Abstract
Mucolipidosis type IV is a lysosomal storage disorder caused by the loss or dysfunction of the mucolipin-1 (TRPML1) protein. It has been suggested that TRPML2 could genetically compensate (i.e., become upregulated) for the loss of TRPML1. We thus investigated this possibility by first studying the expression pattern of mouse TRPML2 and its basic channel properties using the varitint-waddler (Va) model. Here, we confirmed the presence of long variant TRPML2 (TRPML2lv) and short variant (TRPML2sv) isoforms. We showed for the first time that, heterologously expressed, TRPML2lv-Va is an active, inwardly rectifying channel. Secondly, we quantitatively measured TRPML2 and TRPML3 mRNA expressions in TRPML1−/− null and wild-type (Wt) mice. In wild-type mice, the TRPML2lv transcripts were very low while TRPML2sv and TRPML3 transcripts have predominant expressions in lymphoid and kidney organs. Significant reductions of TRPML2sv, but not TRPML2lv or TRPML3 transcripts, were observed in lymphoid and kidney organs of TRPML1−/− mice. RNA interference of endogenous human TRPML1 in HEK-293 cells produced a comparable decrease of human TRPML2 transcript levels that can be restored by overexpression of human TRPML1. Conversely, significant upregulation of TRPML2sv transcripts was observed when primary mouse lymphoid cells were treated with nicotinic acid adenine dinucleotide phosphate, or N-(2-[p-bromocinnamylamino]ethyl)-5-isoquinoline sulfon-amide, both known activators of TRPML1. In conclusion, our results indicate that TRPML2 is unlikely to compensate for the loss of TRPML1 in lymphoid or kidney organs and that TRPML1 appears to play a novel role in the tissue-specific transcriptional regulation of TRPML2.
Keywords: TRP ion channel, TRPML2, Mucolipidosis type IV, Mucolipin-2, Lysosomal storage disorder, TRPML1 knock out mouse
Introduction
Mucolipidosis type IV (ML-IV) is a human lysosomal storage disorder caused by dysfunction of the mucolipin-1 (TRPML1) protein [1-3]. Clinical manifestations of ML-IV include neuromotor retardation, retinal degeneration, corneal opacity, and gastric abnormality [4]. Studies on ML-IV patient fibroblast cells revealed that abnormal TRPML1 results in overacidified lysosomal compartments and the formation of large vacuoles due to lipid accumulation [5, 6]. These observations indicate that TRPML1 may be involved in endosomal–lysosomal biogenesis [7-10] and the regulation of lysosomal pH [11-13].
TRPML1 belongs to the transient receptor potential (TRP) superfamily of ion channels. The TRPML subfamily consists of TRPML1, TRPML2, and TRPML3 proteins. TRPML1 expression is detected in a wide variety of mouse tissues [14]. TRPML3 is detected in melanosomes of melanocytes as well as in hair cells [15] while TRPML2 is expressed in lymphoid (A20 mature B lymphocyte, EL-4 T lymphocyte) and myeloid (5T33 myeloma) cell lines [16]. In mice, two alternatively spliced variants of TRPML2 or Mucolipin-2 (Mcoln2) gene, referred to as long variant (TRPML2lv) and short variant (TRPML2sv), have been deposited in Genbank (Accessions NM_026656 and NM_001005846, respectively). In humans (HsTRPML2 or MCOLN2; Genbank Accession NM_153259), only one TRPML2 isoform consisting of 566 amino acid sequence (Genbank Accession NM_694991) has been detected, which happens to be the same amino acid (AA) sequence length as mouse TRPML2lv protein (Genbank Accession NP_080932). The existence of mouse TRPML2lv and its functional significance is not clear. This was complicated by a report that TRPML2lv is inactive upon introduction of the varitint-waddler (Va)mutation[17] while TRPML1, TRPML2sv, and TRPML3 containing the Va mutation were reported to be constitutively active [17-21]. Note, however, that if mouse TRPML2lv protein is inactive, then the human TRPML2 protein might also be inactive by virtue of its size and AA similarity with the mouse TRPML2lv counterpart. Nevertheless, the current study addresses this apparent inconsistency.
Subcellular localization studies of heterologously expressed mouse or human TRPML proteins suggest that all three members may play a role in endocytic and exocytic signaling events [8, 10, 22-24]. In addition, heterologously expressed TRPML protein subunits interact with each other [23, 24]. There is now evidence that endogenous TRPML subunits only partially colocalize within lysosomal and extralysosomal compartments [25], which indicates that each TRPML protein likely exists as mostly homomeric channels. In addition, heteromeric subunit interactions between endogenous TRPML members are detected in native tissues albeit they are very limited [25]. It remains to be seen if these heteromeric channels are functional and whether they confer novel physiological properties in native tissues. Notwithstanding, genetic complementation of Cup-5−/− null Caenorhabditis elegans using human TRPML1 or TRPML3 produces a complete phenotypic rescue indicating species conservation of TRPML function [26, 27]. Likewise, Song et al. (2006) proposed that the lack of observable phenotype in chicken DT40 B lymphocytes with targeted TRPML1 knockout [22] and in lymphocytes of ML-IV patients [1] could be directly attributed to the genetic compensation (i.e., upregulation of expression) and functional substitution by TRPML2. Moreover, Thompson et al. (2007) suggested that the absence of hyperacidification of late endosomal and lysosomal compartments following RNA interference (RNAi) of mouse TRPML1 in RAW264.7 macrophages could be due to either residual levels of endogenous TRPML1 due to incomplete knockdown or simply the “redundancy” afforded by the presence of endogenous TRPML2 [10]. We therefore hypothesized that, if TRPML2 could functionally replace the loss or dysfunction of TRPML1 protein, then, the expression pattern of TRPML2 would likely be altered upon loss of TRPML1 protein. For example, genetic compensation by TRPC3 (i.e., increased TRPC3 mRNA expression) has been observed in TRPC6−/− knockout mice's vascular smooth muscle cells [28]. In addition, RNAi knockdown of TRPC1 results in compensatory upregulation of TRPC6 mRNA expression in A7r5 vascular smooth muscle cells [29]. Therefore, we set out to investigate the relative mRNA expression patterns of TRPML2 and TRPML3 to provide evidence for, or against, the possibility of genetic compensation by TRPML2 or TRPML3 using the recently reported TRPML1−/− knockout mouse model for ML-IV [30, 31].
Materials and methods
Animals
The animal protocol (Protocol #07-R05) used in this study was approved under the guidelines for animal research set by the IACUC committee at California State University Fullerton. Organ and tissue samples were taken from six TRPML1−/− null mice and six Wt littermates (8–10 weeks old; C57/BL6). The TRPML1−/− knockout mouse model of ML-IV is an excellent model of the human ML-IV pathology and was described in two recent publications [30, 31]. Samples from brain, cerebellum, eye, tongue, thymus, heart, lung, liver, kidney, stomach, spleen, pancreas, small intestine, colon, bladder, and muscle were dissected, quickly frozen, and stored at −80°C until use.
RNA isolation and real-time quantitative polymerase chain reaction (QPCR) analysis
Total RNA was isolated from tissues of TRPML1−/− null mice and their wild-type littermates using TRIzol reagent according to a standard protocol given by the manufacturer (Invitrogen). Superscript III (Invitrogen) was used for the RT reaction and a standardized concentration of 1 μg total RNA of each sample was included in every reactions. A regular PCR analysis using mouse or human GAPDH was performed for all cDNA samples prior to running the real-time QPCR experiment to ensure that the RT reaction worked. Real-time QPCR was done in triplicate using a Sensimix Plus SYBR Supermix kit (Quantace) for the iCycler iQ5 detection system (Bio-Rad). A standard curve was included in all experimental runs using pooled mouse or human cDNAs. We used 18S ribosomal RNA and cyclophilin B housekeeping (HK) genes for mouse and human QPCR analyses, respectively. These housekeeping genes are common internal normalization controls to correct for variations in the levels of input RNA and RT efficiency. They are widely used normalization controls because of their robust and stable expression in many tissues or cell lines [32-34]. Due to high transcript numbers of the HK gene, the RT samples for 18S and cyclophilin B were diluted 1000-fold and 100-fold, respectively, prior to every QPCR run to obtain a whole number upon normalization. The normalized values are presented as transcript ratio between the gene of interest and HK gene [35].
Note that the QPCR primers for TRPML2sv would also detect TRPML2lv transcripts, since both isoforms share the 3′ untranslated region (UTR) of the gene (Fig. S1). Hence, we subtracted the normalized values of TRPML2lv from TRPML2sv to obtain the overall quantity of TRPML2sv transcripts. The mouse (Mm) and human (Hs) primer sequences are found in Supplemental Table S1 and the cDNA map is illustrated in Supplemental Fig. S1.
Site-directed mutagenesis
To create the varitint-waddler (Va) mutant models, we generated point mutants of mouse TRPML1, TRPML2lv, TRPML2sv, and TRPML3 cDNAs with yellow fluorescent protein (YFP) C-terminal fusion using the Quick Change II mutagenesis kit (Stratagene). A proline residue was substituted on the TM5 domain of each TRPML protein, which corresponded to the A419P position of the TRPML3-Va mouse mutation [15]. The introduction of a proline residue on the TM5 domain produces a kink, hinge or swivel [36] on the alpha helical structure that makes the Va mutant channel constitutively active [17, 19-21]. The following TRPML-Va mutants were used in calcium imaging experiments and electrophysiology: TRPML1-V432P, TRPML2lv-A424P, TRPML2sv-A396P, and TRPML3-A419P. All expression vectors were sequence-verified prior to use in any experiments.
Calcium imaging and electrophysiology
Human embryonic kidney (HEK)-293 cells were seeded on a glass cover slip at low density (10-20% confluency), 24 h prior to transfection with GeneJammer (Stratagene). The cells were then prepared for either ratiometric calcium imaging or electrophysiology 15-20 h following transfection as recently described [19]. We used the transfected cells by 20 h following treatment, since the TRPML-Va mutant proteins are cytotoxic over time due to their constitutive channel activity [17, 19-21]. Cells expressing the wild type TRPML, or TRPML-Va mutant proteins were distinguished by fluorescence through the YFP tag. For the calcium imaging study, cells were incubated with 2μM Fura-2-AM, washed twice with PBS buffer, and placed on the iMIC platform for analysis using a Poly-chrome V monochromator (TILL Photonics). We initially calibrated the measurements using cells expressing wild type TRPML3 or TRPML3-Va mutant protein in the presence of ionomycin (a calcium-releasing compound). Subsequently, the F340/F380 ratio for each sample was acquired, and the data were analyzed using the Excel spreadsheet application software. For the electrophysiology recordings, whole-cell currents were recorded with an Alembic Instruments VE-2 amplifier with 100% series resistance compensation, and acquired with JClamp analysis software. Standard bath solution contained (in mM): 138 NaCl, 5.4 KCl, 2 MgCl2, 2 CaCl2, 10 HEPES, and 10 D-glucose; pH 7.4. Standard pipette solution contained (in mM): 140 CsCl, 10 HEPES, 3 ATP-Na, 1 BAPTA, and 2 MgCl2; pH 7.2. Series resistance was 1.8±0.4 MΩ after compensation, and capacitance was 5.5±0.7 pF. Voltage steps were used at increments of 20 mV, and current densities were analyzed at −80 mV.
Primary and cell line culture
All cultured cells were grown and maintained in a standard humidified 37°C incubator, with 5% CO2. HEK-293 cell lines were obtained from American Type Culture Collection (Manassas, Virginia). HEK-293 cells were cultured in Dulbecco's Minimum Essential Medium with high glucose, L-glutamine, and sodium pyruvate (Invitrogen). The culture media were supplemented with 10% fetal bovine serum (FBS; Omega Scientific) and 1X penicillin/streptomycin solution (Cellgro Mediatech).
Primary mouse lymphoid cells were obtained from spleens of adult mice (Swiss Webster) using an established protocol for isolation of splenocytes [37] with modifications appropriate for our study. The lymphoid cells were cultured in RPMI-1640 supplemented with 10% FBS, and 1X penicillin/streptomycin and gentamicin solutions.
RNA interference (RNAi) and cell transfection
RNAi-mediated knock down of endogenous human TRPML1 [11] or murine TRPML1 [10] mRNA expression has been successfully reported. We designed three short-hairpin RNA (shRNA) oligonucleotides targeting human TRPML1 using the Block-IT RNAi Designer web algorithm (Invitrogen). We performed a BLAST search of the human TRPML1 shRNA sequences to ensure specificity for the target gene. The synthesized oligonucleotides were then annealed and ligated into an expression vector containing the human U6 promoter for RNA Pol III according to the manufacturer's recommendations (Invitrogen). The oligonucleotide sequences and their cDNA location can be found in Supplemental Table S2 and Fig. S1,respectively.
HEK-293 cells were transfected with HsTRPML1 shRNA-1208 (2μg) using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). The treated cells were harvested at 48 h post-transfection. Total RNA was isolated and processed for standard RT-PCR and real-time QPCR to assess the endogenous levels of HsTRPML1, HsTRPML2 and HsTRPML3 as described herein.
To test the specificity of the RNAi-mediated knock down of endogenous HsTRPML1, we treated HEK-293 cells with HsTRPML1 shRNA-1208 and simultaneously co-expressed a plasmid driven by a CMV promoter that over-expresses HsTRPML2 (a kind gift from Dr. Rosa Puertollano, NIH/ NHLBI) or HsTRPML1. The pCMV-HsTRPML1, or pCMV-HsTRPML2 vector to shRNA concentration ratio was varied (1:2 and 1:4) in order to show any differential effects between treatments. The treated cells were harvested at 48 h post-transfection. Total RNA was isolated and processed for standard RT-PCR and real-time QPCR to measure the over-expressed levels of HsTRPML1, HsTRPML2 and HsTRPML3 upon RNAi treatment.
To rescue the decrease of endogenous HsTRPML2 upon RNAi knock down of HsTRPML1, we treated HEK-293 cells with HsTRPML1 shRNA-1208 and simultaneously co-expressed a pCMV-HsTRPML1 construct. The pCMV-HsTRPML1 vector to shRNA ratio was varied (1:2 and 1:4) to show any rescuing effects on endogenous HsTRPML2 transcript levels. The treated cells were harvested at 48 h post-transfection. Total RNA was isolated and processed for standard RT-PCR and real-time QPCR in order to quantify the endogenous HsTRPML2 levels.
Drug treatments
Nicotinic acid adenine dinucleotide phosphate (NAADP) and the protein kinase A (PKA) inhibitor, N-(2-[p-bromocinnamylamino]ethyl)-5-isoquinolinesulfonamide (H89), were purchased from Sigma. NAADP is a well-known endogenous second messenger molecule that releases calcium from lysosomes through two-pore channels (TPC), TRPM2, and TRPML1 channels [35, 38-46]. H89 is a potent and widely used selective inhibitor of cyclic AMP-dependent protein kinase A [47] shown to inhibit the PKA-mediated phosphorylation of TRPML1 protein, which consequently increases TRPML1 ion channel activity [48].
Primary mouse lymphoid cells were incubated with either 1μM NAADP or 10μM H89 for one hour. Following treatment, the cell suspension was washed with fresh media, spun at 1000 rpm for 5 minutes, re-suspended with fresh media, and cultured for 24 h prior to total RNA extraction. The specific NAADP and H89 concentrations used in this study were recently reported to increase TRPML1 channel activation and lysosomal calcium release [44, 48]. Untreated cells were used as negative control for data comparison and statistical analysis.
Western blot analysis
Western blot experiments were performed as described previously [49]. Human anti-TRPML1 polyclonal antibodies were a kind gift from Dr. Kirill Kiselyov (Univ. of Pittsburgh), while human anti-TRPML2 polyclonal antibody was commercially obtained from Sigma (St. Louis, MO).
Results
Mouse TRPML2lv and TRPML2sv isoforms are differentially expressed
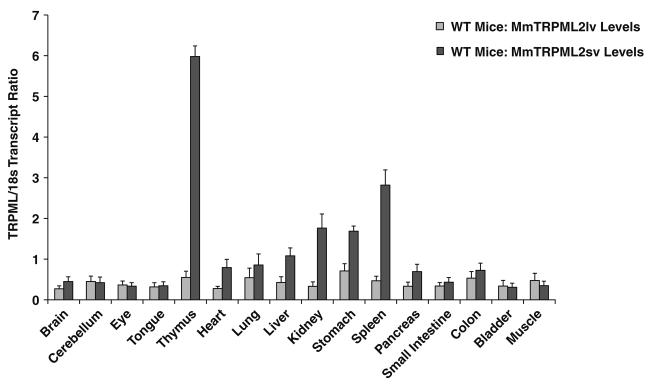
We detected and confirmed two mouse TRPML2 isoforms in wild type (WT) mice – an alternatively spliced long variant TRPML2 (TRPML2lv), and a short variant TRPML2 (TRPML2sv) (Fig. 1). Real-time QPCR analyses revealed that TRPML2lv mRNA expression was very low across all tissues and organs examined (Fig. 1). On the other hand, TRPML2sv transcripts were detected in a tissue-specific expression pattern, with transcript levels predominantly higher in lymphoid (thymus and spleen) and kidney organs followed by heart, lung, liver and stomach (Fig. 1).
Fig. 1.
Real-time QPCR analyses of mouse (Mm) TRPML2lv and TRPML2sv from organs and tissues of wild type (WT) mice. The transcript ratio of TRPML2lv were very low compared with TRPML2sv. The transcript ratio of TRPML2sv showed a tissue-specific expression pattern with normalized values markedly higher in lymphoid (thymus and spleen) and kidney organs followed by heart, lung, liver, and stomach. Data are represented as means ± SEM, N=6 wild-type mice. The samples were processed and analyzed as described in the Materials and methods section
Our in silico analysis of the TRPML2lv mRNA showed that this longer isoform is alternatively spliced (Supplemental Fig. S2), and a new exon 1 with a new open-reading frame is created upstream of the 5′-untranslated region (5′-UTR) of the shorter TRPML2sv isoform. Meanwhile, the exon 1 of the shorter TRPML2sv isoform is its 5′-UTR, and the start codon is found on exon 2 (Supplemental Fig. S2). Both isoforms share the same exons starting from exon 2.
Mouse TRPML2lv is an active, inwardly rectifying cation channel
We wanted to address the previous report that the TRPML2lv isoform was inactive when heterologously expressed as a Va mutant protein. Since no known ligands are available to gate wild-type TRPML proteins, we created Va mutants of all TRPML proteins (Fig. 2a) and analyzed their basic physiological properties using ratiometric calcium imaging and electrophysiology techniques.
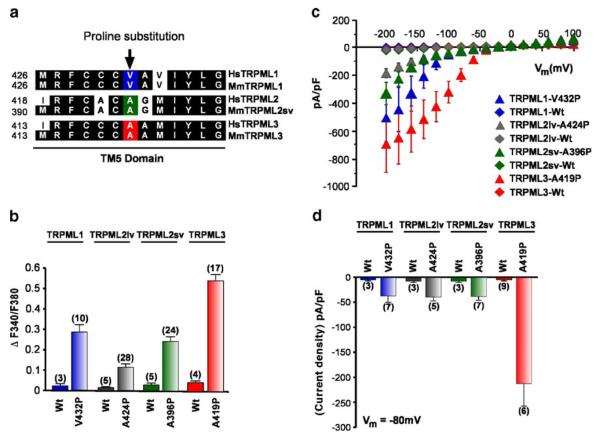
Fig. 2.
Constitutive channel activity of TRPML varitint-waddler (Va) mutant proteins. a Sequence alignment (clustal W) of the inner half of the TM5 domain of the three TRPML proteins. Conserved amino-acid residues are highlighted in black. Arrow indicates the position of the proline substitution from the varitint-waddler mutation A419P in TRPML3 and equivalent positions for TRPML1 and TRPML2 proteins. b Ca2+-imaging experiments showing intracellular Ca2+ levels of HEK-293 cells expressing either wild-type TRPML1, TRPML2lv (long variant), TRPML2sv (short variant), or TRPML3, and TRPML-Va mutant versions TRPML1-V432P, TRPML2lvA424P, TRPML2sv-A396P, and TRPML3-A419P. All experiments were performed 15-20 h after transfection due to cytotoxic effect of the mutation. Data are represented as means ± SEM, N=number in parenthesis. c Steady-state current-voltage plots of constitutively active whole-cell currents elicited by TRPML1-V432P, TRPML2svA396P, TRPML2lv-A424P, and TRPML3-A419P mutant proteins compared with their respective wild-type versions in response to 10 ms voltage steps from a holding potential of +10 mV between −200 mV and +100 mV in 20 mV incremental steps and normalized by cell capacitance (pF). All TRPML-Va mutants showed inward rectification while wild-type TRPMLs did not elicit any response. d Average inward current densities at −80 mV of all TRPML-Va mutant and wild-type proteins shown in panel c and normalized by pF. The TRPML3-Va mutation showed higher current density compared with the other TRPML-Va mutants. Data are represented as means ±SEM, N=numbers in parenthesis. All experimental procedures are outlined in the Materials and methods section
Ratiometric calcium imaging showed that heterologously expressed TRPML1-V432P, TRPML2lv-A424P, TRPML2svA396P, and TRPML3-A419P mutant proteins result in marked increase of intracellular calcium ion concentration [Ca2+]i (Fig. 2b). Heterologously expressed, Wt TRPML proteins did not produce any appreciable increase in intracellular [Ca2+]i levels (Fig. 2b). The levels of calcium influx measured between cells expressing each TRPML-Va mutant proteins were distinct and in the order TRPML3 >> TRPML1 > TRPML2sv > TRPML2lv. The calcium imaging data showed that TRPML2lv-Va is constitutively active as opposed to the previous report that it was inactive [17].
In addition to the above observations, we performed whole-cell patch clamp recordings of HEK-293 cells expressing wild-type or TRPML-Va mutant proteins. All TRPML-Va mutant proteins showed inwardly rectifying currents (Fig. 2c). Consistent with our calcium imaging data, no wild-type TRPML-expressing cells showed any detectable channel activity. Similarly, the electrophysiological property of TRPML2lv-Va was consistent with the calcium imaging data. TRPML2lv-Va was in fact constitutively active albeit the current density was relatively lower than TRPML2sv-Va. On the other hand, the average current densities of TRPML1-Va, TRPML2lv-Va, and TRPML2sv-Va were virtually the same at, or near, the negative resting membrane potential of −80 mV while TRPML3-Va exhibited a strikingly higher current density (Fig. 2d). Note that although the TRPML2lv-Va expressing cells showed a relatively lower calcium influx in our ratiometric calcium imaging experiments (Fig. 2b); our electrophysiological analysis did not show a significant difference between TRPML2lv-Va and those of TRPML2sv-Va or TRPML1-Va current densities at, or near, the negative resting membrane potentials (see Fig. 2c, d).
Quantitative analyses of TRPML2 and TRPML3 mRNA expression in TRPML1−/− mice
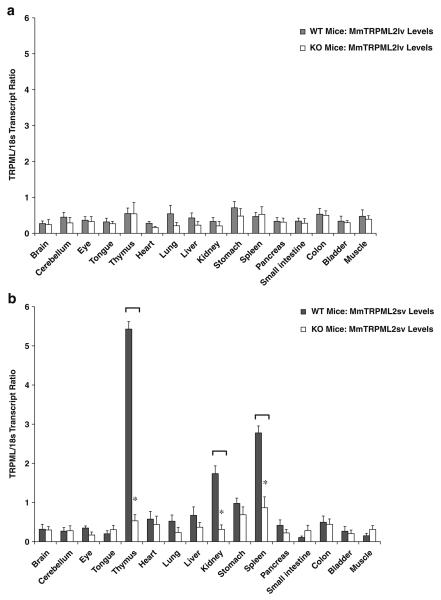
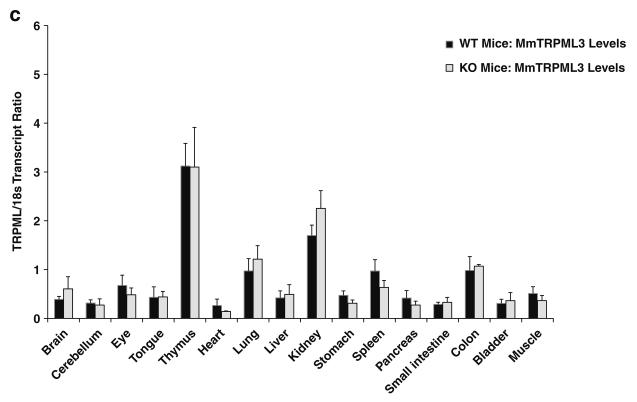
To study the possibility of genetic compensation, we performed real-time QPCR analyses of mouse TRPML2lv, TRPML2sv, and TRPML3 expression patterns from 16 different adult mouse tissues of TRPML1−/− null mice and their wild-type littermates. Standard RT-PCR (data not shown) and real-time QPCR showed that the relative levels of TRPML2lv transcript did not vary regardless of whether the organ and tissue samples came from TRPML1−/− mice or their wild-type littermates (Fig. 3a). It was surprising to find, however, that the relative TRPML2sv transcript levels in thymus, spleen, and kidney were significantly reduced in TRPML1−/− mice but not in wild-type littermate controls (Fig. 3b). The relative TRPML3 transcript levels also showed tissue-specific expression patterns but were not significantly different between TRPML1 −/− and wild-type littermate mice (Fig. 3c). Hence, we focused our efforts characterizing the apparent need for the presence or activity of TRPML1 −/− protein in the tissue-specific expression of TRPML2sv using RNAi and drug compounds on both human and mouse cell culture paradigms.
Fig. 3.
Real-time QPCR analyses of a TRPML2lv, b TRPML2sv, and c TRPML3 mRNA expression levels from various organs and tissues taken from TRPML1−/− null mice and their wild-type littermates. In a, TRPML2lv transcript levels consistently showed low transcript ratio across all samples obtained from both TRPML1−/− mice and wild-type littermates. In b, TRPML2sv levels exhibited a predominantly tissue-specific expression in wild-type but were significantly reduced in lymphoid and kidney organs of TRPML1−/− mice. In c, TRPML3 transcripts showed a similar tissue-specific distribution pattern, although no significant changes were observed in TRPML3 transcript levels between TRPML1−/− mice and wild-type littermates. Real-time QPCR analyses were performed as described in the Materials and methods section. Data are represented as means ±SEM, N=6 TRPML1−/− mice, and N=6 wild-type mice. *p<0.005, Student's t-test, two-tailed
RNA interference of endogenous human TRPML1 results in a reversible reduction of endogenous human TRPML2 transcript levels
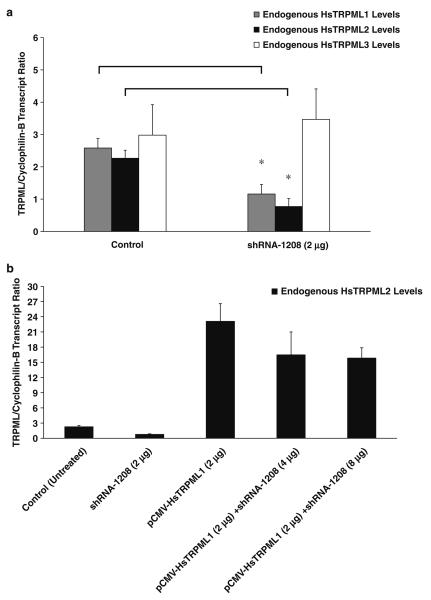
To further verify our in vivo observations, we used RNAi to disrupt the expression of endogenous human TRPML1 (HsTRPML1) in cultured HEK-293 cells. We initially designed three short-hairpin RNA (shRNA) vectors for TRPML1 (see Fig. S1), and found that they all successfully knocked down endogenous HsTRPML1 by 48 h posttransfection (data not shown). However, the shRNA-1208 construct was the most efficient knocking down the endogenous HsTRPML1 (Fig. 4a) and was therefore used in all experiments in the study. Similar to our in vivo findings, the endogenous human TRPML2 (HsTRPML2) expression levels were concomitantly reduced in RNAi-treated HEK-293 cells (Fig. 4a) and HeLa cells (data not shown). The endogenous HsTRPML3 levels were not affected by the RNAi (Fig. 4a).
Fig. 4.
RNAi-mediated knockdown of endogenous HsTRPML1 and subsequent rescue of endogenous HsTRPML2 mRNA expression levels. a Real-time QPCR analyses of control (untreated) and RNAi-treated samples. Endogenous HsTRPML1 expression levels were markedly reduced upon shRNA-1208 treatment. A concomitant decrease of HsTRPML2 transcripts was also observed but not with HsTRPML3 transcripts. Data are represented as means ±SEM, N =3 independent trials. *p< 0.05, Student's t test, two-tailed. b Real-time QPCR of endogenous HsTRPML2 transcript levels upon coexpression of pCMV HsTRPML1 with varying concentrations of shRNA-1208 (4μg and 8μg). The decrease of endogenous HsTRPML2 levels upon RNAi treatment was rescued by overexpressing a pCMV-HsTRPML1 construct. Data are represented as means ± SEM, N=3 independent trials. Real-time QPCR reactions were performed as described in the Materials and methods section
Although these in vitro observations were already in complete agreement with our mouse in vivo data, we wanted to ensure that the shRNA only specifically targeted the HsTRPML1 transcripts, and more importantly, did not target HsTRPML2 or HsTRPML3 transcripts. We therefore coexpressed a pCMV promoter-driven vector (pCMV-X, where X represents HsTRPML1, HsTRPML2, or HsTRPML3 cDNA) together with shRNA-1208 to verify that the knockdown effect is specific to HsTRPML1 transcripts (Supplementary Fig. S4). In three parallel experiments, we initially coexpressed two micrograms (2μg) of pCMV-HsTRPML1, pCMV-HsTRPML2, or pCMV-HsTRPML3 together with two micrograms (2μg) shRNA-1208. We then varied the amounts of shRNA by twofold (4μg) and fourfold (8μg) greater than the initial pCMV-X concentration. Coexpression of pCMV-HsTRPML1 plasmid with twofold and fourfold the amounts of shRNA-1208 significantly decreased the levels of overexpressed HsTRPML1 transcripts when compared with the overexpressed single HsTRPML1 treatment (Fig. S4). On the other hand, co-expression of pCMV-HsTRPML2 vector with shRNA-1208 did not show any significant changes in the overexpressed HsTRPML2 transcript levels even at fourfold higher the shRNA-1208 concentrations (Fig. S4). A similar nonsignificant data were seen in the overexpressed HsTRPML3 transcript levels upon co-transfection of pCMV-HsTRPML3 with shRNA-1208 (Fig. S4).
Our data so far suggested that the presence of TRPML1 influences the expression level of TRPML2 for both in vivo and in vitro paradigms. We then investigated if the reduction of endogenous HsTRPML2 transcripts could be rescued by the reintroduction of HsTRPML1 following RNAi treatment by overexpressing a pCMV-HsTRPML1 plasmid in HEK-293 cells. Actually, the reduction of endogenous HsTRPML2 transcripts was restored and increased by the reintroduction and overexpression of HsTRPML1 (Fig. 4b). The normalized value of endogenous HsTRPML2 transcripts is about two units in control cells, and the RNAi treatment reduces this number to about one half the amount in control cells. When we overexpressed pCMV-HsTRPML1, the normalized value of HsTRPML2 transcripts increased to about 23 units. Co-expression of pCMV-HsTRPML1 with twofold and fourfold greater concentrations of shRNA-1208 did not significantly reduce the normalized values of endogenous HsTRPML2 transcripts.
We performed Western blots using anti-HsTRPML1 and HsTRPML2 polyclonal antibodies to verify the respective protein levels in our experiments. Unfortunately, the human anti-TRPML1 antibody we obtained was unable to detect endogenous protein (data not shown). Likewise, the commercially available anti-HsTRPML2 antibody did not detect the endogenous protein (data not shown).
In summary, these results not only validated our in vivo observations but also showed that the reduction of endogenous levels of either mouse or human TRPML2 transcripts was closely dependent on the presence of mouse or human TRPML1 protein, respectively.
TRPML1 activators, NAADP and H89, increase mouse TRPML2sv expression levels
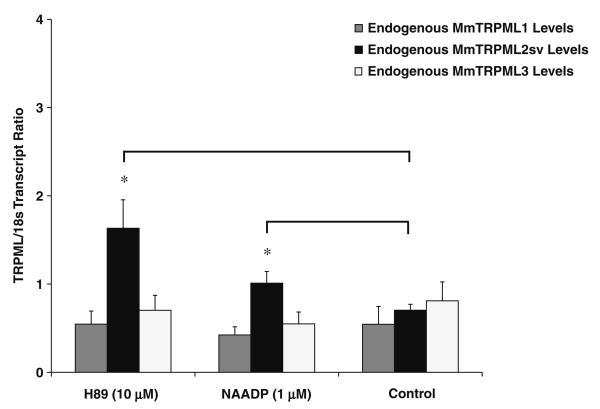
Our in vivo data indicated that mouse TRPML1 might be intimately involved in the tissue-specific transcriptional regulation of mouse TRPML2sv. This observation was further supported by our findings on cultured human cells using RNAi. In addition, we also showed that the transcript levels of human TRPML2 covaried with human TRPML1 expression and that human TRPML2 expression can be rescued and increased by the reintroduction and over-expression of human TRPML1 (Fig. 4a, b). Thus, we hypothesized that, if the reintroduction of TRPML1 rescued TRPML2 expression, then, the presence and/or activity of TRPML1 could also potentially affect the expression levels of TRPML2sv transcripts. We therefore set out to study the effect of two compounds, NAADP and H89, which purportedly activate endogenous TRPML1 channels [44, 48]. Note that TRPML1 channel activation by NAADP was shown to induce lysosomal calcium release [44, 48]. We used mouse primary lymphoid (splenocyte) cells due to the tissue-specific expression of TRPML2sv in the spleen (Fig. 1). Our results showed that both compounds significantly increased the relative transcript levels of TRPML2sv when compared with controls (Fig. 5). In contrast, no significant changes were observed on either mouse TRPML1 or TRPML3 transcript levels upon NAADP or H89 treatment. These findings supported our hypothesis and suggested that the presence and, possibly, the activity of TRPML1 protein impact the tissue-specific expression of TRPML2sv in lymphoid cells.
Fig. 5.
Effects of H89 and NAADP on TRPML2sv expression in primary mouse lymphoid (splenocyte) cells. The transcript ratios of mouse TRPML2sv (MmTRPML2sv) following incubation with H89 (10μM) or NAADP (1μM) were significantly increased when compared with untreated controls. No effects were observed upon analyses of mouse TRPML1 (MmTRPML1) and TRPML3 (MmTRPML3) expression levels upon treatment of the same compounds. Data are represented as means ± SEM, N=5 independent trials. *p<0.05 , Student's t test, two-tailed. Real-time QPCR reaction was performed as described in the Materials and methods section
Discussion
In this study, we confirmed the presence of two mouse TRPML2 isoforms, TRPML2lv (long variant) and TRPML2sv (short variant), in all tissues and organs examined (Fig. 1). The mechanism behind the production of the alternatively spliced TRPML2lv is unclear (Fig. S2). Interestingly, our expression analysis of the mouse TRPML2 isoforms provided more questions than answers but certainly opened new avenues for research. Firstly, why do both MmTRPML2lv and HsTRPML2 proteins have the same AA length and yet the first 28 AA of their N-termini show no conservation (Fig. S2)? Secondly, do the nonconserved 28 AA sequence at the N-terminus of MmTRPML2lv and HsTRPML2 play distinct functions? Thirdly, does a short HsTRPML2 isoform exist in humans, and does it exhibit a tissue-specific pattern as well since MmTRPML2sv transcripts were predominantly expressed in lymphoid and kidney organs? Lastly, do the low levels of TRPML2lv transcripts across many tissues and organs serve a functional purpose? It would be important to know if the native protein levels of both mouse TRPML2 isoforms specifically correspond to the differential transcript levels we observed in this study. Unfortunately, no known working antibodies for either mouse TRPML2 isoforms are available, and our attempts to create polyclonal antibodies were not successful.
Our calcium imaging and electrophysiological analyses of the TRPML2lv with the Va mutation showed constitutive activity with inward rectification. These observations negated the previous report that the TRPML2lv isoform was inactive [17]. Our results made sense since other TRPML-Va mutant members have been shown to exhibit similar channel properties [17, 19-21, 50, 51]. Moreover, TRPML2lv is the same size as the human TRPML2 protein (Fig. S3), and there is no evidence that its human counterpart is a nonfunctional channel. One interesting feature of the constitutively active TRPML2lv-Va mutant was its lower responsiveness when compared with the other TRPML-Va mutants at hyperpolarizing membrane potentials (Fig. 2c). The reason behind the decreased activity of TRPML2lv-Va mutant is enigmatic, but a structural or functional alteration brought about by the additional 28 AA in its N-terminus could explain this effect. Perhaps, a protein interaction domain within the additional AA at the N-terminus region might exist that could allow a binding partner to modulate its channel activity, just as we observed for TRPV4 channel and its protein interactor, PACSIN3 [49, 52]. On the other hand, differential channel responsiveness resulting from heteromeric TRPV subunit interactions is also a well-known phenomenon [53]. This observation appears to be true also for intergroup TRP subunit interactions such as those observed between TRPP2 (a close relative of the TRPML subfamily) and TRPC1 [54]. Future investigations should provide some insights on whether heteromeric TRPML channels confer distinct channel properties in native tissues in vivo.
The relatively high transcript levels of native TRPML2sv in thymus, spleen, and kidney indicate that it plays an important role in these organs. This observation is particularly relevant in the case of Bruton's tyrosine kinase (BTK)-defective immune cells where TRPML2sv transcript levels appear to be downregulated [16]. The BTK protein is a nonreceptor tyrosine kinase involved in lymphocyte development and maturation, and abnormal BTK function causes the chromosome X-linked agammaglobulinemia disorder in humans and X-linked immunodeficiency in mice [55]. Indeed, heterologously expressed human TRPML2 has been shown to colocalize with major histocompatibility complex class I (MHC-I) and CD59 antigen [56]. Mouse TRPML1, on the other hand, has been reported to influence MHC-II internalization in macrophages [10]. Note that overexpression of human TRPML2 enhances the ADP ribosylation factor 6-mediated recycling of glycosyl phosphatidyl inositol-anchored proteins (GPI-APs) [56]. GPIAPs serve many functions ranging from enzymatic to cell adhesion to antigenic signals in cells [57]. Taken together, these data imply that TRPML2sv is very likely to be involved in immune cell development, differentiation, and antigen processing.
We expected that an increased expression of TRPML2 or TRPML3 would be observed in tissues or organs that do not exhibit a phenotypic defect upon loss of TRPML1. Contrary to this hypothesis, our results showed that the transcript levels of the TRPML2sv isoform were significantly downregulated in a tissue-specific manner. Therefore, our data did not support the possibility of gene compensation by TRPML2 particularly in lymphoid (spleen and thymus) and kidney organs of the TRPML1−/− mice. As mentioned earlier, no specific antibody for mouse TRPML2sv is currently available. Hence, it is impossible to ascertain if the tissue-specific protein levels of TRPML2sv were abnormally reduced in these TRPML1−/− mice as well. Notwithstanding, RNAi knockdown of endogenous human TRPML1 (HsTRPML1) from HEK-293 cells concomitantly produced a conspicuous reduction of endogenous human TRPML2 (HsTRPML2) transcript levels (Fig. 4a). This observation supported our in vivo findings. It also suggested that the overall amount of HsTRPML1 in cells, whether endogenous or overex-pressed, closely impacts the expression of endogenous HsTRPML2. The lack of commercially available antibody for human TRPMLs that detect endogenous protein levels prevented us from verifying the RNAi knockdown. The human anti-TRPML1 antibody we obtained only detected overexpressed protein but not endogenous protein. The human antiTRPML2 antibody we commercially purchased also failed to detect the endogenous protein. Nevertheless, our RNAi data not only strongly supported our in vivo mouse observations but also revealed for the first time that TRPML1 may play a novel role in regulating the tissue-specific expression of TRPML2.
To further understand the possible signaling mechanism of TRPML1-mediated transcriptional coregulation of TRPML2sv in lymphoid cells, we studied the effects of NAADP and H89 on primary mouse lymphocytes since both compounds have been recently shown to activate TRPML1 ion channels [44, 48]. Indeed, our results suggested that TRPML1 activation and subsequent calcium release from lysosomes and other organellar stores could be responsible for the increase in TRPML2sv expression upon NAADP or H89 exposure. This is because H89 inhibits the PKA-mediated inactivation of TRPML1 channels, which consequently, increases the activity of the TRPML1 channel [48]. We speculate that the enhanced TRPML1 channel activity could potentially affect intracellular calcium flux just as NAADP influences the TRPML1-mediated lysosomal calcium release [44, 45]. Note, however, that NAADP was also recently reported to mediate lysosomal calcium release via the TRPM2 [38, 41, 58] or TPC2 channels [35, 39, 46]. The initial localized calcium burst from lysosomes may then trigger a widespread calcium release from other calcium stores, which makes NAADP a “universal calcium-release trigger” [40]. Hence, it is very likely that the increase in TRPML2sv expression upon NAADP or H89 incubation is due to downstream calcium signaling events linked to protein kinase C (PKC) activation. This is because primary mouse lymphoid cells exposed to phorbol 12-myristate 13-acetate (PMA; our unpublished result) or BTK-defective lymphoid cell lines exposed to PMA plus ionomycin [16] results in enhanced TRPML2sv expression. PMA is a phorbol ester that mimics the action of diacylglycerol and a potent activator of PKC while ionomycin induces intracellular calcium mobilization that is important in PKC function. Note that the PKC signal transduction pathway is critical for antigen-receptor processing and gene transcription in B and T lymphocytes [59, 60], and at the same time, TRPML2 appears to be important in B-cell development and function [16]. Noteworthy is that neither TRPML1 nor TRPML3 mRNA expression levels were affected by NAADP or H89 (Fig. 5). This implies that the signaling pathway that impinges upon the transcriptional regulation of TRPML1 or TRPML3 is independent of NAADP- or H89-mediated effect on TRPML2sv expression. A case in point, it was recently reported that the transcription factor EB is involved in the transcriptional activation of several lysosomal genes including TRPML1 but not TRPML2 or TRPML3 [61]. What makes this report very interesting is that it clearly shows that the transcription of various lysosomal genes is highly coordinated and regulated by a genetic program that specifies lysosomal biogenesis and function [61].
As a final note, although no significant changes were observed in the tissue-specific expression of mouse TRPML3 across all samples studied (Fig. 3c), we could not rule out the possibility that TRPML3 might substitute for the loss of TRPML1 function. This is because recent reports showed that TRPML3, just like TRPML1 [62, 63], colocalizes within endosomal–lysosomal compartments and plays a role in autophagy [64, 65]. Intriguingly, given that TRPML1 is widely expressed in mouse or human tissues and organs (our unpublished data) and that TRPML2 and TRPML3 have tissue-specific expression, why does the loss of TRPML1 in mice or humans only produce phenotypic defects in certain cell types but not across many cell types? Is there an as-yet-to-be-discovered protein that substitutes for the loss of TRPML1 function in these cell types? Additional studies in this area would likely provide answers to these salient questions.
Concluding remarks
Our study showed for the first time that TRPML1 confers a novel and a more intimate role in the tissue-specific expression of TRPML2. We also showed that TRPML2sv is expressed in a tissue-specific pattern and that the hypothesis that TRPML2 could genetically compensate for the loss of TRPML1 was not supported by our findings. Meanwhile, we also provide new evidence that when the Va mutation is present, the longer TRPML2lv isoform is constitutively active just like with TRPML1, TRPML2sv, and TRPML3. Finally, the association of human TRPML2 with MHC protein complexes [10, 56], the tissue-specific expression of mouse TRPML2sv in lymphoid organs, as well as the tissue-specific TRPML1-mediated transcriptional regulation of TRPML2 expression all point toward a crucial function for TRPML2 in the immune system. Future research should reveal the biological significance of TRPML2.
Supplementary Material
Acknowledgments
We are grateful for the technical support given by Tony Ricci, Michael Schnee, Simone Jörs, and James Mull. We also thank Breck Wheelock for his help on tissue dissection and primary mouse lymphoid cell culture. We thank Dr. Rosa Puertollano (NIH/NHLBI) for providing the human TRPML2 expression plasmid and Dr. Kirill Kiselyov for the human anti-TRPML1 polyclonal antibodies. MPC is partly supported by the Howard Hughes Medical Institute under the HHMI Undergraduate Research Program and a recipient of the California State University Program for Education and Research in Biotechnology (CSUPERB) Faculty-Student Collaborative Research Grant and CSUF Junior Faculty Intramural Research Grant. M. A. S. is a Coppel Graduate Student Science scholar and is a recipient of a research grant from the CSUF Associated Students Inc. JAE is a Howell-CSUPERB scholar, and a recipient of the NagelCSUPERB travel award and the CSUF Associated Students Inc. research grant. SH is funded by NIH grant (DC004563). SAS is funded by NIH grant (NS39995).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00424-009-0716-5) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Bargal R, Goebel HH, Latta E, Bach G. Mucolipidosis IV: novel mutation and diverse ultrastructural spectrum in the skin. Neuropediatrics. 2002;33:199–202. doi: 10.1055/s-2002-34496. [DOI] [PubMed] [Google Scholar]
- 2.Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Jr, Bove C, Kaneski CR, Nagle J, Bromley MC, Colman M, Schiffmann R, Slaugenhaupt SA. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 4.Amir N, Zlotogora J, Bach G. Mucolipidosis type IV: clinical spectrum and natural history. Pediatrics. 1987;79:953–959. [PubMed] [Google Scholar]
- 5.Bach G, Cohen MM, Kohn G. Abnormal ganglioside accumulation in cultured fibroblasts from patients with mucolipidosis IV. Biochem Biophys Res Commun. 1975;66:1483–1490. doi: 10.1016/0006-291x(75)90526-4. [DOI] [PubMed] [Google Scholar]
- 6.Tellez-Nagel I, Rapin I, Iwamoto T, Johnson AB, Norton WT, Nitowsky H. Mucolipidosis IV. Clinical, ultrastructural, histochemical, and chemical studies of a case, including a brain biopsy. Arch Neurol. 1976;33:828–835. doi: 10.1001/archneur.1976.00500120032005. [DOI] [PubMed] [Google Scholar]
- 7.Kiselyov K, Chen J, Rbaibi Y, Oberdick D, Tjon-Kon-Sang S, Shcheynikov N, Muallem S, Soyombo A. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280:43218–43223. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- 8.LaPlante JM, Sun M, Falardeau J, Dai D, Brown EM, Slaugenhaupt SA, Vassilev PM. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol Genet Metab. 2006;89:339–348. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Pryor PR, Reimann F, Gribble FM, Luzio JP. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 2006;7:1388–1398. doi: 10.1111/j.1600-0854.2006.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson EG, Schaheen L, Dang H, Fares H. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 2007;8:54. doi: 10.1186/1471-2121-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miedel MT, Rbaibi Y, Guerriero CJ, Colletti G, Weixel KM, Weisz OA, Kiselyov K. Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. J Exp Med. 2008;205:1477–1490. doi: 10.1084/jem.20072194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raychowdhury MK, Gonzalez-Perrett S, Montalbetti N, Timpanaro GA, Chasan B, Goldmann WH, Stahl S, Cooney A, Goldin E, Cantiello HF. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum Mol Genet. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 13.Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, Kiselyov K. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J Biol Chem. 2006;281:7294–7301. doi: 10.1074/jbc.M508211200. [DOI] [PubMed] [Google Scholar]
- 14.Falardeau JL, Kennedy JC, Acierno JS, Jr, Sun M, Stahl S, Goldin E, Slaugenhaupt SA. Cloning and characterization of the mouse Mcoln1 gene reveals an alternatively spliced transcript not seen in humans. BMC Genomics. 2002;3:3. doi: 10.1186/1471-2164-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Palma F, Belyantseva IA, Kim HJ, Vogt TF, Kachar B, Noben-Trauth K. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc Natl Acad Sci U S A. 2002;99:14994–14999. doi: 10.1073/pnas.222425399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindvall JM, Blomberg KE, Wennborg A, Smith CI. Differential expression and molecular characterisation of Lmo7, Myo1e, Sash1, and Mcoln2 genes in Btk-defective B-cells. Cell Immunol. 2005;235:46–55. doi: 10.1016/j.cellimm.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, Delling M, Li L, Dong X, Clapham DE. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc Natl Acad Sci U S A. 2007;104:18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm C, Cuajungco MP, van Aken AF, Schnee M, Jors S, Kros CJ, Ricci AJ, Heller S. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 2007;104:19583–19588. doi: 10.1073/pnas.0709846104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Muallem S. Gain-of-function mutation in TRPML3 causes the mouse varitint-waddler phenotype. J Biol Chem. 2007;282:36138–36142. doi: 10.1074/jbc.C700190200. [DOI] [PubMed] [Google Scholar]
- 21.Nagata K, Zheng L, Madathany T, Castiglioni AJ, Bartles JR, Garcia-Anoveros J. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci U S A. 2008;105:353–358. doi: 10.1073/pnas.0707963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Y, Dayalu R, Matthews SA, Scharenberg AM. TRPML cation channels regulate the specialized lysosomal compartment of vertebrate B-lymphocytes. Eur J Cell Biol. 2006 doi: 10.1016/j.ejcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Venkatachalam K, Hofmann T, Montell C. Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J Biol Chem. 2006;281:17517–17527. doi: 10.1074/jbc.M600807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeevi DA, Frumkin A, Bach G. TRPML and lysosomal function. Biochim Biophys Acta. 2007;1772:851–858. doi: 10.1016/j.bbadis.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Zeevi DA, Frumkin A, Offen-Glasner V, Kogot-Levin A, Bach G. A potentially dynamic lysosomal role for the endogenous TRPML proteins. J Pathol. 2009 doi: 10.1002/path.2587. [DOI] [PubMed] [Google Scholar]
- 26.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 27.Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci U S A. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich A, Mederos YSM, Gollasch M, Gross V, Storch U, Dubrovska G, Obst M, Yildirim E, Salanova B, Kalwa H, Essin K, Pinkenburg O, Luft FC, Gudermann T, Birnbaumer L. Increased vascular smooth muscle contractility in TRPC6−/− mice. Mol Cell Biol. 2005;25:6980–6989. doi: 10.1128/MCB.25.16.6980-6989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selli C, Erac Y, Kosova B, Tosun M. Post-transcriptional silencing of TRPC1 ion channel gene by RNA interference upregulates TRPC6 expression and store-operated Ca(2+) entry in A7r5 vascular smooth muscle cells. Vascul Pharmacol. 2009;51:96–100. doi: 10.1016/j.vph.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Micsenyi MC, Dobrenis K, Stephney G, Pickel J, Vanier MT, Slaugenhaupt SA, Walkley SU. Neuropathology of the Mcoln1(−/−) knockout mouse model of mucolipidosis type IV. J Neuropathol Exp Neurol. 2009;68:125–135. doi: 10.1097/NEN.0b013e3181942cf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venugopal B, Browning MF, Curcio-Morelli C, Varro A, Michaud N, Nanthakumar N, Walkley SU, Pickel J, Slaugenhaupt SA. Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. Am J Hum Genet. 2007;81:1070–1083. doi: 10.1086/521954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295:17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 33.Riccio A, Medhurst AD, Mattei C, Kelsell RE, Calver AR, Randall AD, Benham CD, Pangalos MN. mRNA distribution analysis of human TRPC family in CNS and peripheral tissues. Brain Res Mol Brain Res. 2002;109:95–104. doi: 10.1016/s0169-328x(02)00527-2. [DOI] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 35.Brailoiu E, Churamani D, Cai X, Schrlau MG, Brailoiu GC, Gao X, Hooper R, Boulware MJ, Dun NJ, Marchant JS, Patel S. Essential requirement for two-pore channel 1 in NAADP-mediated calcium signaling. J Cell Biol. 2009;186:201–209. doi: 10.1083/jcb.200904073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuajungco MP, Samie MA. The varitint-waddler mouse phenotypes and the TRPML3 ion channel mutation: cause and consequence. Pflugers Arch. 2008;457:463–473. doi: 10.1007/s00424-008-0523-4. [DOI] [PubMed] [Google Scholar]
- 37.Beeton C, Chandy KG. Preparing T cell growth factor from rat splenocytes. J Vis Exp. 2007:402. doi: 10.3791/402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beck A, Kolisek M, Bagley LA, Fleig A, Penner R. Nicotinic acid adenine dinucleotide phosphate and cyclic ADPribose regulate TRPM2 channels in T lymphocytes. FASEB J. 2006;20:962–964. doi: 10.1096/fj.05-5538fje. [DOI] [PubMed] [Google Scholar]
- 39.Calcraft PJ, Ruas M, Pan Z, Cheng X, Arredouani A, Hao X, Tang J, Rietdorf K, Teboul L, Chuang KT, Lin P, Xiao R, Wang C, Zhu Y, Lin Y, Wyatt CN, Parrington J, Ma J, Evans AM, Galione A, Zhu MX. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459:596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guse AH, Lee HC. NAADP: a universal Ca2+ trigger. Sci Signal. 2008;1:re10. doi: 10.1126/scisignal.144re10. [DOI] [PubMed] [Google Scholar]
- 41.Lange I, Yamamoto S, Partida-Sanchez S, Mori Y, Fleig A, Penner R. TRPM2 functions as a lysosomal Ca2+-release channel in beta cells. Sci Signal. 2009;2:ra23. doi: 10.1126/scisignal.2000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singaravelu K, Deitmer JW. Calcium mobilization by nicotinic acid adenine dinucleotide phosphate (NAADP) in rat astrocytes. Cell Calcium. 2006;39:143–153. doi: 10.1016/j.ceca.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Steen M, Kirchberger T, Guse AH. NAADP mobilizes calcium from the endoplasmic reticular Ca(2+) store in T-lymphocytes. J Biol Chem. 2007;282:18864–18871. doi: 10.1074/jbc.M610925200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, Jin S, Yi F, Li PL. TRP-ML1 Functions as a Lysosomal NAADP-Sensitive Ca(2+) Release Channel in Coronary Arterial Myocytes. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Li PL. Reconstitution and characterization of a nicotinic acid adenine dinucleotide phosphate (NAADP)-sensitive Ca2+ release channel from liver lysosomes of rats. J Biol Chem. 2007;282:25259–25269. doi: 10.1074/jbc.M701614200. [DOI] [PubMed] [Google Scholar]
- 46.Zong X, Schieder M, Cuny H, Fenske S, Gruner C, Rotzer K, Griesbeck O, Harz H, Biel M, Wahl-Schott C. The two-pore channel TPCN2 mediates NAADP-dependent Ca(2+)-release from lysosomal stores. Pflugers Arch. 2009 doi: 10.1007/s00424-009-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 48.Vergarajauregui S, Oberdick R, Kiselyov K, Puertollano R. Mucolipin 1 channel activity is regulated by protein kinase A-mediated phosphorylation. Biochem J. 2008;410:417–425. doi: 10.1042/BJ20070713. [DOI] [PubMed] [Google Scholar]
- 49.Cuajungco MP, Grimm C, Oshima K, D'Hoedt D, Nilius B, Mensenkamp AR, Bindels RJ, Plomann M, Heller S. PACSINs bind to the TRPV4 cation channel. PACSIN 3 modulates the subcellular localization of TRPV4. J Biol Chem. 2006;281:18753–18762. doi: 10.1074/jbc.M602452200. [DOI] [PubMed] [Google Scholar]
- 50.Dong XP, Wang X, Shen D, Chen S, Liu M, Wang Y, Mills E, Cheng X, Delling M, Xu H. Activating mutations of the TRPML1 channel revealed by proline scanning mutagenesis. J Biol Chem. 2009 doi: 10.1074/jbc.M109.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Soyombo AA, Muallem S. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 2008;27:1197–1205. doi: 10.1038/emboj.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D'Hoedt D, Owsianik G, Prenen J, Cuajungco MP, Grimm C, Heller S, Voets T, Nilius B. Stimulus-specific modulation of the cation channel TRPV4 by PACSIN 3. J Biol Chem. 2008;283:6272–6280. doi: 10.1074/jbc.M706386200. [DOI] [PubMed] [Google Scholar]
- 53.Cheng W, Yang F, Takanishi CL, Zheng J. Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. J Gen Physiol. 2007;129:191–207. doi: 10.1085/jgp.200709731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai CX, Giamarchi A, Rodat-Despoix L, Padilla F, Downs T, Tsiokas L, Delmas P. Formation of a new receptor-operated channel by heteromeric assembly of TRPP2 and TRPC1 subunits. EMBO Rep. 2008;9:472–479. doi: 10.1038/embor.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan WN, Alt FW, Gerstein RM, Malynn BA, Larsson I, Rathbun G, Davidson L, Muller S, Kantor AB, Herzenberg LA, et al. Defective B cell development and function in Btk-deficient mice. Immunity. 1995;3:283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 56.Karacsonyi C, Miguel AS, Puertollano R. Mucolipin-2 localizes to the Arf6-associated pathway and regulates recycling of GPI-APs. Traffic. 2007;8:1404–1414. doi: 10.1111/j.1600-0854.2007.00619.x. [DOI] [PubMed] [Google Scholar]
- 57.Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cell Mol Life Sci. 2001;58:1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lange I, Penner R, Fleig A, Beck A. Synergistic regulation of endogenous TRPM2 channels by adenine dinucleotides in primary human neutrophils. Cell Calcium. 2008;44:604–615. doi: 10.1016/j.ceca.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isakov N, Mally MI, Altman A. Mitogen-induced human T cell proliferation is associated with increased expression of selected PKC genes. Mol Immunol. 1992;29:927–933. doi: 10.1016/0161-5890(92)90131-g. [DOI] [PubMed] [Google Scholar]
- 60.Martin P, Duran A, Minguet S, Gaspar ML, Diaz-Meco MT, Rennert P, Leitges M, Moscat J. Role of zeta PKC in B-cell signaling and function. EMBO J. 2002;21:4049–4057. doi: 10.1093/emboj/cdf407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 62.Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, Slaugenhaupt SA. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2008 doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 63.Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet. 2008;17:2723–2737. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim HJ, Soyombo AA, Tjon-Kon-Sang S, So I, Muallem S. The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10:1157–1167. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martina JA, Lelouvier B, Puertollano R. The calcium channel mucolipin-3 is a novel regulator of trafficking along the endosomal pathway. Traffic. 2009;10:1143–1156. doi: 10.1111/j.1600-0854.2009.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.