the nucleus.
We next examined Notch activity in migrating neurons using a Notch signaling-dependent reporter constructs. In utero electroporation of the Rbpj-bp-reporter construct enabled us to monitor the acute on/off level of Rbpj dependent transcription activated by Notch signaling (Kohyama et al., 2005). The reporter expression confirmed Notch activity in post-mitotic migrating neurons in both the IZ and CP in wild-type, as well as in mitotic neuronal progenitor cells in the VZ/SVZ as previously reported (Kohyama et al., 2005; Ohtsuka et al., 2006; Figure S2). In contrast, the reporter activity was barely detected in Reeler (Figure S3).
These results suggest that Notch signaling is active in migrating neurons but is significantly reduced in Reelin signaling deficient cortex. As additional confirmation we performed quantitative RT-PCR, and found reduced Hes1 and Hes5 transcription (downstream target genes of Notch signaling) in Reeler while the transcription level of Notch1 was comparable to wild-type (Figure S3). Taken together, these findings suggest that Reelin signaling via Dab1 may regulate nuclear Notch ICD levels, and thereby active Notch signaling, by mechanisms distinct from transcriptional regulation or cleavage processing of the Notch receptor.
Notch ICD and Dab1 interact during cerebral cortical development
We next tested whether the Reelin and Notch signaling pathways interact during cortical development. Using multiple antibodies against Dab1 (see antibody list in supplemental experimental procedures), we were able to co-immunoprecipitate the p110, a non-phosphorylated form (Redmond et al., 2000) of Notch1 ICD from E18.5 neocortical lysate (Figure 1J). Conversely, Dab1 co-immunoprecipitated with Notch1 using multiple antibodies against Notch1 intracellular domain (data not shown, see antibody list in supplemental experimental procedures). This is consistent with previous reports that Drosophila Disabled interacts with Notch via the phosphotyrosine-binding (PTB) domain (Giniger, 1998; Le Gall and Giniger, 2004) and that the Dab1 PTB domain preferentially binds non-phosphorylated proteins (Howell et al., 2000). These results indicate that Dab1 and Notch1 ICD physically interact during mouse cortical development and served as the foundation for our hypothesis that a Reelin-Notch signaling interaction may be involved in neuronal migration in the cortex.
Notch signaling is indispensable for proper radial migration in the cerebral cortex
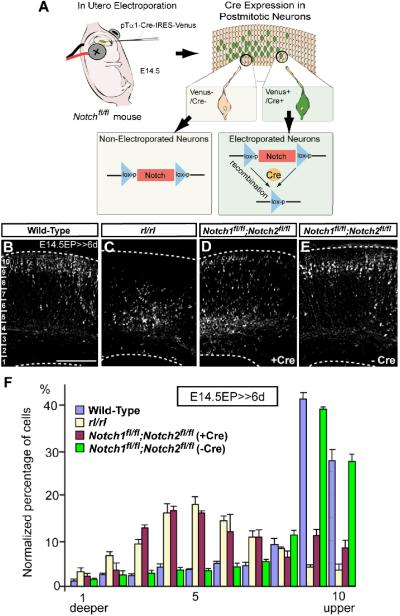
To test whether Notch signaling has a functional role in migration, we systematically deleted Notch genes within post-mitotic migrating neurons. Because Notch also plays important developmental roles in Cajal-Retzius cells in the MZ and VZ/SVZ neural progenitor cells (Yoon and Gaiano, 2005; Louvi and Artavanis-Tsakonas, 2006), it was crucial to preclude any secondary effects of Notch deletion in these cell populations. We therefore took advantage of the Cre/loxP system in combination with in utero electroporation to delete Notch in post-mitotic migrating neurons within the neocortex. As depicted in Figure 2A, we electroporated a construct including Cre recombinase and Venus (an enhanced yellow fluorescent protein [EYFP] variant) reporter under the T alpha 1 alpha-tubulin promoter (pTα1-Cre-IRES-Venus) into E14.5 cortex of floxed Notch mice. The Tα1 promoter activates predominantly in post-mitotic neurons (Gloster et al., 1994), allowing us to delete Notch in post-mitotic neurons by Cre-mediated recombination (Figure 2A). Some reports claim that the Tα1 promoter also drives gene expression in small population of mitotic neuronal progenitor cells (Sawamoto et al., 2001; Gal et al., 2006), while other groups detected expression only in postmitotic neurons (Gloster et al., 1994; Coksaygan et al., 2006). In our system, Cre/LoxP recombination was activated almost exclusively in postmitotic neurons, which was confirmed by a series of experiments (Figure S4). In the supplemental text, we further discuss the differences between the experimental systems used in our previous (Gal et al., 2006) and current report.
Figure 2. Notch signaling is required for proper radial migration of cortical neurons.
(A) Schematic representation of Notch genomic deletion in the migrating neurons. (B-E) Venus immunostaining 6 days post-electroporation with pTα1-Cre-IRES-Venus (B-D) or pTα1-IRES-Venus (E) into indicated mice. Bar (μm) = 100. Dashed lines indicate pial and ventricular surfaces. (F) The graphs indicate quantification of the distribution of Venus+ neurons in the 10 bins dividing the whole thickness of the cortex as indicated in B in each genotype. The data represent the mean ± SEM of 4 brains each from independent experiments. p<0.0001, p<0.0001, and p>0.05 for rl/rl, Notch1fl/fl;Notch2fl/fl(+Cre) and Notch1fl/fl;Notch2fl/fl(−Cre), respectively compared with wild-type by Two-sample Kolmogorov-Smirnov test (K-S test). F(9,54)=107.40, p<0.0001, F(9,45)=54.92, p<0.0001, and F(9,54)=0.78, p>0.05, respectively by Repeated Measures ANOVA.
Deletion of Notch1 by introduction of pTα1-Cre-IRES-Venus into floxed Notch1 homozygote (Notch1fl/fl) resulted in a similar distribution of Venus+ neurons in the cortex as wild-type, floxed Notch1 and/or Notch2 (closest paralog of Notch1) heterozygotes (Figure S5, data not shown). Similarly, Notch2 deletion (Notch2fl/fl) did not affect neuronal positioning (Figure S5). To eliminate possible compensatory effects of single Notch deletion, we next deleted Notch1 and Notch2 simultaneously. In contrast to wild-type cortex in which most Venus+ neurons reached the upper CP (Figures 2B and 2F), many Venus+ neurons in Notch1fl/fl; Notch2fl/fl brains were abnormally located within the lower CP and IZ (Figures 2D, 2F). Although direct comparison between the cases in Reeler (Figure 2C) and Notch1fl/fl; Notch2fl/fl brains (Figure 2D) is not strictly adequate (Notch deletion by electroporation in a sparse population among normal cells v.s. the Reeler mutant.), Figure 2F implies the similarity between the migration defect by Notch deficiency and that in Reeler. This dispositioning effect in Notch1fl/fl; Notch2fl/fl brains did not appear to be due to defects in neuronal/glial differentiation, progenitor proliferation rate or the pattern of apoptotic cell death (Figures S6 and S7). Additionally, Reelin expression in the MZ also was not affected (Figure S7) and introduction of pTα1-IRES-Venus (without Cre) showed no effect in Notch1fl/fl; Notch2fl/fl brains (Figures 2E and 2F).
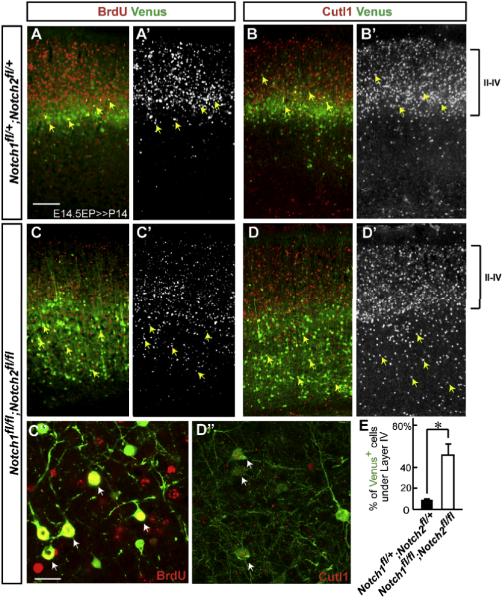
We next examined the distribution of the electroporated neurons at postnatal day 14 (P14) when neurons have settled into their final position. Most (over 90%) Venus+ neurons electroporated with a Cre expression plasmid at E14.5 were located within Layers II-IV in control (Notch1 fll+; Notch2 fl/+) neocortex (Figures 3A and 3B). In contrast, fewer than half of Venus+ neurons were found within these layers, instead over 50% of Venus+ neurons beneath layer IV in Notch1 fl/fl; Notch2 fl/fl cortex (Figures 3C, 3D and 3E). BrdU labeling of Venus+ cells 24h after electroporation confirmed similar results (Figures 3A, 3A’ and 3C-3C”). Further, Notch-deleted neurons expressing Cutl1 (also known as Cux1), a marker for layers II-IV, were abnormally positioned in deep neocortical layers in Notch1 fl/fl; Notch2 fl/fl brains compared to control (Figures 3B, 3B’ and 3D-3D”). While the percentage of BrdU or Cutl1 positive cells in Venus-expressing neurons was similar between heterozygote and homozygote brains (BrdU: heterozygote 27.45%±0.78, homozygote 26.15%±0.51, p=n.s. by Student’s t-test, Cutl1: heterozygote 41.92%±1.06, homozygote 39.66%±0.43, p=n.s. by Student’s t-test; the data represent the mean ± SEM of 5 brains each), Venus expression was undetectable in some displaced BrdU+ or Cutl1+ neurons suggesting that arrested neurons might cause the arrest of adjacent/nearby migrating neurons. These results demonstrate that Notch is required for neuronal migration in the neocortex, and the migration defect produced by the loss of Notch signaling results in a laminar displacement of neurons postnatally.
Figure 3. Notch deletion causes laminar displacement of later-born neurons.
(A-D”) Venus (green) immunostaining with BrdU or Cutl1 (red or white) staining in indicated mutants at P14. Note that Cutl1 analysis was performed in the dorsal somatosensory region where endogenous Cutl1 expression in lower layers is normally absent. BrdU+/Venus+ or Cutl1+/Venus+ neurons in the Notch deleted cortex (C-D’, arrows) are abnormally positioned in lower layers than in the control cortex (A-B’, arrows). (C”, D”) Higher magnification views of BrdU+/Venus+ and Cutl1+/Venus+ double-labeled neurons around layer V, respectively (arrows). Bars (μm) = 100 (A-D”), 10 (C”, D”’). (E) Quantification of the distribution of Venus+ neurons under Layer IV in the Notch1 fl/+; Notch2 fl/+ and Notch1 fl/fl; Notch2 fl/fl cortex. The data represent the mean ± SEM of 4 brains each from independent experiments. * p<0.005, Student’s t-test.
Notch signaling is required for proper morphology of migrating neurons
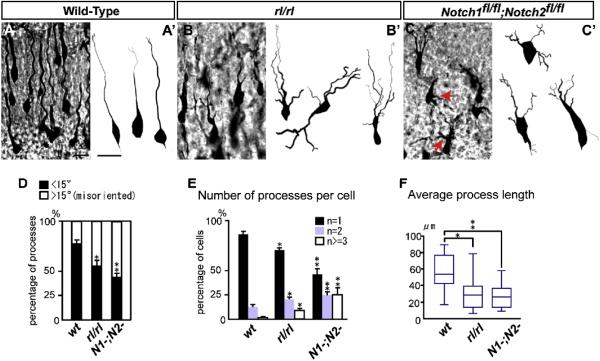
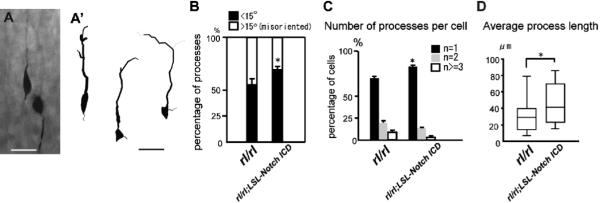
The abnormal morphology of Reelin signal-deficient neurons has been documented and is suspected to contribute the disrupted positioning of these neurons (Pinto-Lord et al., 1982; Sanada et al., 2004; Olson et al., 2006). We next examined whether Notch signal-deficient migrating neurons exhibit similar morphological defects to those observed in Reeler. Venus-labeled neurons in wild-type extended a long process toward the MZ (Figures 4A, and 4A’), however, as previously described, neurons in Reeler retained stunted, bi- or multifarious leading processes (Figures 4B, and 4B’). Similarly, Notch-targeted neurons exhibited shorter, multiple and inconsistently oriented processes protruding directly from the cell soma (Figures 4C-4F). Thus Reelin and Notch signaling-deficient migrating neurons share similar migratory and morphological abnormalities.
Figure 4. Morphological defects in migrating neurons after loss of Notch signaling.
(A-C) Venus immunostaining revealed migrating neuronal morphology 3 days post-electroporation with pTα1-Cre-IRES-Venus in wild-type, Reeler and Notch1 fl/fl; Notch2 fl/fl mice. Red arrows in (C) indicate ectopic primary processes. (A’-C’) 3D reconstruction of Venus+ migrating neurons in mice of each genotype 3 days post-electroporation. (D) Percentage of primary processes (directly protruded from cells) oriented normally (The ‘oriented normally’ primary processes were defined according to their angle towards the pial surface within ±15°) and abnormally. * p<0.01, **p<0.001, Student’s t-test comparing with wt. (E) Percentage of cells with one (black), two (blue) or >3 (white) processes per cell. * p<0.05, **p<0.01, Student’s t-test comparing corresponding bins to wt. (F) Box plots of the average primary process length per cell (total process length/number of the primary processes). *p<0.05, **p<0.01, Mann-Whitney’s U test. A total of 120 cells/genotype from four brains (different litters) were analyzed for orientation (D) and number of processes (E). The box plots of primary process length (F) were obtained for each genotype (n=30 each from 3 brains {different litters}). Bars (μm) = 10.
Forced expression of Notch ICD mitigates the migration defect caused by Reelin signal deficiency
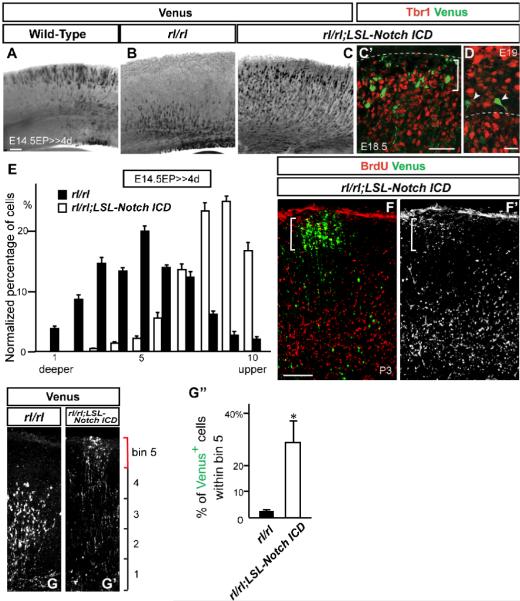
The above results showed phenotypic similarities between Reelin and Notch deleted neurons as well as reduction of Notch ICD in Reeler brains, but a functional interaction between these signaling remains to be examined. Thus, to examine a potential interaction between Reelin and Notch signaling, we tested whether forced expression of Notch ICD in migrating neurons can affect the Reeler phenotype. Here we used the same method as gene deletion experiments, in which post-mitotic neuron enriched Cre/LoxP recombination was confirmed (Figure 2, 3, S3, S4). Thus, we electroporated pTα1-Cre-IRES-Venus into wild-type, Reeler, and mice with a compound background of Reeler and Loxp-Stop-Loxp-Notch ICD (rl/rl;LSL-Notch ICD), in which Notch ICD expression can be induced after Cre/LoxP recombination. In wild-type mice 4 days post-electroporation, Venus+ neurons migrated into the upper layers in the CP (Figure 5A). As expected, Venus+ neurons were arrested in deeper layers in Reeler (Figure 5B). Strikingly, in rl/rl;LSL-Notch ICD mice, significantly fewer electroporated neurons were arrested in deeper cortical layers compared to Reeler (Figures 5B, 5C, and 5E). Instead, a significant number of Venus+ neurons in which Notch ICD was replenished in the Reeler background migrated into the upper layers (Figure 5C). The transcription factor Tbr1, which is strongly expressed in deeper layers (primarily subplate and layer VI) and Cajal-Retzius cells in wild-type, are abnormally located in the upper layers in Reeler neocortex (Hevner et al., 2003). Venus+ electroporated neurons in Reeler cortex migrated past these Tbr1+ neurons (Figure 5C’), and some were found within the most superficial superplate (SPP). Farther migration of Notch ICD replenished neurons than Reelin signal deficient neurons can be observed as early as 3.5 days post electroporation, suggesting that replenishment of Notch ICD might mitigate the slower migration of Reelin signal-deficient neurons (Sanada et al., 2005; Figure S8). These neurons remained in the upper layers even in the postnatal cortex (P3) (Figures 5F-5G”). BrdU injection at E15.5 revealed that BrdU+ neurons outside the electroporated region distributed in lower layers of the Reeler cortical plate, while the Venus+/BrdU+ electroporated neurons reached upper layers (Figures 5F, and 5F’).
Figure 5. Replenishing of Notch ICD mitigates neuronal migration defects in Reeler.
(A-D) Immunostaining for Venus (black, green) and Tbr1 (red) in cortical slices of indicated genotypes 4 days (A-C’) or 4.5 days (D) post-electroporation (with pTα1-Cre-IRES-Venus). (D) Higher magnification view around the SPP. Dashed lines in (C’) and (D) indicate the pial surface and the border between the SPP and CP, respectively. Bracket in (C’) shows the SPP region. Note that electroporated Venus+ cells did not change their fate to Tbr1+ early-born neurons (C’, D). (E) Quantification of neuronal distribution showed significantly more cells in upper CP of rl/rl;LSL-Notch ICD compared to rl/rl cortex (K-S test, p<0.0001; Repeated Measures ANOVA, F(9,54)=18.91, p<0.0001). The data represent the mean ± SEM of 6 brains each. (F, F’) Immunostaining for Venus (green) and BrdU (red, white) in P3 cortical slices of rl/rl;LSL-Notch ICD electroporated with pTα1-Cre-IRES-Venus. Note that Venus+/BrdU+ cells (indicated by bracket) located over BrdU+ cells in surrounding lower layers. (G, G’) Venus immunostaining in P3 cortical slices of rl/rl and rl/rl;LSL-Notch ICD electroporated with pTα1-Cre-IRES-Venus. (G”) Quantification of Venus+ neurons located in the upper part of the CP (within the bin 5 indicated by a red bracket in G, G’. Entire thickness of the cortex was subdivided into 5 bins.) The data represent the mean ± SEM of 3 brains each. *p<0.05, Student’s t-test. Bars (μm) = 100 (A-C’, F-G’), 20 (D).
One characteristic of Reelin signaling deficient neurons is that terminally positioned neurons exhibit abnormally oriented dendrites (Pinto-Lord et al., 1982; Pinto-Lord and Caviness, 1979). Similarly, we found that, at 4.5 days post-electroporation (E19) and at P3, more mature Notch ICD introduced neurons in the SPP also displayed abnormally orientated dendrites (arrowheads in Figure 5D, and data not shown). In contrast, overexpression of Notch ICD mitigated the morphological defects typical of migrating Reelin-signal deficient neurons, and reduced the number of multifarious leading process (at 3 days after electroporation, Figures 6A-6D). Further, many of these electroporated migrating neurons exhibited a long process that oriented towards the MZ (compare Figures 6A, 6A’ with Figure 4B, 4B’). We did not observe obvious phenotypes in neuronal distribution, neurogenesis, and radial glial morphology by overexpression of Notch ICD in LSL-Notch ICD (Figure S9 and S10). Thus, these results indicate that in Reeler background, Notch ICD plays a significant role in neuronal migration, but likely not in determination of final dendritic orientation.
Figure 6. Replenishing of Notch ICD mitigates morphological defects in Reeler.
(A) Venus+ neurons 3 days post-electroporation with pTα1-Cre-IRES-Venus in rl/rl;LSL-Notch ICD mice. Left neuron in (A) showed rescued morphology with processes that were more pial-oriented compared to rl/rl (e.g. Fig. 4B). (A’) 3D reconstruction of Venus+ neurons (compare with rl/rl in Fig.4B’). (B-D) Quantification of direction of primary processes (B), primary process number per cell (C) and average primary process length per cell (D) for the electroporated neurons in mice of indicated genotypes. *p<0.05, Student’s t-test (B, C), *p<0.05, Mann-Whitney’s U test (D). A total of 120 cells/genotype from four independent experiments were analyzed for orientation (B) and number of processes (C). The box plots of primary process length (D) were obtained for each genotype (n=30 each from 3 independent experimental sets). For better comparison with Reeler, duplicated data from Figure 4 were included. Bar (μm) = 10.
Replenishing Notch activity mitigates neuronal migration defects induced by disrupted Dab1 signaling
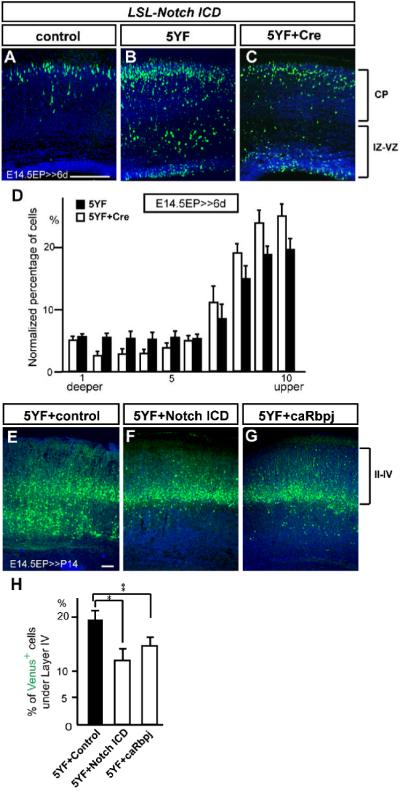
To rule out the possibility that the above alleviation effects appear only in the Reeler background where all cells (in addition to the electroporated cells) lack exposure to Reelin, we next tested whether Notch replenishment can mitigate the migration defect cell-autonomously within a wild-type background. Since Dab1 is a critical mediator of Reelin signaling and Dab1-null mice display a similar phenotype to Reeler (Sheldon et al., 1997; Howell et al., 1997; Ware et al., 1997), we electroporated a dominant negative mutant of Dab1 (5YF, a Reelin signal-insensitive mutant) (Howell et al., 2000; Keshvara et al., 2001) into LSL-Notch ICD cortex (no exogenous Notch ICD is introduced without Cre recombinase). 5YF was sufficient to induce a migration defect (Figures 7A and 7B, K-S test between 5YF and vector only control yields p<0.001; ANOVA, F(9,36)=12.61, p<0.0001.) and served as a cell-autonomous model of Reelin-signal deficiency as reported previously (Sanada et al., 2004). We were able to mitigate the 5YF-mediated migration defect by simultaneous introduction of Notch ICD through Cre-mediated recombination (Figures 7C, and 7D). Fewer neurons were located near the IZ (bins 2-5 in Figure 7D) while more neurons reached the upper CP compared to 5YF alone (bins 7-10 in Figure 7D). The mitigating effect appeared to be Reelin-pathway specific since Notch ICD overexpression was unable to mitigate the displacement of neurons lacking MEK kinase 4 (MEKK4; Sarkisian et al., 2006), a signaling pathway considered independent of Reelin (Figure S11). The mitigating effect by Notch signaling activity was further examined in P14 brains by another approach: electroporation was performed at E14.5 with pTα1-Cre-IRES-Venus, 5YF and either CALSL-Notch ICD or CALSL-caRbpj (constitutively active form of Rbpj). Through Cre-mediated recombination, expression of Notch ICD or caRbpj was driven from the CALSL plasmid (Matsuda and Cepko, 2007) in migrating neurons (data not shown), and we confirmed that both Notch ICD and caRbpj can mitigate the positioning defect of neurons caused by 5YF (Figures 7E-7H). Together, these results demonstrate that Notch ICD and Rbpj are involved in Reelin-Dab1 signaling-mediated control of neuronal migration.
Figure 7. Notch ICD mitigates radial migration defects induced by a dominant-negative form of Dab1.
(A-C) Venus+ neurons were detected by immunohistochemistry 6 days after electroporation with pCDNA3.1 empty (A) or containing Dab1 5YF (B, C) plasmid with pTα1-IRES-Venus (A, B) or pTα1-Cre-IRES-Venus (C) plasmid in LSL-Notch ICD mice. Note that 5YF arrested many neurons beneath the CP which was mitigated by overexpression of Notch ICD. (D) Quantification of neuronal distribution in the LSL-Notch ICD cortex electroporated with indicated plasmids (K-S test, p<0.05; Repeated Measures ANOVA, F(9,126)=5.24, p<0.05). The data represent the mean ± SEM of 6 brains each. (E-G) Venus+ neurons were detected by immunohistochemistry at P14 after E14.5 electroporation with pCALSL empty (E), containing Notch ICD (F) or caRbpj (G) with pTα1 -Cre-IRES-Venus and pCDNA3.1-Dab1 5YF plasmids in wild-type mice. Note that compared to control (E), overexpression of Notch ICD (F) or caRbpj (G) resulted in fewer neurons located beneath layer IV. (H) Percentages of Venus+ neurons below layer IV in P14 wild-type cortex electroporated with indicated plasmids. The data represent the mean ± SEM of 10, 8, and 11 brains, respectively. *,**p<0.01, 0.05, by Student’s t-test. Bars = 100 μm.
Reelin stimulated Dab1 blocks degradation of Notch ICD
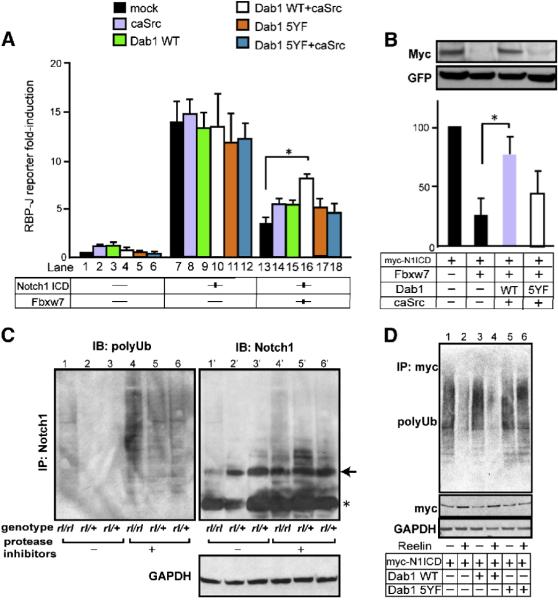
Based on the findings that nuclear Notch1 ICD is reduced in Reeler and that Dab1 binds Notch1 ICD (Figure 1), we investigated possible molecular mechanisms by which the Reelin-Dab1 pathway regulates levels of nuclear Notch ICD. Notch ICD is known to be degraded by a proteasome pathway via various E3 ubiquitin ligases (Lai, 2002), and Dab1 has been shown to inhibit the function of these ligases (Park et al., 2003). Furthermore, Dab1 is an adaptor protein that can control subcellular protein trafficking (Stolt and Bock, 2006; Honda and Nakajima, 2006; Hoe et al., 2006), which is a critical step in protein degradation. Thus, we hypothesized that Reelin-Dab1 signaling may regulate this degradation pathway by stabilizing and/or controlling the levels of Notch ICD. To assess the influence of Reelin-Dab1 signaling on Notch activity via the proteasome pathway, we measured Rbpj luciferase reporter expression levels in Cos-7 cells. Reporter expression is induced by introduction of Notch ICD, the level of which can be controlled by introduction of Fbxw7 (also known as Sel-10 or Cdc4), an adapter molecule of E3 ligase that leads to degradation of Notch ICD (Gupta-Rossi et al., 2001; Oberg et al., 2001; Wu et al., 2001). Consistent with previous reports, transfection with Notch ICD led to robust induction of luciferase expression (Figure 8A compare lane 7 with 1), which was reduced by co-introduction of Fbxw7 (Figure 8A, lane 13). Cotransfection of wild-type Dab1 with the constitutively active form of Src kinase (caSrc), a condition that recapitulates Reelin signal-stimulated Dab1 activation in vitro (Bock and Herz, 2003), significantly blocked the reduction of reporter activity due to Fbxw7 (Figure 8A, compare lane 16 with 13). In contrast, the 5YF mutant of Dab1 did not elicit enhanced reporter activity (Figure 8A, p=n.s. between lane 13 and 18). Dab1 did not affect the reporter activities when Fbxw7 was not transfected (Figure 8A, lane 1-12). These results suggest that Reelin-stimulated Dab1 might protect Notch ICD from Fbxw7-induced degradation. Additionally, Fbxw7-mediated reduction of Notch ICD levels and enhancement of its polyubiquitination in Cos-7 cells was significantly inhibited in the presence of wild-type but not the 5YF mutant form of Dab1 (Figure 8B, data not shown, n=5). Thus, our in vitro experiments indicate that Reelin-Dab1 signaling can inhibit Notch ICD degradation through the Fbxw7-mediated pathway.
Figure 8. Stimulated Dab1 blocks Notch ICD degradation.
(A) Effects of active Dab1 on Notch ICD activity. Relative Rbpj luciferase activity of each condition is compared to lane 1 set as 1.0. The indicated values are the mean ± SEM of three experiments performed in triplicate. *p<0.05, Student’s t test. (B) Cos-7 cells were transfected with indicated plasmids, and the expression of Notch1 ICD and GFP (as an internal control, reprobed on the same membrane) were analyzed by IB. The graphs show the relative intensity of the Notch1 ICD against the left lane after normalization with GFP as a transfection control. The data represents the mean ± SEM of 3 independent experiments. *p<0.01 paired t-test. Transfection of caSrc without Dab1 or Dab1 without caSrc constructs gave equivalent results to Dab1 5YF with/without caSrc. Transfection of caSrc with/without Dab1 constructs did not alter both endogenous and exogenous Fbxw7 expression levels (data not shown). (C) IB of lysates from rl/rl and rl/+ brain slices against polyubiquitin (FK2) and Notch1 after immunoprecipitation with a Notch1 antibody. IB against GAPDH was obtained from the flow-through of IP. The samples in the left 3 lanes were obtained from freshly prepared cortical slices, and the samples in right 3 lanes were from slices cultured with proteasome inhibitors. Note the decrease of precipitated non-polyubiquitinated Notch1 in Reeler (lanes 1’ and 4’), consistent with the increase of polyubiquitinated Notch1 (lanes 1 and 4). Arrow indicates p100-120 Notch1 ICD bands. Asterisk shows IgG bands. (D) IB of lysates from cultured cortical neurons transfected with the indicated plasmids against polyubiquitin (FK2) and myc after IP with a myc antibody. The transfected neurons were exposed to Reelin containing- or mock medium for 6 hours with protease inhibitors before collection. IB against GAPDH was obtained from the IP flow-through.
The experiments described above have the advantage of ability to precisely control the activities of both Reelin and Notch signaling, but the system is relatively artificial. Thus, to gain further evidence, we next examined whether the ubiquitination of Notch is actually affected by Reelin deficiency during cortical development. Slices were prepared from wild-type and Reeler brains. We then made lysates from non-cultured slices or slices cultured for 4 hours in the presence of proteasome inhibitors to allow accumulation of the polyubiquitinated proteins by inhibiting their degradation, immunoprecipitated Notch1 ICD and immunoblotted using an anti-polyubiquitin antibody. Consistent with results in Cos-7 cells, we observed a noticeable increase of polyubiquitin bands in Reeler both with and without protease inhibitors (Figure 8C, compare lanes 1 and 4 with lanes 2, 3 and 5, 6 respectively; n>3/each genotype). Since we were unable to determine whether Notch1 ICD was specifically polyubiquitylated in this system (because we could not obtain enough precipitate from brain lysates using either anti-Notch1 ICD (Val 1744) or anti-polyubiquitin antibodies), we confirmed specific ubiquitination of nuclear Notch1 ICD by using Notch1 ICD transfected cortical neurons in vitro (Figure 8D, n=3). Stimulation by Reelin-containing medium significantly reduced polyubiquitinated Notch1 ICD (compare lane 2 with 1) and this was inhibited by addition of Dab1 5YF (lane 6) but not wild-type Dab1 (lane 4). Altogether, these data suggest that Notch1 ICD is likely targeted by the Reelin signaling pathway during cortical development and suggests that Reelin-Dab1 signaling prevents Notch ICD degradation.
Discussion
Despite over half a century of research since the first report of Reeler the underlying pathogenetic mechanisms still remain unclear. Disruptions in several developmental processes have been proposed to cause the phenotype. First, the actin-cytoskeleton of Reelin-signaling deficient neurons is abnormally organized which may lead to disruption of the leading process and subsequent failed migration (Pinto-Lord and Caviness, 1982; Sanada et al., 2004; Olson et al., 2006). Second, Reelin-deficient neurons may fail to detach from radial glia at the appropriate position as a result of increased neuron-glia adhesion due to abnormally high levels of α3 integrin (Sanada et al., 2004). Additionally, the aberrantly superficial positioning of early-generated neurons may physically obstruct the migration of later-born neurons thereby giving rise to an inverted lamination of the cortex (Pinto-Lord and Caviness, 1979; Tabata and Nakajima, 2002).
To explore Notch’s potential role in migration, we employed a methodology that could circumvent the complications introduced by traditional knockout and transgenic strategies which are unable to discriminate among Notch deletion defects due to proliferation, differentiation, or apoptosis. Using this approach, we provide the first evidence that the morphology and migration of post-mitotic neurons is regulated by Notch signaling whose activity is likely under the control of the Reelin-Dab1 pathway. We observed that both the nuclear Notch ICD expression and Notch ICD activity-dependent Rbpj-mediated transcription typical of wild type migrating neurons were significantly reduced in Reeler cortex (Figure 1 and Figure S3). These findings are consistent with a report that the expression of the Notch target gene, Strawberry Notch, is also downregulated in Reeler (Baba et al., 2006).
To explore the consequence of the observed reductions in active Notch ICD and downstream signaling, we deleted Notch in migrating neurons and observed morphological and migration defects similar to those of Reelin-signal deficient neurons (Figures 2-4). Furthermore, introducing Notch ICD or caRbpj mitigated migration defects observed in Reelin-Dab1 signaling deficient neurons in wild-type (or LSL-Notch ICD) background (Figure 7) as well as those in the Reeler background (Figures 5,6, and Figure S8). Thus Notch signaling appears to play a cell-autonomous role during neuronal migration. Finally, replenishing Notch ICD was able to alleviate migrating neuronal morphology but was not sufficient to reorient the dendrites of matured neurons in Reeler (Figures 5 and 6). This suggests that Notch signaling is required during migration but not during final maturation stages which include somal and dendritic orientation. Alternatively, disruption of the latter may be secondary to the abnormal formation of the internal plexiform zones in Reeler as previously suggested (Pinto-Lord and Caviness, 1979; Tabata and Nakajima, 2002). Consistent with this physical barrier hypothesis, we (this study, data not shown) and others (Sanada et al., 2004; Olson et al., 2006) did not observe neurons with inverted dendrites after cell-autonomous reduction of Reelin-Dab-1 signaling by 5YF or Dab1 shRNA introduction. Whether the superficial positioning of early-born neurons in the Reeler cortex forms a physical barrier to migration is unclear, however reintroduction of Notch ICD enabled later-born Reelin signal-deficient neurons to migrate past the abnormally superficial band of early-born neurons (including subplate neurons) to reach the upper layers (Figure 5). Thus it is possible that the positioning of later-born neurons in Reeler is due to a cell-autonomous migration defect rather than physical obstruction. Interestingly, similar alleviation effects can be achieved by expressing Reelin in Reeler VZ cells (Magdaleno et al., 2002), suggesting that migrating neurons may require Reelin stimulation and Notch activation much earlier than previously suspected (i.e. before they arrive in the cortical plate). These results support the model that Reelin signaling works as “an instructive signal” (D’Arcangelo et al. 1997) to engage cytoskeletal remodeling events critical to neuronal migration.
Both Notch and Reelin-signaling deficient neurons exhibit processes with disrupted morphology (Figure 4). These morphological defects may be a result of premature terminal differentiation of dendrites. However, given that neuronal maturation correlates with an increase of Notch ICD (Figure 1, Sestan et al., 1999), premature terminal differentiation by Notch reduction/deletion is unlikely. Alternatively, the transition from the multipolar to bipolar stage - a critical step during proper neuronal migration (LoTurco and Bai, 2006) - might be impaired. While common transitional defects occur mostly within the SVZ to IZ and our Notch-targeted defects were observed mainly in the lower CP, these differences may simply reflect our methodological approach for gene knockdown (e.g. T alpha1 promoter driven Cre/loxP system vs U6 promoter driven shRNA). Nevertheless, given that Reelin can regulate actin dynamics in neurons (e.g. Suetsugu et al., 2004; Chen et al., 2004), the morphological defects seen in Reelin- or Notch-deficient migrating neurons most likely reflect specific disruptions of the leading process. Recent study has shown that Notch ligands are specifically displayed by intermediate progenitors in the SVZ and young neurons in the IZ during the period of neurogenesis and neuronal migration (Yoon et al., 2008). Given the significant migration arrest of Notch-deficient neurons in the IZ to lower CP (Figure 2), an intriguing possibility is that Notch ligands displayed in the IZ is critical for radial movement of neurons.
Our finding of a dose-dependent Notch deletion effect on radial neuronal migration (Notch1 fl/fl; Notch2 fl/+ < Notch1 fl/fl; Notch2 fl/fl most severe defect; see Figure S5) supports our model that reduced Notch activity, rather than a complete loss of its activity, can lead to migration defects in Reeler. In agreement with previous reports (Yoon and Gaiano 2005; Louvi and Artavanis-Tsakonas, 2006), this finding also supports the pleiotropy of Notch signaling in an activity level-dependent manner. Therefore Notch processing and activity is precisely controlled at various stages (Bray, 2006), and we now implicate Reelin signaling in this regulation. Our results suggest that Reelin signaling may govern the level of nuclear Notch ICD levels by affecting Notch ICD degradation. We showed that Notch polyubiquitination/degradation is increased in Reeler cortex, and that degradation of Notch ICD through the Fbxw7-mediated proteasome pathway is inhibited by activated Dab1 in vitro (Figure 8). However, Reelin signaling also promotes Dab1 degradation (Armaud et al., 2003; Bock et al., 2004, Kuo et al., 2005; Feng et al., 2007), which at first glance does not fit to our model. Interestingly, studies have shown that Dab1 could function in the trafficking of some molecules (Honda and Nakajima, 2006; Hoe et al., 2006), suggesting that Dab1 could potentially serve to traffic Notch ICD, thereby sequestering it away from the degradation pathway. It is also noteworthy that alteration of Notch intracellular distribution (trafficking) can significantly affect its degradation rate and activity (Mukhejee et al., 2005). Fbxw7-mediated Notch ICD ubiquitination can occur specifically within the nucleus (Gupta-Rossi et al., 2001). We did not determine whether Fbxw7 mediates Notch degradation during neuronal migration, but did observe specific expression of Fbxw7 in migrating neurons (K.H-T and P.R. unpublished data). Therefore, Reelin-Dab1 signaling may facilitate the trafficking of Notch ICD to reduce its degradation via Fbxw7 in the developing neocortex.
Additional mechanisms could also control the ubiquitin-mediated degradation of Notch ICD. For example, a complex of Numb and Itch E3 ubiquitin ligase mediates lysosomal degradation of Notch ICD in the cytoplasm (McGill and McGlade, 2003). Both Numb and Dab1 contain a PTB domain that exhibit similar binding and functional properties (Lai. 2002). Thus Dab1 and Numb may compete for Notch binding thereby regulating degradation. Alternatively, inhibition of Notch ICD degradation may be achieved by direct binding of Dab1 to E3 ubiquitin ligases to block its ubiqutination activity (Park et al., 2003), or simply competing for the same E3 ligases for its degradation with Notch ICD. Although Notch cleavage processing was not significantly affected in Reeler (Figure 1), we did not formally examine the possible effects of Reelin signaling on Notch processing. Therefore a weaker interaction between Notch and Reelin signaling may exist at this level, similar to the mechanism whereby Reelin-Dab1 signaling promotes APP processing and trafficking (Hoe et al., 2006).
Although our study showed an interaction of Notch and Reelin signaling pathways in the control of radial migration of cortical neurons, it remains to be examined whether the defect of this interaction underlies Reeler phenotypes besides impaired neuronal migration, such as radial glial dysmorphology (Dulabon et al., 2000; Forster et al., 2002; Jossin and Goffinet, 2003; Hartfuss et al., 2003) and neuronal invasion into the layer I (Trommsdorff et al., 1999; Hack et al., 2007). Considering that Notch and Reelin signalings directly control the expression of BLBP, a radial glial gene (Hartfuss et al., 2003; Anthony et al., 2005), radial glial development might be regulated by the interaction between Reelin and Notch signaling. During the review period of this paper, a study reported that a Dab1-Notch ICD interaction and Reelin-dependent increase of Notch1 are reproducible in a human neural progenitor cell line (Keilani and Sugaya, 2008), supporting this possibility. Future studies will test these possibilities using total brain-specific deletion of Notch and determine whether it reproduces Reeler phenotypes in other various aspects of brain development.
Experimental Procedures
Mice
Reeler and Scrambler mice were purchased from Jackson Laboratory. The tissues and lysates of Vldlr/ApoER2 double KO mice were generous gifts from Drs. A. Goffinet and Y. Jossin. Generation and genotyping of floxed Notch1, floxed Notch2, LSL-Notch ICD (also known as CALSL-NICD(H)) and LSL-Gfp (transgene includes loxP-flanked STOP cassette followed by Gfp, Jackson Laboratory) mouse lines were described previously. A list of references for floxed mice is available as Supplemental data. Animals were handled according to protocols approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine.
Quantitative RT-PCR
Total RNA was isolated from freshly dissected brain tissue by using the Rneasy plus kit (Qiagen) and cDNA was synthesized by using SuperScript First-strand synthesis system for RT-PCR with random hexamer primers (Invitrogen). GAPDH levels were detected by Taqman rodent GAPDH control reagents and used for normalization. Thermocycling was carried out by using the Applied Biosystems 7900 system and monitored by SYBR Green I dye detection. All reactions were performed in triplicate from 4 brains each.
In utero electroporation
In utero electroporation was performed at E14.5 as previously described (Sarkisian et al., 2006). A list of DNA solutions used for injection is available as Supplemental data. All control experiments were performed using empty vectors at the same concentrations. All BrdU labeling was performed 24h after electroporation according to previous studies (Sarkisian et al., 2006).
Immunohistochemistry and data analysis
Immunohistochemistry was performed with the previously described methods (Sarkisian et al., 2006). A list of antibodies is available as Supplemental data. Electroporated neurons around the somatosensory medial cortical region were counted for all positioning analysis. Tracing of the morphology, and quantification of orientation, number of processes, and length of the processes protruded from Venus+ neurons were done by the Neurolucida system (MicroBrightfield). In these analyses, we chose neurons located in the lower CP close to the IZ, and excluded terminal branches of processes in the MZ for quantification. Axons were excluded from both tracing and quantification. In box plot analysis, the line in the box, and the upper and lower edge of the box indicate the median, and the 25th and 75th percentiles, respectively. Error bars indicate the 5th and 95th percentiles.
Immunoprecipitation and immunoblotting
Protein samples from E18.5 or P0 mouse brain were harvested and used for immunoprecipitation and immunoblotting using a standard protocol. The antibodies used for immunoprecipitations and immunoblots are listed in Supplemental data. Analysis of band intensity was performed as previously described (Sarkisian et al., 2006).
Luciferase assay
Subconfluent Cos-7 cells were transiently transfected with plasmids at 50ng (caSrc), 200 ng (pGL2-8xCBF-luc and phRL) or 500 ng (myc-Notch ICD, Fbxw7 and Dab1 constructs) per well into 12 well plates. An equal amount of control construct (pCDNA3.1 empty vector) was transfected in mock experiments. The cells were subjected to the assay using Dual-Luciferase Reporter Assay system (Promega) 1 day after transfection. For the detection of luciferase activity, TD-20/20 (Turner Designs) was used.
Ubiquitination assay
By using Fugene6 (Roche), subconfluent Cos-7 cells were transiently transfected with the plasmids containing Dab1, Notch1 ICD, Fbxw7 and pCDNA3.1-HA-Ub, pCAG-GFP, and harvested 48 hours later. Proteasome inhibitors MG-132 and clasto-lactacystin β-Lactone (Calbiochem) were added at 10 μM 6 hours before harvest. For slice culture of E18.5 cerebral cortex, chopped slices at 300 μm thickness were incubated on the membrane floating in the Neurobasal medium with proteasome inhibitors for 4 hours. Cortical neurons were prepared for primary culture from dissected E18.5 cortex, and transfected with the plasmids using amaxa Nucleofector Kit (Lonza). Reelin containing-or mock medium was prepared from 293T cells transfected with pCrl or pCDNA-EGFP, respectively, as described (Honda and Nakajima, 2006), and applied with the proteasome inhibitors to the culture 2 days after passage.
Supplementary Material
Acknowledgements
We thank Drs. H. Okano, A. Goffinet, Y. Jossin, A. N. Gaiano, Miyawaki, F.D. Miller, A. Israël, J. Nye, R. Baron, S.D. Hayward, L.H. Tsai, and K. Kamon for providing the samples and plasmids, Jue Bao for technical assistance and A. Ghosh, C.Y. Kuan, J. Breunig for discussion. This work was supported by an Epilepsy Foundation of America postdoctoral fellowship (M.R.S.), the Kavli Institute for Neuroscience at Yale and the National Institute of Health (P.R. and N.Š.).
Footnotes
Additional information related to DNA constructs is included in the Supplemental data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaud L, Balif BA, Cooper JA. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol. Cell Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Baba K, Dekimoto H, Muraoka D, Agata K, Terashima T, Katsuyama Y. A mouse homologue of Strawberry Notch is transcriptionally regulated by Reelin signal. Biochem. Biophys. Res. Commun. 2006;350:842–849. doi: 10.1016/j.bbrc.2006.09.135. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Arnaud L, Arthur WT, Guris D, Imamoto A, Cooper JA. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr.Biol. 2004;14:606–610. doi: 10.1016/j.cub.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Bar I, Lambert de Rouvroit C, Goffinet AM. The Reelin signaling pathway in mouse cortical development. Eur. J. Morphol. 2000;38:321–325. doi: 10.1076/ejom.38.5.321.7361. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell. Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Bielas SL, Gleeson JG. Cytoskeletal-associated proteins in the migration of cortical neurons. J. Neurobiol. 2004;58:149–159. doi: 10.1002/neu.10280. [DOI] [PubMed] [Google Scholar]
- Bock HH, Jossin Y, Liu P, Forster E, May P, Goffinet AM, Herz J. Phosphatidylinositol 3-Kinase Interacts with the adaptor protein Dab1 in Response to Reelin signaling and is Required for normal cortical lamination. pp. 38772–38779. [DOI] [PubMed]
- Bock HH, Herz J. Reelin activates SRC family tyrosine kinase in neurons. Curr. Biol. 2003;13:18–26. doi: 10.1016/s0960-9822(02)01403-3. [DOI] [PubMed] [Google Scholar]
- Bock HH, Jossin Y, May P, Bergner O, Herz J. Apolipoprotein E receptors are required for reelin-induced proteasomal degradation of the neuronal adaptor protein Disabled-1. J.Biol.Chem. 2004;279:33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr., Rakic P. Mechanisms of cortical development: a view from mutations in mice. Annu. Rev. Neurosci. 1978;1:297–326. doi: 10.1146/annurev.ne.01.030178.001501. [DOI] [PubMed] [Google Scholar]
- Chen K, Ochalski PG, Tran TS, Sahir N, Schubert M, Pramatarova A, Howell BW. Interaction between Dab1 and CrkII is promoted by Reelin signaling. J. Cell Sci. 2004;117:4527–4536. doi: 10.1242/jcs.01320. [DOI] [PubMed] [Google Scholar]
- Coksaygan T, Magnus T, Cai J, Mughal M, Lepore A, Xue H, Fischer I, Rao MS. Neurogenesis in Talpha-1 tubulin transgenic mice during development and after injury. Exp. Neurol. 2006;197:475–85. doi: 10.1016/j.expneurol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. J. Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G. Reelin mouse mutants as models of cortical development disorders. Epilepsy Behav. 2006;8:81–90. doi: 10.1016/j.yebeh.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster E, Tielsch A, Saum B, Weiss KH, Johanssen C, Graus-Porta D, Muller U, Frotscher M. Reelin, Disabled 1, and beta 1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc. Natl. Acad. Sci. USA. 2002;99:13178–13183. doi: 10.1073/pnas.202035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster E, Jossin Y, Zhao S, Chai X, Frotscher M, Goffinet AM. Recent progress in understanding the role of Reelin in radial neuronal migration, with specific emphasis on the dentate gyrus. Eur J Neurosci. 2006;23:901–909. doi: 10.1111/j.1460-9568.2006.04612.x. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israël A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Hack I, Hellwig S, Jughans D, Brunne B, Bock HH, Zhao S, Frotscher M. Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons. Development. 2007;34:3883–3891. doi: 10.1242/dev.005447. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J. Neurosci. 2006;26:1045–56. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E. A role for Abl in Notch signaling. Neuron. 1998;20:667–681. doi: 10.1016/s0896-6273(00)81007-7. [DOI] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD. The T alpha 1 alpha-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci. 1994;14:7319–7330. doi: 10.1523/JNEUROSCI.14-12-07319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss E, Forster E, Bock HH, Hack MA, Leprince P, Luque JM, Herz J, Frotscher M, Gotz M. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development. 2003;130:4597–4609. doi: 10.1242/dev.00654. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev. Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Tran TS, Matsuoka Y, Howell BW, Rebeck GW. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J. Biol. Chem. 2006;281:35176–35185. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- Honda T, Nakajima K. Mouse Disabled1 (DAB1) is a nucleocytoplasmic shuttling protein. J. Biol. Chem. 2006;281:38951–38965. doi: 10.1074/jbc.M609061200. [DOI] [PubMed] [Google Scholar]
- Hong SE, Shugart YY, Huang DT, Shahwan SA, Grant PE, Hourthane JO, Martin ND, Walsh CA. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat. Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Howell BW, Lanier LM, Frank R, Gertler FB, Cooper JA. The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol. 1999;19:5179–5188. doi: 10.1128/mcb.19.7.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr. Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Ogawa M, Metin C, Tissir F, Goffinet AM. Inhibition of SRC family kinases and non-classical protein kinases C induce a reeler-like malformation of cortical plate development. J. Neurosci. 2003;23:9953–9959. doi: 10.1523/JNEUROSCI.23-30-09953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Goffinet AM. Reelin signals through Phosphatidylinositol 3-Kinase and Akt to control cortical development and through mTor to regulate dendritic growth. Moc. Cell Biol. 2007;27:7113–7124. doi: 10.1128/MCB.00928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatani S, Tabata H, Nakajima K. Neuronal migration in cortical development. J. Child. Neurol. 2005;20:274–279. doi: 10.1177/08830738050200040201. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci. 2008;30:36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- Keilani S, Sugaya K. Reelin induces a radial glial phenotype in human neural progenitor cells by activation of Notch-1. BMC Dev. Biol. 2008;8:69. doi: 10.1186/1471-213X-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshvara L, Benhayon D, Magdaleno S, Curran T. Identification of reelin-induced sites of tyrosyl phosphorylation on disabled 1. J.Biol.Chem. 2001;276:16008–16014. doi: 10.1074/jbc.M101422200. [DOI] [PubMed] [Google Scholar]
- Kohyama J, Tokunaga A, Fujita Y, Miyoshi H, Nagai T, Miyawaki A, Nakao K, Matsuzaki Y, Okano H. Visualization of spatiotemporal activation of Notch signaling: live monitoring and significance in neural development. Dev. Biol. 2005;286:311–325. doi: 10.1016/j.ydbio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kuo G, Arnaud L, Kronstad-O’Brien P, Cooper JA. Absence of Fyn and Src causes a reeler-like phenotype. J.Neurosci. 2005;25:8578–8586. doi: 10.1523/JNEUROSCI.1656-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Protein degradation: four E3s for the notch pathway. Curr. Biol. 2002;12:R74–78. doi: 10.1016/s0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM. Neuronal migration. Mech. Dev. 2001;105:47–56. doi: 10.1016/s0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- Le Gall M, De Mattei C, Giniger E. Molecular separation of two signaling pathways for the receptor, Notch. Dev Biol. 2008;313:556–567. doi: 10.1016/j.ydbio.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall M, Giniger E. Identification of two binding regions for the suppressor of hairless protein within the intracellular domain of Drosophila notch. J Biol Chem. 2004;279:29418–29426. doi: 10.1074/jbc.M404589200. [DOI] [PubMed] [Google Scholar]
- LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 2006;29:407–413. doi: 10.1016/j.tins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Curran T. Brain Development: integrins and the Reelin pathway. Curr. Biol. 2001;11:R1032–1035. doi: 10.1016/s0960-9822(01)00618-2. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Keshvara L, Curran T. Rescue of ataxia and preplate splitting by ectopic expression of Reelin in reeler mice. Neuron. 2002;33:573–586. doi: 10.1016/s0896-6273(02)00582-2. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U S A. 2007;16:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signaling by non-visual beta-arrestin. Nat. Cell Biol. 2005;7:1159–1161. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Oberg C, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol. Cell Neurosci. 2006;31:109–122. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Olson EC, Kim S, Walsh CA. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J. Neurosci. 2006;26:1767–1775. doi: 10.1523/JNEUROSCI.3000-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Hamanaka H, Ohshima T, Watanabe N, Mikoshiba K, Nukina N. Inhibition of ubiquitin ligase Siah-1A by disabled-1. Biochem. Biophys. Res. Commun. 2003;302:671–678. doi: 10.1016/s0006-291x(03)00247-x. [DOI] [PubMed] [Google Scholar]
- Pramatarova A, Ochalski PG, Chen K, Gropman A, Myers S, Min KT, Howell BW. Nck beta interacts with tyrosine-phosphorylated disabled 1 and redistributes in Reelin-stimulated neurons. Mol. Cell Biol. 2003;23:7210–7221. doi: 10.1128/MCB.23.20.7210-7221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramatarova A, Chen K, Howell BW. A genetic interaction between the APP and Dab1 genes influences brain development. Mol. Cell Neurosci. 2008;37:178–186. doi: 10.1016/j.mcn.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Lord MC, Caviness VS., Jr. Determinants of cell shape and orientation: a comparative Golgi analysis of cell-axon interrelationships in the developing neocortex of normal and reeler mice. J. Comp. Neurol. 1979;187:49–69. doi: 10.1002/cne.901870104. [DOI] [PubMed] [Google Scholar]
- Pinto-Lord MC, Evrard P, Caviness VS., Jr. Obstructed neuronal migration along radial glial fibers in the neocortex of the reeler mouse: a Golgi-EM analysis. Brain Res. 1982;256:379–393. doi: 10.1016/0165-3806(82)90181-x. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of the cerebral cortical areas. Science. 1988;8:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- Rice DS, Sheldon M, D’Arcangelo G, Nakajima K, Goldowitz D, Curran T. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- Rice DS, Curran T. Role of the Reelin signaling pathway in central nervous system development. Ann. Rev. Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- Šestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron. 2004;42:197–211. doi: 10.1016/s0896-6273(04)00222-3. [DOI] [PubMed] [Google Scholar]
- Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, Torii M, Flavell RA, Rakic P. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Yamamoto A, Kawaguchi A, Yamaguchi M, Mori K, Goldman SA, Okano H. Direct isolation of committed neuronal progenitor cells from transgenic mice coexpressing spectrally distinct fluorescent proteins regulated by stage-specific neural promoters. J. Neurosci. Res. 2001;65:220–227. doi: 10.1002/jnr.1145. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D’Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Soriano E, Del Rio JA. The cells of cajal-retzius: still a mystery one century after. Neuron. 2005;46:389–394. doi: 10.1016/j.neuron.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Stolt PC, Bock HH. Modulation of lipoprotein receptor functions by intracellular adaptor proteins. Cell Signal. 2006;18:1560–1571. doi: 10.1016/j.cellsig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Tezuka T, Morimura T, Hattori M, Mikoshiba K, Yamamoto T, Takenawa T. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem. J. 2004;384:1–8. doi: 10.1042/BJ20041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Neurons tend to stop migration and differentiate along the cortical internal plexiform zones in the Reelin signal-deficient mice. J. Neurosci. Res. 2002;69:723–730. doi: 10.1002/jnr.10345. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of the cerebral cortical areas. Science. 1988;8:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Walsh CA, Goffinet AM. Potential mechanisms of mutations that affect neuronal migration in man and mouse. Curr. Opin. Genet. Dev. 2000;10:270–274. doi: 10.1016/s0959-437x(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware ML, Fox JW, Gonzalez JL, Davis NM, Lambert de Rouvroit C, Russo CJ, Chua SC, Jr., Goffinet AM, Walsh CA. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- Wines-Samuelson M, Handler M, Shen J. Role of presenilin-1 in cortical lamination and survival of Cajal-Retzius neurons. Dev. Biol. 2005;277:332–346. doi: 10.1016/j.ydbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind Bomb 1-Expressing Intermediate Progenitors Generate Notch Signaling to Maintain Radial Glial Cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Zaki M, Shehab M, El-Aleem AA, Abdel-Salam G, Koeller HB, Ilkin Y, Ross ME, Dobyns WB, Gleeson JG. Identification of a novel recessive RELN mutation using a homozygous balanced reciprocal translocation. Am. J. Med. Genet. A. 2007;143:939–944. doi: 10.1002/ajmg.a.31667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.