Abstract
The cataract-associated Pro23 to Thr (P23T) mutation in human γD-crystallin (HGD) has a variety of phenotypes and is geographically widespread. Therefore there is considerable interest in understanding the molecular basis of cataract formation due to this mutation. We showed earlier [Pande, et al. (2005) Biochemistry 44, 2491-2500] that the probable basis of opacity in this case is the severely compromised, retrograde solubility and aggregation of P23T relative to HGD. The dramatic solubility change occurs even as the structure of the mutant protein remains essentially unchanged in vitro. We proposed that the retrograde solubility and aggregation of P23T were mediated by net hydrophobic, protein-protein interactions. Based on these initial findings for P23T and related mutants, and the subsequent finding that they show atypical phase behavior, we concluded that the protein clusters formed in solutions of the mutant proteins were held together by net hydrophobic, anisotropic interactions. Here we show, using chemical probes, that the surface hydrophobicities of these mutants are inversely related to their solubility. Furthermore, by probing the isolated N-terminal domains of HGD and P23T directly, we find that the increase in surface hydrophobicity of P23T is localized in the N-terminal domain. Modeling studies suggest the presence of sticky patches on the surface of the N-terminal domain that could be engaged in forming protein clusters via hydrophobic protein-protein interactions. This work thus provides direct evidence for the dominant role played by net hydrophobic and anisotropic protein-protein interactions in the aggregation of P23T.
Keywords: hydrophobicity, crystallin, fluorescence, cataract, mutation, bis-ANS, Nile Red, aggregation, self-association
The cataract-associated, Pro23 to Thr (P23T) mutation in human γD-crystallin (HGD), is geographically widespread and phenotypically heterogeneous (1-5). In an earlier study we showed that the solubility of the mutant protein in vitro is severely compromised, while the protein structure remains largely unaltered (6). More significantly, we found that, in contrast to the wild type, the mutant protein showed a retrograde solubility, (i.e. the solubility had an inverse dependence on temperature and increased upon lowering the temperature). These results clearly suggested that the insoluble phase is the result of the self-aggregation of the mutant protein mediated predominantly by net hydrophobic interactions. This behavior of P23T has a parallel with that of sickle cell hemoglobin, HbS (7). More recently, using NMR spectroscopy, we found that the effects of the P23T mutation were localized largely in the N-terminal domain and are likely to result in the creation of hydrophobic patches on the surface of the protein (8).
Other laboratories have compared the structures of HGD and P23T in solution as well. Synchrotron-radiation CD studies also showed minor spectral changes between the wild-type and mutant proteins which led the authors to speculate that these changes somehow lead to the compromised solubility of the mutant (9). More recently, a comprehensive NMR study by Jung et al. (10), points to local conformational and dynamic differences between the proteins, particularly those associated with the His22 residue, leading the authors to conclude that these small changes are in some way responsible for initiating the aggregation of the mutant protein. However, neither one of these studies explains the retrograde solubility change in P23T and other related mutants (6), which we believe is the key factor essential to understanding how this mutation leads to light scattering and cataract.
In order to address this issue definitively, we examined the surface hydrophobicities of HGD and P23T using two unrelated extrinsic dyes, bis-ANS and Nile Red (NR) (Fig. 1) , both commonly used as fluorescent probes of surface hydrophobicity in proteins (11). The data presented here, show that the surface hydrophobicity of P23T is indeed higher than that of HGD, and provide new, compelling experimental evidence for the hydrophobic protein-protein interactions and retrograde solubility exhibited by the mutant. We have also compared the surface hydrophobicities of the N-terminal domains of HGD and P23T, and find that the net increase in hydrophobicity in P23T resides primarily on the surface of its N-terminal domain.
FIGURE 1.
Chemical structures of bis-ANS and Nile Red.
MATERIALS AND METHODS
Cloning, Expression and Purification of recombinant proteins
Overexpression and purification of recombinant wild type HGD and its three mutants P23T, P23V and P23S has been reported earlier (6, 12). Primers were designed (synthesized by MSG Operon) to introduce a stop-codon at position 83 of the HGD and P23T sequence to express only their N-terminal domains, Nt-HGD and Nt-P23T respectively. In the resulting plasmids, the nucleotide sequences were confirmed in-house at the Biomolecular Core facility. The truncated proteins were expressed and purified in the same manner as the parent proteins. The purity of the recombinant proteins was verified using electrospray ionization mass spectrometry at the Center for functional Genomics at the University at Albany. The masses of Nt-HGD and Nt-P23T were 9,640 and 9,630 ± 1 Da respectively.
The concentration of HGD and its three mutants P23T, P23V and P23S was determined using the same extinction coefficient, 41.4 mM-1cm-1 (i.e. E280 for a 0.1% solution is 2.1) (13) at 280 nm as before (6). For Nt-HGD and Nt-P23T, an extinction coefficient of 2.25 at 280 nm for a 0.1% solution was used based on ProtParam tool (14) implemented in the EXPASY proteomics server (www.expasy.ch).
Fluorescence Spectra
These were recorded in a Jovin-Yvon Fluorolog-3 spectrometer using an excitation wavelength of 390 nm (for bis-ANS binding) and 540 nm (for NR binding). Excitation and emission slits were set to 5 nm; spectra were measured using a protein concentration of 0.1 mg/ml in 0.1 M phosphate buffer, pH 7.0. Stock solutions of the proteins were made in 20 mM acetate buffer, pH 4.5, and diluted with phosphate buffer to adjust the pH to 7 just before recording the spectra. Stock solutions of bis-ANS and NR were prepared in methanol and the final alcohol concentration was maintained below 7% v/v when the reagents were mixed with the proteins. Concentrations of bis-ANS and NR were measured using extinction coefficients of 16.8 mM-1 cm-1 at 385 nm (15) and 45 mM-1cm-1 at 552 nm (16) respectively. For a fixed protein concentration of 0.1 mg/ml (~5 μM), the concentrations of bis-ANS and NR were varied from 5 to 500 μM and 5 to 100 μM respectively.
Measurement of the temporal change in molecular size using static and dynamic light scattering
Stock solutions of Nt-HGD and Nt-P23T (~1.0 mg/ml) in 20 mM acetate buffer, pH 4.5 were diluted into 100 μl of 0.1 M phosphate buffer, pH 7. Increase in intensity of the Rayleigh scattering was monitored as a function of time in a Zetasizer-Nano analyzer (Malvern instruments) at 25°C, with toluene as a scattering standard. Two aliquots of the protein solution were removed at various time intervals: one aliquot was quickly placed in a Dynapro dynamic light scattering instrument (Wyatt Instruments) to measure the hydrodynamic radius (Rh), and the second was immediately mixed with bis-ANS solution and the fluorescence spectrum recorded as described above.
Circular Dichroism (CD) Spectra
CD spectra were recorded on a Jasco model J-815 spectropolarimeter. HGD, Nt-HGD and Nt-P23T solutions at concentrations of 1.2 mg/ml in 20 mM acetate buffer, pH 4.5, were used to measure the near-UV CD spectra using a 10 mm path length cuvette. Far-UV CD spectra were obtained using 0.1 mg/ml protein solutions in 5 mM acetate buffer, pH 4.5, in a 1 mm path length cuvette. Near- and far-UV CD spectra were normalized with respect to protein concentration and the concentration of peptide bonds respectively.
Computation of surface hydrophobicity
The solution structures (20 models) obtained from NMR studies of the P23T mutant (10) were downloaded from the Protein Data Bank (PDB ID: 2KFB). The accessible surface area (ASA) of all the amino acid residues in the twenty structures was calculated using NACCESS (17). In a similar way the ASA of all the amino acid residues was calculated for HGD using its crystal structure (PDB ID: 1HK0) (18). By multiplying the ASA of the hydrophobic amino acid residues (Ile, Leu, Tyr, Phe, Val, Trp, and Met) with their hydrophobicity index, the total surface hydrophobicity of the twenty structures of P23T and that of HGD was calculated (19). Two P23T structures that scored the highest in N-terminal hydrophobicity were structure numbers 3 and 19 of Jung et al (10). The surface patches on the protein surface of structure 3 of P23T and on HGD were located computationally by using the SHARP2 protein server (20, 21). SHARP2 is a web-based server for the prediction of protein interaction sites on the protein surface. A number of surface patches are defined based on a selectable parameter set. We used only 2 parameters – hydrophobicity and solvation potential, and set them to high and low respectively. In this manner we restricted the patches to those that would be hydrophobic and prefer to remain in a protein-protein interface rather than exposed to the solvent at the protein surface.
RESULTS
Fluorescent dyes such as ANS are commonly used to determine the surface hydrophobicity of proteins. While ANS is the most popular reagent used, its derivative bis-ANS generally shows better fluorescence characteristics (22). These dyes are known to bind to hydrophobic patches on the protein surface (11) largely by means of their aromatic moieties (23). However, some of these, such as ANS and bis-ANS also carry electrostatic charges, and thus have the potential to interact with charged groups on the protein surface. Therefore, in order to eliminate the effect of charge-charge interactions between the dye and the protein of interest, we also used, besides bis-ANS, a neutral dye known as Nile Red (NR), to determine surface hydrophobicity of our proteins. The structures of both dyes used in this study are shown in (Fig. 1)
Bis-ANS binding profiles of HGD, P23T, P23S and P23V
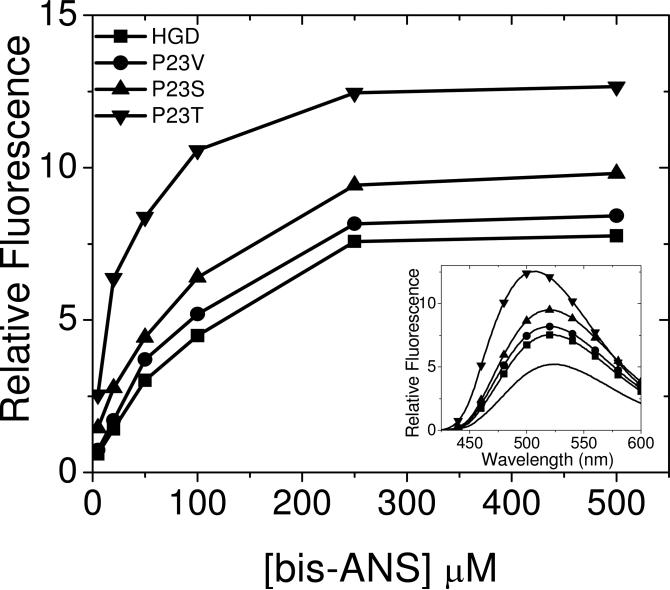
In Fig. 2 we plot the fluorescence intensity at 515 nm — the emission-maximum of bis-ANS — in mixtures containing various mutants of HGD (P23T, P23V and P23S), as a function of increasing concentrations of reagent. As expected, HGD shows the lowest emission intensity, followed by P23V, P23S and P23T, in ascending order of fluorescence emission. Clearly, the fluorescence emission of P23T is significantly higher than that of all the other mutants. These data further highlight the inverse relationship between the solubility of these mutants determined earlier (6), and the bis-ANS fluorescence profiles shown here: The P23T mutant with the lowest solubility has the highest fluorescence emission, while P23V with the highest solubility (approaching that of the wild type) has the lowest fluorescence emission, closest to that of the wild-type. From the plots shown in Fig. 2, we also obtain the lowest concentration of bis-ANS that is sufficient to attain the maximum fluorescence intensity in all the mutants — i.e. the so-called saturation-concentration of bis-ANS — and note that this concentration is comparable for all four proteins.
FIGURE 2.
Relative fluorescence intensity at the emission maximum, 515 nm (excitation at 390 nm) for mixtures of bis-ANS with human γD-crystallin and its mutants (P23V, P23S and P23T) at various bis-ANS concentrations. Protein concentration kept constant at 0.1 mg/ml (~5 μM) in 0.1 M phosphate buffer, pH 7.0. Symbols represent the data and lines are merely visual guides. Inset shows the corresponding fluorescence emission spectra of the mixtures at a bis-ANS concentration of 250 μM. The dotted line is the emission spectrum of bis-ANS alone.
The inset to Figure 2 shows the emission spectra, clearly revealing that the P23T mutant has a substantially higher emission intensity compared to the others, as well as a larger blue-shift of the emission maximum. It follows therefore that the surface of the cataract-associated mutant P23T is much more hydrophobic than that of the other mutants or the wild-type. Setting the increase in fluorescence emission of P23T arbitrarily at 100% relative to bis-ANS alone, we estimate that the percent emission values of P23S, P23V and HGD are about 59%, 44% and 34% respectively. The inset to Fig. 2 also shows that the emission maxima of the other two mutants P23S and P23V do not show a blue-shift relative to that of HGD, even though their emission intensities are higher. We note that the emission of P23V is closest to that of HGD, as is its solubility determined earlier (6, 24).
Nile Red binding profiles of HGD, P23T, P23S and P23V
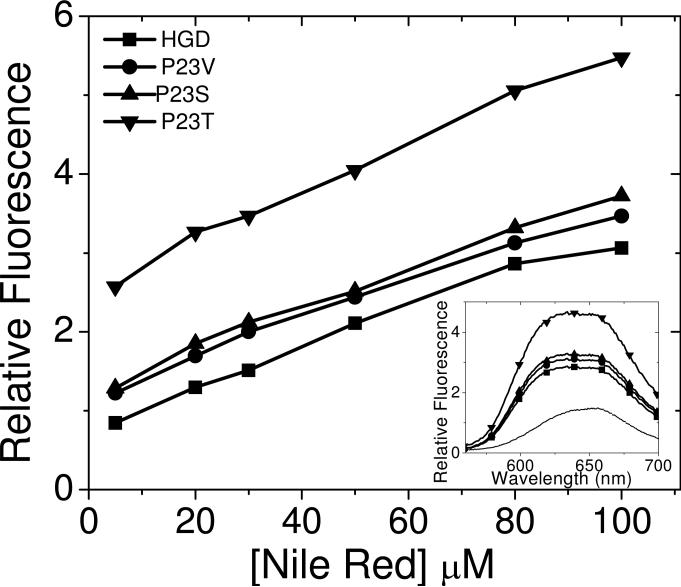
Although NR has been used to probe surface hydrophobicity in proteins (11, 25, 26), it is not as popular as ANS and bis-ANS. We chose NR because it is not charged and thus likely to show different binding characteristic to the hydrophobic sites on the protein surface. Moreover, since these methods — although time-tested, are nevertheless primarily empirical — we reasoned that the collective weight of evidence using different chemistries would be more definitive. The data using NR as the extrinsic dye are shown in Fig. 3. Once more we observe that — consistent with the bis-ANS data shown in Fig. 2 — the intensity of the fluorescence emission is inversely related to protein solubility, and again the P23T mutant stands out with a much higher fluorescence intensity than the wild type and other mutants. We did not observe saturation of binding even at the highest concentration of NR used here. However, we were unable to go to concentrations higher than those shown in Fig. 3, as NR precipitates a few minutes after mixing.
FIGURE 3.
Relative fluorescence intensity at the emission maximum, 650 nm (excitation at 540 nm) for mixtures of Nile Red with human γD-crystallin and its mutants (P23V, P23S and P23T) at various Nile Red concentrations. Protein concentration kept constant at 0.1 mg/ml (~5 μM) in 0.1 M phosphate buffer, pH 7.0. Symbols represent the data and lines are merely visual guides. Inset shows the corresponding fluorescence emission spectra of the mixtures at a Nile-red concentration of 80 μM. The dotted line is the emission spectrum of Nile Red alone.
The inset to Fig. 3 shows that the emission spectra are broad. A contributing factor to this broadening could be the unbound NR in solution which shows significantly higher emission intensity around 660 nm (see dotted curve in Fig. 3, inset). Furthermore, it is also likely that NR emission is more sensitive than bis-ANS in binding to sites with different hydrophobicities. Such broad spectra are also observed in the case of tubulin polymerization, and are apparently a result of binding to different sites (27). However, the significance of the data shown in Fig. 3 is that, just as with bis-ANS, the emission of P23T bound to NR is significantly higher than that of the others. Here again, if we arbitrarily set the increase in fluorescence emission of P23T at 100% relative to NR alone, we estimate that the percent emission values of P23S, P23V and HGD are about 56%, 50% and 44% respectively, which are comparable to those obtained with bis-ANS, shown in Fig. 2.
Bis-ANS fluorescence of Nt-HGD and Nt-P23T
In order to understand how the mutation of Pro23 to Thr leads to an increase in surface hydrophobicity, we refer to our recent NMR studies (8). In that study we showed that it is primarily the local, small changes in the N-terminal domain of HGD that are likely to lead to the formation of hydrophobic patches on the surface of P23T. Therefore, if that were indeed the case, we hypothesized that we should also observe a difference in hydrophobicity in the isolated N-terminal domains of HGD and P23T — just as in the full-length proteins.
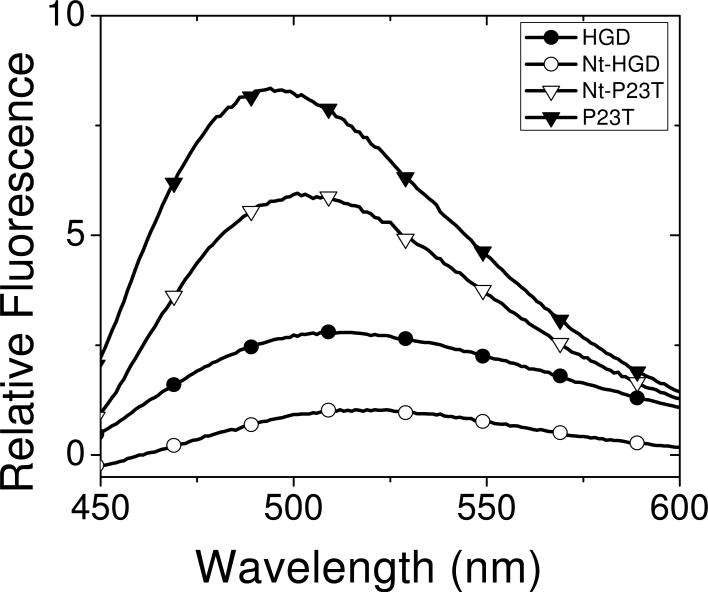
To test this hypothesis, we reacted the N-terminal domains of HGD and P23T (Nt-HGD and Nt-P23T respectively) with bis-ANS. The results shown in Fig. 4 compare the emission spectra of the two N-terminal domains and their corresponding parent proteins when bound to bis-ANS. Here the spectra are shown after subtracting the emission spectrum of bis-ANS alone, which allows us to compare the magnitude of the fluorescence change in all four cases. From the difference spectrum [HGD minus Nt-HGD], we can estimate the contribution of Ct-HGD to the fluorescence intensity. Similarly, from the difference spectrum [P23T minus Nt-P23T], we can estimate the contribution of Ct-P23T. From such simple subtraction methods, the data in Fig. 4 show that of the total difference in fluorescence intensity between HGD and P23T, about 10% is contributed by the C-terminal domain, and the remaining 90% by the N-terminal domain. These conclusions are based on our assumption that the fluorescence arising from the N-terminal domain alone, does not change from that observed in the intact proteins.
FIGURE 4.
Fluorescence emission spectra of HGD, Nt-HGD, P23T, and Nt-P23T in mixtures with bis-ANS. Spectra are corrected for bis-ANS by subtracting its contribution. Protein concentration was about 5μM and bis-ANS concentration was 250μM in each case. Excitation wavelength was 390 nm.
It should be noted that Nt-P23T rapidly aggregates (for details, see Fig. 6), even at low concentrations, e.g. 0.1 mg/ml, at pH 7 and ambient temperature. Therefore for these and other experiments, we stored it at pH 4.5 in low ionic strength buffer (20 mM Acetate) and raised the pH of the solution to 7 just before use by mixing with 0.1M phosphate buffer at pH 7.0.
FIGURE 6.
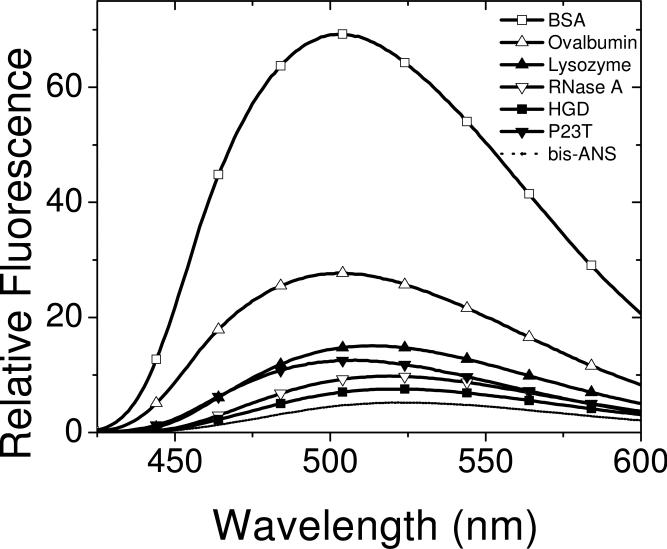
Fluorescence emission spectra (excitation at 390 nm) for mixtures of bis-ANS with bovine serum albumin (BSA), albumin from chicken egg white (ovalbumin), lysozyme from chicken egg white, ribonuclease A from bovine pancreas (RNase A), and HGD and P23T at protein concentrations of ~ 5 μM and bis-ANS concentration of 250 μM in 0.1 M phosphate buffer pH 7.0. Dotted line shows the emission spectrum of 250 μM bis-ANS alone in the same buffer.
Structures of Nt-HGD and Nt-P23T
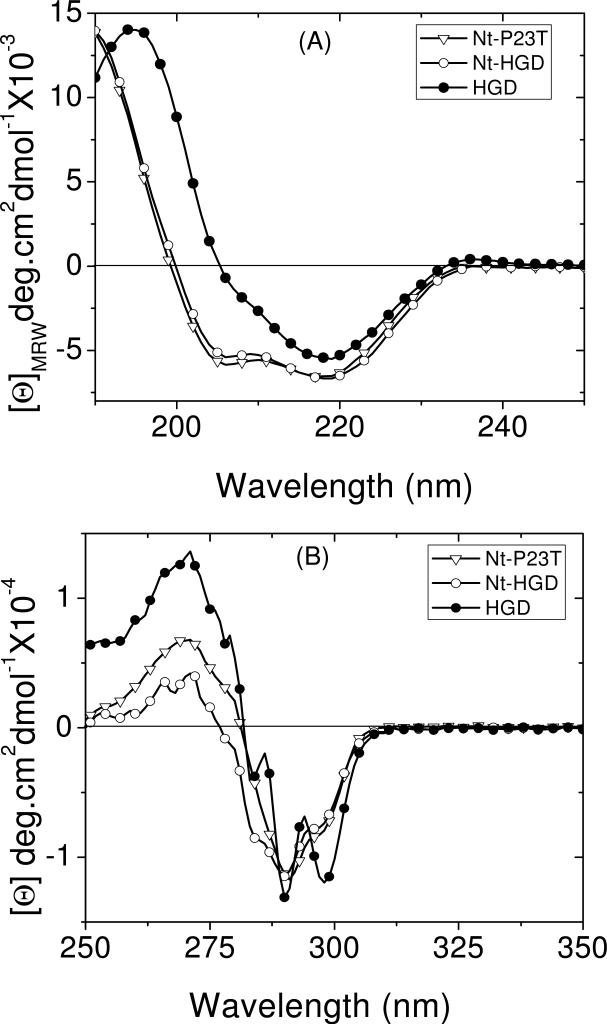
From the above discussion it is evident that the difference between the bis-ANS fluorescence of Nt-P23T and Nt-HGD is comparable to that observed in the parent proteins (Figs. 2 and 4), which clearly suggests that the difference arises directly from the mutation. This conclusion is further strengthened by measuring the far- and near-UV CD spectra of the two proteins (Figs. 5A & B respectively). For the CD measurements the samples were prepared in pH 4.5 buffer to prevent the aggregation of Nt-P23T which is significant at pH 7. The HGD spectrum shown here was measured under identical conditions for reference.
FIGURE 5.
(A) Far-UV and (B) near-UV CD spectra of Nt-HGD, Nt-P23T and HGD at pH4.5. Protein concentrations were 0.1 mg/ml in 5 mM acetate buffer for the far-UV CD measurements, and 1.2 mg/ml in 20 mM acetate buffer for the near-UV CD measurements.
Both far- and near-UV spectra show that the N-terminal domains of the two proteins have nearly identical structures. Mills et al (28) have reported the far-UV CD spectrum of Nt-HGD but their sample (unlike ours) contains an N-terminal His-tag. Our far-UV CD spectrum of Nt-HGD appears to be visually comparable to their spectrum. The differences in our far-UV CD spectra of Nt-HGD and Nt-P23T are minor, and comparable to the small differences reported by Evans et al (9) for intact HGD and P23T. Since NMR studies (10) show that the two proteins have nearly identical structures, it is reasonable to conclude that the differences observed in our far-UV CD spectra of Nt-HGD and Nt-P23T are also not significant. In general, the near-UV CD spectrum is more sensitive to changes in tertiary structure and side-chain rearrangements in proteins than the far-UV CD spectrum. Thus, the difference we observe in the near-UV CD between these proteins may reflect the minor changes in structure that were observed in the NMR spectra of the parent proteins (8, 10). Overall therefore, it is reasonable to conclude that there is no significant structural change between Nt-HGD and Nt-P23T beyond what is expected from the structures of the parent proteins.
Defining a measure of surface hydrophobicity
We also compared the fluorescence emission spectra of bis-ANS binding to several well-studied proteins, with those of HGD and P23T (Fig. 6). This enabled us to obtain a more quantitative estimate of the magnitude of the surface hydrophobicity-difference between HGD and P23T. From the data shown in Fig. 6 we deduce that the difference in surface hydrophobicity between HGD and P23T (about 9%) is comparable to that between ribonuclease A (RNase A) and lysozyme (about 8%), when the increase in fluorescence emission for bovine serum albumin (BSA) (with the largest magnitude of fluorescence emission in Fig. 6) is arbitrarily set to 100% relative to bis-ANS alone. All the reference proteins used here, namely, RNase A, lysozyme, ovalbumin and BSA have been tested earlier for surface hydrophobicity using ANS (29). Thus, our data is in good agreement with this published work. Thus, a comparison with the hydrophobicity data of some well studied proteins provides an empirical measure of the increase in the surface hydrophobicity of P23T.
Correlation of the increase in hydrodynamic radius (Rh) and scattering intensity of Nt-P23T with the fluorescence emission intensity due to bis-ANS binding
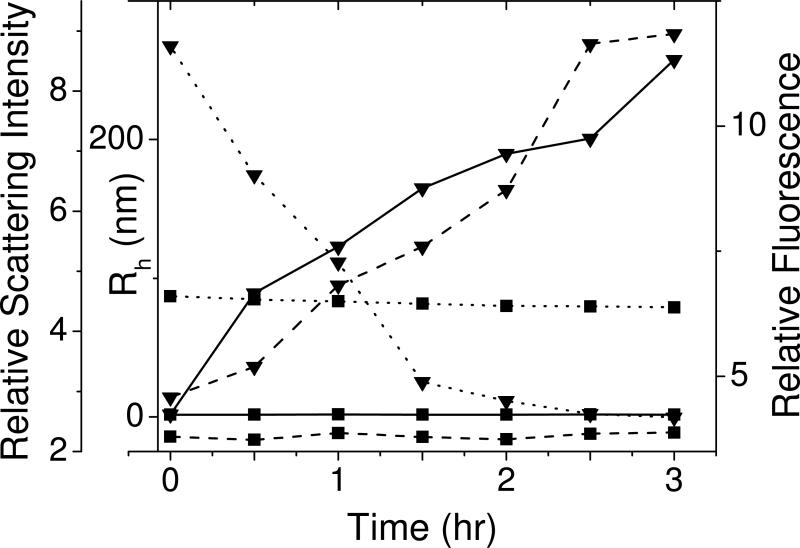
Fig. 7 shows that at pH 7, Nt-P23T, aggregates over time resulting in an increase in the light scattering intensity. This leads to an increase in the hydrodynamic radius of the clusters from ~1 nm to ~250 nm in 3 hours. Here, as in our earlier paper (6), we use “cluster” and “aggregate” interchangeably to describe the solid phase. Larger aggregates are formed from the smaller clusters and may become irreversible — a phenomenon well understood in colloidal systems (30). As the aggregate size grows there is a corresponding drop in the intensity of the bis-ANS fluorescence. Notably, throughout this period, the Nt-HGD solution does not register any change: The hydrodynamic radius corresponds to that of the monomeric protein, and the bis-ANS fluorescence remains virtually constant. These results vividly demonstrate that the same hydrophobic surface patches in Nt-P23T that bind to bis-ANS, are also involved in the self-aggregation of the N-terminal domain. This is clearly shown by the fact that as the protein aggregates grow in size and number, the hydrophobic sites for bis-ANS binding available on the surface of Nt-P23T decrease. Eventually, as the aggregate size grows sufficiently large, the protein aggregates precipitate. The temporal changes observed in Nt-P23T solutions are highly significant since they explain why the bis-ANS fluorescence of Nt-P23T is lower in intensity than that of Nt-HGD for the same protein concentration, beyond a certain time point.
FIGURE 7.
Temporal change in hydrodynamic radius (left-axis, solid line), light scattering intensity (left-axis, dashed line) and fluorescence emission, (emission maximum, 515 nm, excitation at 390 nm, right-axis, dotted line) for bis-ANS mixtures with Nt-HGD (■) and Nt-P23T (▼) in 0.1 M phosphate buffer pH 7.0. Bis-ANS and protein concentrations are 250 μM and 0.1 mg/ml (5 μM) respectively.
Mapping hydrophobic patches on the protein surface
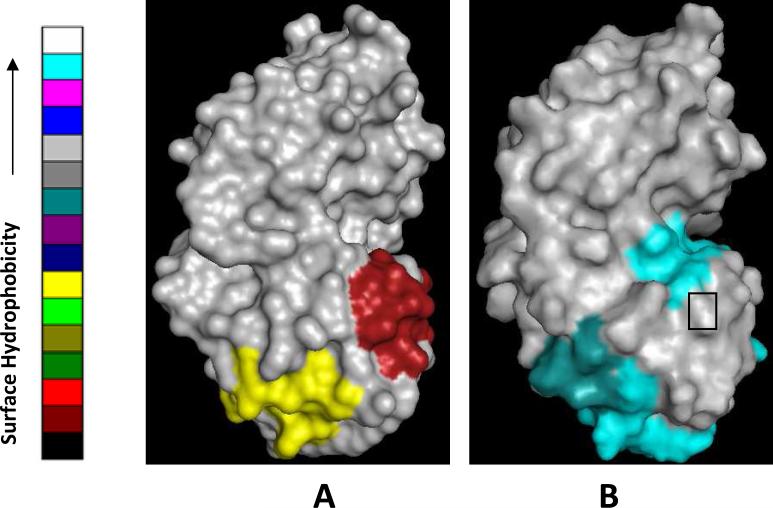
In Fig. 8 are shown the models for HGD and P23T based on the X-ray- (18) and NMR- (10) derived structures respectively. The colored patches displayed on their surfaces were computed using the SHARP2 server (20, 21) as already described (Materials and Methods). In Fig. 8A, the yellow patch on HGD corresponds to a moderately hydrophobic region based on the surface hydrophobicity scale on the left, and includes the residues K2, T4, H15, Y16, E17, C18, R36, and the red patch corresponds to low surface hydrophobicity and includes the residues P23, N24, Q47, P48, N49, Y50, R76. In Fig. 8B, the contiguous teal and cyan patch near the bottom shows a considerable increase in the level of hydrophobicity in the P23T mutant and covers the residues, K2, T4, Y6, F11, Q12, G13, R14, H15, Y16, E17, S20, R36, whereas the smaller cyan patch, which also corresponds to a higher level of hydrophobicity, includes the residues Y45, Y50, G52, L53, Q54, F56, R58, M69, R76, R79 (residues are numbered as in the HGD crystal structure). Thus, the putative potentially-hydrophobic patches in HGD reorganize and extend to cover a larger surface area and may represent the “sticky patches” engaged in the formation of protein clusters and solid precipitate. Fig.8 also shows that residue 23, which is a Pro in HGD and part of the red patch of weaker hydrophobicity (Fig. 8A), is replaced by a Thr in P23T (Fig. 8B), and is at the bottom of the smaller teal patch, just outside the region of considerably higher hydrophobicity. We should clarify that this computation is presented here only as a model to show how this mutation could lead to aggregation driven by hydrophobic protein-protein interactions and drastically lowered solubility. We do not claim to have identified the exact location of the “sticky patches” on the surface of P23T.
FIGURE 8.
Surface representation of (A) the crystal structure of HGD (PDB ID: 1HK0) and (B) the NMR structure 3 of P23T (PDB ID: 2KFB). The potentially “sticky” patches on the surface of HGD (shown in red and yellow in (A) and described earlier (8)), appear to extend over a wider surface and become more hydrophobic in P23T (B). The models are depicted here according to the surface hydrophobicity color-scale. In (B), the small rectangle at the bottom of the smaller cyan patch marks the location of residue 23. Figures were made in PyMol (32).
DISCUSSION
Several laboratories have explored the basis for cataract formation due to the P23T mutation in human γD-crystallin (6, 9, 10). It is evident from all of these studies that despite the dramatic lowering in solubility, the protein structure remains folded and undergoes very minor changes due to the mutation. In our earlier investigations (6, 24) we noted that P23T as well as related mutants of HGD (i.e. P23S and P23V), all showed a retrograde solubility profile in addition to the solubility being significantly compromised. This was in sharp contrast to wild-type HGD, which is soluble to concentrations greater than 400 mg/ml. Based on this work (6), we concluded that the minor structural changes accompanying the mutation had resulted in creating protein clusters that were held together by predominantly hydrophobic interactions. Subsequently, it was observed (24) that an analog of P23T — i.e. the P23V mutant — engages in highly anisotropic protein-protein interactions in the solid aggregated state. This conclusion was derived from the observation that the P23V mutation, while it alters the solubility of the protein and the chemical potential of the solid (i.e. crystal) phase, it does not affect the phase separation properties of the solution phase (i.e. the liquid-liquid phase boundary is indistinguishable from that of the wild type). Thus, the work of McManus et al. (24) suggests that the inter-protein interactions in P23V are anisotropic, in that the orientation effects due to the anisotropy are manifest only in the solid phase but are randomly distributed and averaged out in solution. These arguments should also hold for the P23T and P23S mutants but cannot be experimentally demonstrated, because it has neither been possible to concentrate their aqueous solutions to a high enough concentration to obtain liquid-liquid phase separation, nor has it been possible to suppress aggregation and crystallize them.
We also note that such anisotropic distribution of hydrophobic patches on the protein surface is not readily apparent. Our recent NMR studies (8) showed that the effect of the P23T mutation is localized primarily in the N-terminal domain. Thus, the sticky patches created in the P23T mutant that lead to protein clusters and solid-precipitation are likely to reside mainly in the N-terminal domain. In fact, using the SHARP2 server (21) we concluded that potentially-hydrophobic surface patches already exist in HGD, but are rendered more hydrophobic and hence more effective in promoting the aggregation of P23T (8).
As stated in the Results section, the current study was undertaken to probe the surface-hydrophobicity issue of P23T directly using molecules such as bis-ANS that are sensitive to the surface hydrophobicity of proteins. Bis-ANS is a better alternative to the more commonly used ANS (11, 29) due to its superior fluorescence properties (22). Using bis-ANS we have shown here that the intensity of fluorescence emission of the three mutant proteins, P23T, P23S and P23V, correlates inversely with their solubility profiles that we determined earlier: Higher the surface hydrophobicity, lower the solubility of the mutant proteins at ambient temperature. While this is precisely what we expected based on our earlier work (6, 24), we do not yet understand why the P23T mutant shows the largest increase in surface hydrophobicity. In fact, we previously made several mutations at and adjacent to site 23, in an attempt to address this issue (6), but it still remains unresolved. The close correspondence between our fluorescence data obtained using bis-ANS and Nile Red make a compelling case for the dominant role played by surface hydrophobicity in facilitating the aggregation of P23T, and lead us to conclude that it is indeed the extent of surface hydrophobicity that correlates inversely with protein solubility.
In earlier studies we (8) and others (10) observed that in introducing a mutation at site 23, the C-terminal end of the protein remains minimally affected. Interestingly, while our data showed minor structural perturbation in the C-terminal domain, Jung et al (10), even to their surprise, showed no perturbation whatsoever in the C-terminal. In any event, both studies agreed that, as expected, the main structural change resided in the N-terminal domain. It seemed logical therefore, to examine the N-terminal domains alone (residues 1-82) of HGD and P23T, (i.e. Nt-HGD and Nt-P23T respectively) for changes in surface hydrophobicity. The data showed that as expected, the difference in the surface hydrophobicities of these two proteins observed by bis-ANS fluorescence is closely similar to that in the intact, full-length proteins. More specifically, as we showed in the Results section (see Bis-ANS fluorescence of Nt-HGD and Nt-P23T), it appears that ~90% of the difference in surface hydrophobicity between HGD and P23T can be accounted for by that in the N-terminal domain alone, with a minor portion (about 10%) arising from the C-terminal domain. This is qualitatively consistent with our NMR study (8) which showed that there is a small change in the C-terminal domain as well upon mutation. Thus it is clear that mutation of Pro23 to Thr in HGD leads to an increase in surface hydrophobicity largely in the N-terminal domain.
In the case of Nt-P23T, the increase in surface hydrophobicity makes the protein unstable at pH 7. It rapidly aggregates and forms increasingly large aggregates. We have observed that the larger the cluster-size, the lower the binding to bis-ANS. Thus, the same hydrophobic sites that bind to bis-ANS appear to be responsible for the self-aggregation of the N-terminal domain.
The data shown in Fig. 6 in which we compare the bis-ANS binding to several proteins including HGD and P23T, show that relative to other well-studied proteins, the increase in surface hydrophobicity of P23T is relatively small. Thus, it is not simply the absolute surface hydrophobicity of the mutant protein per se, but the net increase in surface hydrophobicity that is the dominant trigger for the lowering of solubility.
Based on our NMR data (8), we speculated that there are two potentially hydrophobic patches on the surface of HGD, which are likely to engage in hydrophobic interactions in P23T. We examined all 20 recently-available NMR structures of the P23T mutant (10), and compared them to our high-resolution x-ray crystal structure of HGD (18). Specifically, we compared the total surface hydrophobicity and found that by this measure nearly all the NMR structures of P23T show a higher hydrophobicity than HGD. We analyzed one of the structures of P23T with the highest hydrophobicity index for possible hydrophobic patches on the N-terminal domain and found two patches nearly identical to those we reported earlier (8). One patch had a considerably higher hydrophobicity in P23T compared to HGD, while the other showed a significantly increased hydrophobicity, albeit of a smaller magnitude. These data shown in Fig. 8 serve as a reasonable model to understand our experimental data.
In summary, our experimental data published earlier (6, 8) taken together with the work of McManus et al. (24), and the studies reported here, lead us to conclude that (i) the cataract-associated, P23T mutation in HGD leads to a significantly lower and retrograde solubility of the protein, which aggregates, even as its overall structure remains essentially unaltered, (ii) these aggregates are formed by a net increase in the hydrophobic protein-protein interactions that are anisotropic in the solid (crystal) phase, (iii) NMR data suggested the possibility of formation of hydrophobic patches on the surface of the N-terminal domain of P23T relative to HGD, and (iv) chemical probes of surface hydrophobicity have now provided compelling evidence that the N-terminal domain is the major contributor to the net increase in hydrophobicity due to the mutation.
While these data provide a plausible mechanism for lens opacity due to the P23T mutation, we recognize that the question of the phenotypic heterogeneity of the resulting cataract itself is more challenging to explain. Since protein aggregation is triggered by the creation of hydrophobic surface patches in this case, it is conceivable that a variety of distinct nucleation sites in vivo give rise to different aggregate forms depending on where the initial hydrophobic cluster nucleates: For example, at the cell membrane or with other crystallins or non-crystallin protein components within the fiber cell — all due to an increase in surface hydrophobicity. Such interactions could generate a variety of phenotypes. We should also note however that, as in the case of sickle hemoglobin (31), which also exhibits several distinct phenotypes, genetic determinants not linked to the HGD gene could modulate the phenotype and contribute to phenotypic heterogeneity as well.
ACKNOWLEDGEMENT
The authors thank the reviewers for their careful reading of the manuscript and constructive suggestions for improvement.
Abbreviations
- HGD
human γD-crystallin
- CD
circular dichroism
- DTT
dithiothreitol
- P23T
Pro23 to Thr mutant of HGD
- ANS
1-anilinonaphthalene-8-sulfonate
- bis-ANS
4,4’-bisanilinonaphthalene-8-sulfonate
- Rh
hydrodynamic radius
- Nt-HGD
N-terminal domain of HGD
- Nt-P23T
N-terminal domain of P23T
- Ct-HGD
C-terminal domain of HGD
- Ct-P23T
C-terminal domain of P23T
- NR
Nile Red (9-diethylamino-5H-benzo[a]phenoxazine-5-one)
Footnotes
Supported by NIH Grant EY010535
REFERENCES
- 1.Nandrot E, Slingsby C, Basak A, Cherif-Chefchaouni M, Benazzouz B, Hajaji Y, Boutayeb S, Gribouval O, Arbogast L, Berraho A, Abitbol M, Hilal L. Gamma-D crystallin gene (CRYGD) mutation causes autosomal dominant congenital cerulean cataracts. J Med Genet. 2003;40:262–267. doi: 10.1136/jmg.40.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santhiya ST, Shyam Manohar M, Rawlley D, Vijayalakshmi P, Namperumalsamy P, Gopinath PM, Loster J, Graw J. Novel mutations in the gamma-crystallin genes cause autosomal dominant congenital cataracts. J Med Genet. 2002;39:352–358. doi: 10.1136/jmg.39.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shentu X, Yao K, Xu W, Zheng S, Hu S, Gong X. Special fasciculiform cataract caused by a mutation in the gammaD-crystallin gene. Mol Vis. 2004;10:233–239. [PubMed] [Google Scholar]
- 4.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. Autosomal dominant coralliform cataract related to a missense mutation of the gammaD-crystallin gene. Chin Med J (Engl) 2004;117:727–732. [PubMed] [Google Scholar]
- 5.Mackay DS, Andley UP, Shiels A. A missense mutation in the gammaD crystallin gene (CRYGD) associated with autosomal dominant “coral-like” cataract linked to chromosome 2q. Mol Vis. 2004;10:155–162. [PubMed] [Google Scholar]
- 6.Pande A, Annunziata O, Asherie N, Ogun O, Benedek GB, Pande J. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 7.Eaton WA, Hofrichter J. Sickle cell hemoglobin polymerization. Adv Protein Chem. 1990;40:63–279. doi: 10.1016/s0065-3233(08)60287-9. [DOI] [PubMed] [Google Scholar]
- 8.Pande A, Zhang J, Banerjee PR, Puttamadappa SS, Shekhtman A, Pande J. NMR study of the cataract-linked P23T mutant of human gammaD-crystallin shows minor changes in hydrophobic patches that reflect its retrograde solubility. Biochem Biophys Res Commun. 2009;382:196–199. doi: 10.1016/j.bbrc.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, Slingsby C. The P23T cataract mutation causes loss of solubility of folded gammaD-crystallin. J Mol Biol. 2004;343:435–444. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 10.Jung J, Byeon IJ, Wang Y, King J, Gronenborn AM. The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes. Biochemistry. 2009;48:2597–2609. doi: 10.1021/bi802292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharmaceutical Research. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pande A, Pande J, Asherie N, Lomakin A, Ogun O, King JA, Lubsen NH, Walton D, Benedek GB. Molecular basis of a progressive juvenile-onset hereditary cataract. Proc Natl Acad Sci U S A. 2000;97:1993–1998. doi: 10.1073/pnas.040554397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andley UP, Mathur S, Griest TA, Petrash JM. Cloning, expression, and chaperone-like activity of human alphaA-crystallin. J Biol Chem. 1996;271:31973–31980. doi: 10.1074/jbc.271.50.31973. [DOI] [PubMed] [Google Scholar]
- 14.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the Expasy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 15.Yu XC, Margolin W. Inhibition of assembly of bacterial cell division protein FtsZ by the hydrophobic dye 5,5'-bis-(8-anilino-1-naphthalenesulfonate) J Biol Chem. 1998;273:10216–10222. doi: 10.1074/jbc.273.17.10216. [DOI] [PubMed] [Google Scholar]
- 16.Ruvinov SB, Yang XJ, Parris KD, Banik U, Ahmed SA, Miles EW, Sackett DL. Ligand-mediated changes in the tryptophan synthase indole tunnel probed by nile red fluorescence with wild type, mutant, and chemically modified enzymes. J Biol Chem. 1995;270:6357–6369. doi: 10.1074/jbc.270.11.6357. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard SJ, Thornton JM. ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. [Google Scholar]
- 18.Basak A, Bateman O, Slingsby C, Pande A, Asherie N, Ogun O, Benedek GB, Pande J. High-resolution X-ray crystal structures of human gammaD crystallin (1.25 A) and the R58H mutant (1.15 A) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 19.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. Relationship of sidechain hydrophobicity and alpha-helical propensity on the stability of the single-stranded amphipathic alpha-helix. J Pept Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 20.Jones S, Thornton JM. Analysis of protein-protein interaction sites using surface patches. J Mol Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 21.Murakami Y, Jones S. SHARP2: protein-protein interaction predictions using patch analysis. Bioinformatics. 2006;22:1794–1795. doi: 10.1093/bioinformatics/btl171. [DOI] [PubMed] [Google Scholar]
- 22.Rosen CG, Weber G. Dimer formation from 1-amino-8-naphthalenesulfonate catalyzed by bovine serum albumin. A new fluorescent molecule with exceptional binding properties. Biochemistry. 1969;8:3915–3920. doi: 10.1021/bi00838a006. [DOI] [PubMed] [Google Scholar]
- 23.Bothra A, Bhattacharyya A, Mukhopadhyay C, Bhattacharyya K, Roy S. A fluorescence spectroscopic and molecular dynamics study of bis-ANS/protein interaction. Journal of Biomolecular Structure & Dynamics. 1998;15:959–966. doi: 10.1080/07391102.1998.10508216. [DOI] [PubMed] [Google Scholar]
- 24.McManus JJ, Lomakin A, Ogun O, Pande A, Basan M, Pande J, Benedek GB. Altered phase diagram due to a single point mutation in human gamma D-crystallin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16856–16861. doi: 10.1073/pnas.0707412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter M, Oliveira S, Sanders NN, Lucas B, van Hoek A, Hink MA, Visser AJ, De Smedt SC, Hennink WE, Jiskoot W. Sensitive spectroscopic detection of large and denatured protein aggregates in solution by use of the fluorescent dye Nile red. J Fluoresc. 2007;17:181–192. doi: 10.1007/s10895-007-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jee AY, Park S, Kwon H, Lee M. Excited state dynamics of Nile Red in polymers. Chemical Physics Letters. 2009;477:112–115. [Google Scholar]
- 27.Sackett DL, Knutson JR, Wolff J. Hydrophobic surfaces of tubulin probed by time-resolved and steady-state fluorescence of nile red. J Biol Chem. 1990;265:14899–14906. [PubMed] [Google Scholar]
- 28.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins gammaD-crystallin and gammaS-crystallin. Protein Sci. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardamone M, Puri NK. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem J. 1992;282(Pt 2):589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young TM, Roberts CJ. Structure and thermodynamics of colloidal protein cluster formation: comparison of square-well and simple dipolar models. J Chem Phys. 2009;131:125104. doi: 10.1063/1.3238569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinberg MH, Rodgers GP. Pathophysiology of sickle cell disease: role of cellular and genetic modifiers. Semin Hematol. 2001;38:299–306. doi: 10.1016/s0037-1963(01)90023-x. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL. The Pymol Molecular Graphics System. 2008.