Summary
Type VI Secretion Systems (T6SS) have been studied primarily in the context of pathogenic bacterial-host interactions. Recent data suggest, however, that these versatile secretion systems may also function to promote commensal or mutualistic relationships between bacteria and eukaryotes, or to mediate cooperative or competitive interactions between bacteria.
Introduction
The Type VI Secretion System (T6SS) was recognized as a distinct class of bacterial protein secretion system in 2006 with two papers from the Mekalanos laboratory. The first study (Pukatzki et al., 2006) found that the IcmF-associated homologous protein (IAHP) gene cluster of Vibrio cholerae was required for secretion of the proteins Hcp and VgrG and for cytotoxicity toward Dictyostelium amoebae. The second study (Mougous et al., 2006) included structural data that indicated the formation of a secretion apparatus, and provided evidence that the apparatus was functional during chronic lung infection of Pseudomonas aeruginosa in cystic fibrosis patients. Both studies described Type VI Secretion (T6S) in the context of pathogenesis – indeed, the T6SS-encoding genes of V. cholerae were named virulence associated secretion (vas) genes.
Putative T6SS-encoding gene clusters have now been identified in over one fourth of sequenced Gram-negative bacterial genomes (Bingle et al., 2008; Persson et al., 2009; Pukatzki et al., 2009). Many of these T6SS-containing bacteria are known pathogens, and T6SSs have been experimentally shown to play a role in virulence in several cases (see Bingle et al., 2008 and Pukatzki et al., 2009 for specific references). In Burkholderia mallei, T6S is required for virulence in a hamster model and promotes giant multinucleated cell formation and intracellular spread in macrophage cell lines (Burtnick et al. 2010). In Burkholderia cenocepacia, Hcp is required for host cell actin remodeling and confers resistance to predation by Dictyostelium amoebae, and Tn insertions in T6SS-encoding genes were associated with virulence defects in in vivo chronic lung infection models. Deletion of genes encoding T6SS components in Edwardsiella tarda caused a virulence defect in a blue gourami fish host. And when produced inside HeLa cells, Hcp of Aeromonas hydrophila induced apoptosis and T6S contributed to virulence of this pathogen in mice. In some cases, putative T6SS components contribute to virulence, but in a manner that appears to be independent of other T6SS components. In Agrobacterium tumefaciens, for example, Hcp appeared to contribute to tumorigenesis of potato tuber disks, but deletion of the rest of the T6SS-encoding genes had no effect. Similarly, homologues of T6SS-encoding genes in Francisella tularensis were required for growth within macrophages and virulence in mice, but the secretion of those gene products appeared not to require the other putative T6SS-encoding genes (Barker et al 2009). The Agrobacterium and Francisella studies highlight the need for a better understanding of what exactly constitutes a T6SS and how T6SSs and their effectors function mechanistically.
The importance of T6S in pathogenesis is becoming increasingly clear. However, many bacteria with genomes encoding putative T6SSs are not known to be pathogens or even symbionts, suggesting T6S may also function in nonpathogenic bacteria-host interactions or in interactions not involving eukaryotic partners. Several papers, including three published in Cell Host & Microbe in the last year, highlight the potential diversity of functions for T6S.
T6SSs as virulence factors
Understanding the mechanisms underlying T6S and its role in pathogenesis has been challenging because of the lack of identified effector proteins. Although in vitro studies identified two proteins that were secreted in a T6SS-depndent manner, Hcp and VgrG, each was required for the secretion of the other, suggesting that both might be part of the secretion apparatus and not bona fide effectors. V. cholerae produces three VgrG proteins, VgrG-1, VgrG-2, and VgrG-3. These proteins share a conserved N-terminal domain but contain divergent C-termini. The C-terminal domain of VgrG-1 has predicted similarity to the actin crosslinking domain (ACD) of RtxA, a large RTX-containing toxin of V. cholerae. Incubation of purified recombinant VgrG-1 with eukaryotic cell lysates showed that VgrG-1 crosslinks actin, and incubation of J774 cells with various T6SS mutant and parental V. cholerae strains showed that VgrG-1 and other core T6SS components (VasK and Hcp) were necessary to induce actin crosslinking in host cells (Pukatzki et al., 2007). These data suggested an actin crosslinking effector function for VgrG-1, but the requirement of VgrG-1 for secretion confounded its classification as an effector.
Ma et al. (2009) aimed to disentangle the effector and secretory functions of VgrG-1 by comparing V. cholerae strains that contained deletions of either the entire vgrG-1 gene or only the region encoding the C-terminal ACD. Their data showed that the ACD was required for actin crosslinking and cytotoxicity toward J774 cells and Dictyostelium, but not for T6SS-dependent protein secretion. To determine if the ACD was translocated into host cells, J774 cells were infected with V. cholerae strains containing a ß-lactamase reporter fused to or replacing the ACD of VgrG-1. The reporter proteins were detected inside host cells as long as the N-terminal portion of VgrG-1 and known core T6SS components were intact. Together, these data indicated that the conserved N-terminal portion of VgrG-1 is part of the secretion system proper, while the C-terminal ACD functions as an effector. This work was the first to definitively and specifically identify a T6S effector. Interestingly, previous work had noted that several other bacteria encode VgrG orthologues containing divergent C-terminal extensions with diverse predicted functions (Pukatzki et al., 2007). Whether these orthologues represent true effectors, and what their specific functions are, remains to be determined. Identification of additional effectors, their targets and mechanisms of action, and their conservation among T6SSs of different bacteria will be important for understanding the role of T6S in pathogenesis and other types of interactions.
The second important finding to come out of the Ma et al. (2009) study was that T6S only occurs after the bacteria are internalized and that VgrG-1-mediated actin crosslinking inhibits further phagocytic activity. While studies of other bacteria, including Burkholderia pseudomallei, B. cenocepacia, and Salmonella enterica serovar Typhimurium (S. Typhimurium), had also found that expression of T6SS-encoding genes was induced upon uptake into host cells, the resulting impairment of further phagocytic activity indicated by the Ma et al. study was an important new finding. The authors hypothesized that actin crosslinking and resultant inhibition of phagocytosis by early-colonizing bacteria may protect additional “bystander” or late-colonizing bacteria from phagocytosis, i.e., that T6S may mediate altruistic behavior among bacteria – an intriguing idea.
Having shown that translocation of the VgrG-1 ACD requires endocytosis and results in actin crosslinking in host cells in vitro, Ma and Mekalanos went on to investigate the mechanism of action of VgrG-1 and the T6SS in vivo. They first compared fluid accumulation and gene expression in the intestines of mice infected with various T6SS mutant and rtxA deletion strains (Ma and Mekalanos, 2010). (In both Ma et al., 2009 and Ma and Mekalanos, 2010, parental strains were ΔhlyAΔhapA, since these genes encode accessory toxins that would mask effects of T6S on mammalian cells. In some cases, rtxA was also deleted to avoid confounding results, since RtxA also crosslinks actin.) Fluid accumulation and monocyte infiltration in the intestinal lumen and expression of several inflammation-associated genes were decreased in the T6SS mutant-infected mice compared with those infected with parental strains. These phenotypes required not only the core components of the T6SS, but also the ACD of VgrG-1, providing in vivo evidence for the effector function of the ACD. In addition, crosslinked actin was detected in intestines of mice infected with parental strains, but not those infected with strains carrying deletions in any of the T6SS-encoding genes tested. Together, these results suggest that the ACD of VgrG-1 is translocated into and cross-links actin in host cells in vivo, and that this activity leads to inflammation and pathology in the intestines of mice. Ma and Mekalanos took their study a step further to test the hypothesis that early-colonizing bacteria use their T6SSs to facilitate colonization by late-colonizing bacteria. They first “pre-infected” mice with bacteria producing either wild type VgrG-1 or ΔACD VgrG-1, then “super-infected” four hours later with either the same or the opposite strain. They found that pre-infection with bacteria producing wild type VgrG-1, but not with bacteria producing ΔACD VgrG-1, allowed super-infected ΔACD VgrG-1-producing bacteria to grow in the mouse intestines to levels indistinguishable from that of wild type VgrG1-producing bacteria. The data suggest that initial infection by wild type VgrG1-producing bacteria creates a favorable within-host environment for the growth of V. cholerae. Whether this environment is favorable because the phagocytic cells have been inactivated or because of some other aspect of the VgrG1-dependent altered inflammatory response remains to be determined. Nonetheless, the results suggest that the V. cholerae T6SS may function as a virulence factor in both the immediate sense of causing inflammation, and in a longer-term, or ultimate, sense of contributing to within-host growth and probably subsequent transmission.
It is worth noting that the use of Dictyostelium to study V. cholerae pathogenesis was pivotal to the discovery of T6S as a new class of secretion system with a role in virulence. The use of invertebrate models for mimicking human infection processes has been variably successful and a topic of debate. For pathogens with environmental habitats, however, it is likely that some virulence factors used for human infection evolved from and/or still function as factors important during interactions with diverse organisms that facilitate environmental survival (Matz and Kjelleberg, 2005; Matz et al., 2008). For V. cholerae, a role for T6S during interactions with environmental amoeba seems likely based on the clear, measurable phenotype of the T6S mutant in Dictyostelium. The importance of T6S during infection of mammals by V. cholerae is less obvious and may have been missed completely if not for the use of Dictyostelium, since T6S-dependent phenotypes in mammalian hosts or cell lines require the use of V. cholerae strains with deletions in genes encoding accessory toxins (hlyA, hapA, and/or rtxA). As with V. cholerae, many other T6SS-possessing bacterial species can be found free-living in the environment or in association with non-mammalian hosts. While many of these bacteria are studied most intensively due to their potential as facultative pathogens of humans, awareness of their environmental interactions as part of the disease cycle is important and rising. For example, it is clear that understanding the interaction between Yersinia pestis and fleas is critical to understanding how this pathogen causes plague in humans. Burkholderia pseudomallei, a soil saprotroph, is being studied in amoeba, nematodes, a larval insect, and tomato plants with the hope that understanding these interactions will shed light on the mechanisms it uses to cause human melioidosis. Although some factors may be host-specific, it seems likely that others are not and their discovery may be facilitated by the use of non-mammalian models.
T6SS as anti-pathogenesis factors
While several studies support a role for T6S in pathogenesis, a few support an alternate view: that T6S may function to limit bacterial replication or virulence, pushing interactions with hosts away from pathogenesis and toward a commensal or mutualistic state. In fact, one of the first T6SS mutants characterized was one involved in a mutualistic relationship. Rhizobium leguminosarum colonizes the roots of leguminous plants, forming nodules in which it fixes nitrogen. The Rhizobium-plant interaction is highly specific and certain variants of R. leguminosarum form nitrogen fixing nodules only with plants in the clover group. Bladergroen et al. (2003) found that a strain with a deletion mutation in the imp gene cluster (now known to encode a T6SS) was also able to form functional nitrogen-fixing nodules on pea plants. Their data indicated that the imp gene cluster (i.e., the T6SS) was involved in secretion of proteins that block infection of pea plants by R. leguminosarum, thus determining host-specificity of the Rhizobium-plant interaction.
Another example of an anti-pathogenesis role for the T6SS comes from studies with S. Typhimurium. Parsons and Heffron (2005) identified a locus, sciS (now known to encode part of a T6SS) that, when mutated, results in increased replication of S. Typhimurium in macrophages at 24 hours post-inoculation and increased virulence in a mouse model, suggesting the role of sciS, and hence T6SS, in S. Typhimurium is to limit virulence and therefore contribute to long-term colonization.
A recent study of Helicobacter hepaticus further indicates that T6S can function to limit within-host growth and virulence. H. hepaticus is a common symbiont and possible facultative pathogen of rodent gastrointestinal tracts. Using confocal microscopy and gentamicin protection assays, Chow and Mazmanian (2010) showed that H. hepaticus adherence to and internalization by mouse intestinal epithelial cell (IEC) lines was increased for a T6SS mutant compared with the wild type strain. Furthermore, colonization of mouse colon and invasion of IECs in vivo, as determined by 16S qPCR of colon tissue and bacterial CFU counts from gentamicin-treated purified IECs, were increased in the T6SS mutant. Chow and Mazmanian then asked whether the T6SS could reduce the inflammation caused by H. hepaticus under experimentally induced dysbiosis. They defined dysbiosis as an imbalance in the composition of the host-associated microbiota, which can cause some symbiotic bacteria to have pathogenic effects. To produce a model of dysbiosis, they reconstituted T and B cell deficient mice with naïve CD4+CD45Rb high T cells (i.e., T cells from germ-free mice, such that they have not been exposed to bacterial antigens). These T cells react pathogenically to certain gut bacteria, including H. hepaticus, whereas “experienced” T cells from mice with normal gut microbiota tolerate these bacteria. In this dysbiosis model, the T6SS mutant showed higher colonization and induced a stronger inflammatory response compared with wild type H. hepaticus. The authors concluded that the T6SS functions to limit inflammation caused by H. hepaticus, thus possibly reducing negative impacts of dysbiosis on the host.
The data presented by Chow and Mazmanian are compelling. Important next steps include the identification of effectors involved in limiting pathogenesis or mediating mutualism, since the T6S-dependent translocation of effectors into host cells has so far only been explicitly demonstrated in pathogenic interactions. Identification of effectors involved in non-pathogenic interactions will also help determine if the nature of a T6SS-mediated interaction, whether antagonistic or beneficial, is dictated by effectors specific to different types of interactions, or alternatively whether orthologous effectors in different bacteria can mediate both positive and negative interactions. A greater mechanistic understanding would help distinguish whether pathogenic versus non-pathogenic effects result from fundamentally different interactions at the molecular level, or are simply a result of differences in the number of bacteria present within a host. For example, in the H. hepaticus system, does the T6SS prevent inflammation by altering host cell response to the presence of the bacterium, or simply by limiting the number of bacteria?
Differentiating between pathogenic and non-pathogenic interactions can be complicated by the fact that the distinction is often ambiguous and can depend on the context and time frame of the interaction. Time frame can be particularly important since factors that reduce acute harm to a host, as seen for T6S in S. Typhimurium, may in fact promote transmission by enabling a latent sit-and-wait phase. Detection of these various effects will depend on the design and scope of the study. The H. hepaticus system illustrates the importance of ecological context, since the pathological effects of the bacterium are dependent on prior exposure of the host to gut microflora. For H. hepaticus, translating differences in inflammation-associated gene expression to pathology will help elucidate how the T6SS of H. hepaticus ultimately maintains a non-pathogenic relationship with the host. Studies incorporating both immediate and long-term effects of T6S on hosts, as discussed for the Ma and Mekalanos pathogenesis study, above, will be important in further distinguishing antagonistic from harmless or mutualistic effects of T6S.
T6SS in interbacterial interactions
New data present the intriguing possibility that T6S may function to mediate interactions between bacteria rather than between bacteria and their eukaryotic hosts. A review of Myxococcus xanthus extracellular biology alluded to a role for T6S in the formation of fruiting bodies, suggesting T6S may be used for intraspecies microbial cooperation (Konovalova et al. 2010). Alternatively, recent work by Hood et al. (2010) suggests that T6SSs may mediate antagonistic interactions between bacteria. By comparing secretomes of Pseudomonas aeruginosa strains with either disruption or constitutive over-expression of the H1-T6SS gene cluster, the group identified three putative T6SS effectors, which they named Tse1-3 (type six effector 1-3). All three putative effectors were secreted into culture supernatants in a manner requiring the presence of the core T6S components Hcp1 and ClpV1 and deletion of genes encoding negative regulators of H1-T6S (retS which encodes a global regulator that represses transcription of H1-T6SS genes and/or pppA, which encodes a serine-threonine phosphatase that post-translationally represses T6S). However, none of the tse genes was required for the secretion of Hcp, indicating that they are not part of the secretion apparatus and may function solely as effectors.
One of the putative effectors, Tse2, was toxic to yeast, mammalian cells, and bacteria when produced intracellularly. The inability to introduce a deletion mutation in a small open reading frame immediately downstream of tse2 if tse2 was intact (but not if it was also deleted) caused the authors to suspect that the downstream gene, which they named tsi2, encoded a protein conferring immunity to the toxic effects of tse2. In support of this hypothesis, Tsi2 interacted with Tse2 in co-immunoprecipitation experiments, and production of Tsi2 within cells relieved toxicity of Tse2 for both eukaryotes (yeast and mammalian cells) and bacteria.
While intracellular production of Tse2 resulted in toxicity for all cells tested, subsequent experiments exposing target cells to tse2-expressing P. aeruginosa yielded a surprising result: eukaryotic cells were unaffected. Thus, although Tse2 was secreted by P. aeruginosa in a T6S-dependent manner, it appeared not to be translocated into eukaryotic hosts, even when P. aeruginosa was phagocytosed, suggesting that eukaryotes are not the true target of the Tse2 effector. By contrast, under certain culture conditions, the growth of bacterial target cells lacking tse2/tsi2 was severely inhibited by co-culture with Tse2/Tsi2 expressing bacteria. Expression of tsi2 in the target bacteria relieved the inhibition. Although Hood et al. did not show that Tse2 was translocated into target bacteria, the data suggest that Tse2 is a T6S effector that mediates competitive interactions among bacteria.
The study by Hood et al., which used strains of P. aeruginosa engineered to express their T6SSs constitutively and in which interbacterial inhibition occurred only when bacteria were grown on a solid support, raises many questions. When and where might the bacteria deploy their T6SSs to inhibit the growth of neighboring bacteria in nature? Do these systems mediate in both intra- and inter-species inhibition? Can they distinguish self from non-self target bacteria? Do T6SSs encoded by other bacteria function as inter-bacterial competition systems? If so, do they have Tse2/Tsi2 orthologues with different specificities? Do the T6SSs that target bacteria (and therefore must translocate proteins across two bacterial membranes) differ from those that target eukaryotic cells (and therefore must translocate proteins across only one plasma membrane)? Answering these and other questions will be critical to gaining a complete understanding of T6SSs and their potential.
T6SS as mediators of multiple interactions in diverse bacteria
The studies reviewed here showed that T6S functions in pathogenesis in several bacteria, in anti-pathogenesis in a few others, and in inter-bacterial interactions in a third case. However, several bacterial genomes contain multiple T6S-encoding gene clusters that appear not to be paralogues, suggesting the possibility for diverse functions. P. aeruginosa has three predicted T6S-encoding gene clusters, Y. pestis and Photorhabdus luminescens each have four, and B. mallei, B. thailandensis, and B. pseudomallei have four, five, and six, respectively. Many bacteria with the potential to produce several T6SSs have multiple hosts or survive in diverse environmental conditions. Y. pestis interacts intimately with both insects and mammals, and incidentally, at least one of its T6SSs is regulated by changes in temperature comparable to the body temperatures of fleas and mammals (Robinson et al. 2009). P. aeruginosa is both an environmental microbe and facultative pathogen of humans and can persist for years in the lungs of cystic fibrosis patients. Similarly, B. pseudomallei is an environmental soil microbe and facultative human pathogen, causing infections that reportedly can persist for more than 60 years. Given the mounting evidence for divergent functions of the T6SS, the idea that multiple T6SSs within individual bacterial species could mediate various types of interactions with diverse hosts, predators, cooperators or competitors is an enticing possibility that demands further study.
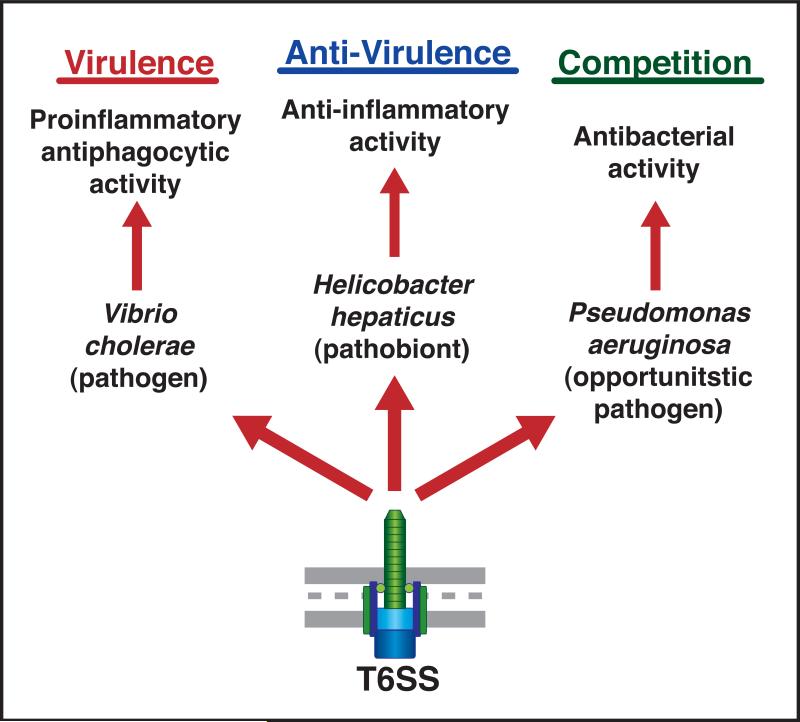
Figure 1. Multifunctionality of Type VI Secretion.
As highlighted by three papers published in the last year in Cell Host & Microbe, Type VI Secretion Systems have been implicated in virulence, commensalism or symbiosis, and interbacterial competition.
Acknowledgements
The authors are supported by grants from the NIH (AI43986 and a subproject of AI65359 to P.A.C.) and NSF (NSF Graduate Research Fellowship to A.J.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected Reading
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ. Burkholderia mallei Cluster 1 Type VI Secretion Mutants Exhibit Growth and Actin Polymerization Defects in RAW 264.7 Murine Macrophages. InfectImmun. 2010;78:88–99. doi: 10.1128/IAI.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Petters T, Sogaard-Andersen L. Extracellular biology of Myxococcus xanthus. FEMS Microbiol Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 2010;107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. Off the hook--how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, Egan S, Steinberg P, Kjelleberg S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One. 2008;3:e2744. doi: 10.1371/journal.pone.0002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun. 2005;73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson OP, Pinhassi J, Rieman L, Marklund B-I, Rhen M, Normark S, Gonzalez JM, Hagstrom A. High abundance of virulence gene homologues in marine bacteria. EnvironMicrobiol. 2009;11:1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009;12:11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Robinson JB, Telepnev MV, Zudina IV, Bouyer D, Montenieri JA, Bearden SW, Gage KL, Agar SL, Foltz SM, Chauhan S, Chopra AK, Motin VL. Evaluation of a Yersinia pestis mutant impaired in a thermoregulated type VI-like secretion system in flea, macrophage and murine models. MicrobialPath. 47:243–251. doi: 10.1016/j.micpath.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]