Abstract
Background
Most of our knowledge about how antiretrovirals and host immune responses influence the HIV-1 protease gene is derived from studies of subtype B virus. We investigated the effect of protease resistance-associated mutations (PRAMs) and population-based HLA haplotype frequencies on polymorphisms found in CRF01_AE pro.
Methods
We used all CRF01_AE protease sequences retrieved from the LANL database and obtained regional HLA frequencies from the dbMHC database. Polymorphisms and major PRAMs in the sequences were identified using the Stanford Resistance Database, and we performed phylogenetic and selection analyses using HyPhy. HLA binding affinities were estimated using the Immune Epitope Database and Analysis.
Results
Overall, 99% of CRF01_AE sequences had at least 1 polymorphism and 10% had at least 1 major PRAM. Three polymorphisms (L10 V, K20RMI and I62 V) were associated with the presence of a major PRAM (P < 0.05). Compared to the subtype B consensus, six additional polymorphisms (I13 V, E35D, M36I, R41K, H69K, L89M) were identified in the CRF01_AE consensus; all but L89M were located within epitopes recognized by HLA class I alleles. Of the predominant HLA haplotypes in the Asian regions of CRF01_AE origin, 80% were positively associated with the observed polymorphisms, and estimated HLA binding affinity was estimated to decrease 19–40 fold with the observed polymorphisms at positions 35, 36 and 41.
Conclusion
Polymorphisms in CRF01_AE protease gene were common, and polymorphisms at residues 10, 20 and 62 most likely represent selection by use of protease inhibitors, whereas R41K and H69K were more likely attributable to recognition of epitopes by the HLA haplotypes of the host population.
Keywords: CRF01_AE, HIV, HLA, polymorphisms, protease, resistance
Introduction
Almost two million people currently live with HIV in south and southeast Asia, and CRF01_AE is responsible for more than 80% of these infections [1]. Antiretroviral therapy has dramatically reduced the mortality associated with AIDS [2]. Mutations associated with resistance to protease inhibitors in the protease gene (pro) can be classified as either ‘major’ or ‘accessory’ [3]. Ongoing HIV replication during protease inhibitor exposure results in the selection for accessory mutations in pro or polymorphisms that often co-exist in wild type circulating strains of HIV-1.
In addition to antiretroviral drug pressure selecting for mutations, host immune responses can also drive genetic changes [4]. Human leukocyte antigen (HLA) alleles determine the cytotoxic T-lymphocyte (CTL) response, which targets specific HIV protein epitopes for immune control [4–7]. Variations in MHC class-I molecules among different human populations can influence HIV evolution [5], and CTL-driven viral escape represents a major determinant of HIVevolution and diversity at both the individual and population levels [8]. We investigated how the observed genetic structure of CRF01_AE pro on a population level has been affected by the introduction of protease inhibitor-based antiretroviral therapy and the frequency of specific HLA haplotypes in Asia.
Material and methods
All HIV-1 pro sequences annotated as CRF01_AE in the Los Alamos National Laboratory (LANL) database were downloaded on April 28, 2008. A modified GARD procedure [9] was used to screen sequences for misclassification. Retained sequences were aligned using a modified Needleman-Wunsch algorithm [10], and all duplicate sequences were removed. HYPERMUT 2.0 and Epitope Location Finder (available at http://hiv.lanl.gov/) were used to detect APOBEC-induced hypermutation and to identify CTL-recognized epitopes [11].
Polymorphisms and PRAMs were identified based on sequence data using the Stanford Resistance Database (http://hivdb.stanford.edu/, May 2008). Additional polymorphisms were defined as those amino acid residues in pro that differed from the subtype B consensus sequence. The prevalence of specific HLA alleles in the populations studied (Thai and Chinese) was obtained from the dbMHC database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/gv/mhc, August 2008). An HLA-subtype B association was assigned if, given the HLA haplotypes in the study population, the amino acid residue observed in the subtype B consensus sequence was present that would be expected with CTL escape. Similarly, an HLACRF01_AE association was assigned if, given the HLA haplotypes in the study population, the amino acid residue observed in the CRF01_AE consensus sequence was present that would be expected with CTL escape. HLA binding affinities were estimated using the Immune Epitope Database and Analysis resource (accessed October 2008). All analyses were performed using SPSS software 11.5 (SPSS Inc., Chicago, Illinois, USA).
Results
Of 1322 retrieved CRF01_AE sequences, 328 duplicated/identical sequences and 121 non-CRF01_AE or putative intra-subtype recombinant sequences were excluded. No hypermutation was observed in the remaining 873 sequences (available at http://www.hyphy.org/pubs/AE/AE.fas.) The majority of sequences (n = 772, 88.4%) were sampled in Asia, including Thailand (21.5%), Vietnam (19.8%), China (16.2%), Cambodia (13.8%), and others (17.1%). In 873 sequences, 638 were annotated with the year of sampling; of those, 168 (26.3%) were sampled between 1990 and 2000, and 470 (73.7%) were between 2001 and 2007. The maximum likelihood estimate of the mean pairwise genetic divergence between pro sequences, based on the fit of the codon model to a neighbor joining tree [12] constructed from the Tamura-Nei [13] distance matrix, was 4.22% (95% profile likelihood confidence interval of 4.05–4.39%), consistent with typical intra-subtype divergence levels reported in the pol coding region (http://hiv.lanl.gov/). Alignment-wide dN/dS was estimated at 0.242 (95% CI = 0.228–0.257), a value congruent with previous studies for selection in HIV-1 pro [14]. Residue sites 12, 13, 63, 70, 74, 82, and 93 (numbered with reference to the HXB2 pro) showed evidence of positive selection with dN/dS (P < 0.05). Ten residue pairs demonstrated evidence (posterior probability >90%) of coevolution (Table 1).
Table 1.
Co-evolving residue pairs in HIV-1 CRF01_AE protease.
| Residue 1 | Residue 2 | Probability 1 →2 | Probability 2 →1 | Posterior probability 1 ↔2 |
|---|---|---|---|---|
| L63‡ | I93‡ | 0.359 | 0.640 | 0.999 |
| I54 | V82 | 0.567 | 0.429 | 0.997 |
| A71 | M89* | 0.340 | 0.654 | 0.995 |
| L10 | I84 | 0.846 | 0.146 | 0.992 |
| P39 | Q61 | 0.734 | 0.228 | 0.962 |
| I64 | L90 | 0.520 | 0.440 | 0.960 |
| K43 | Q61 | 0.790 | 0.168 | 0.958 |
| K43 | R57 | 0.323 | 0.606 | 0.930 |
| M46 | K55 | 0.439 | 0.486 | 0.925 |
| K69* | I72 | 0.550 | 0.372 | 0.923 |
Residues that differed between the CRF01_AE and subtype B consensus sequences in pro.
Residues found to be under positive selection by the single likelihood ancestor counting [24].
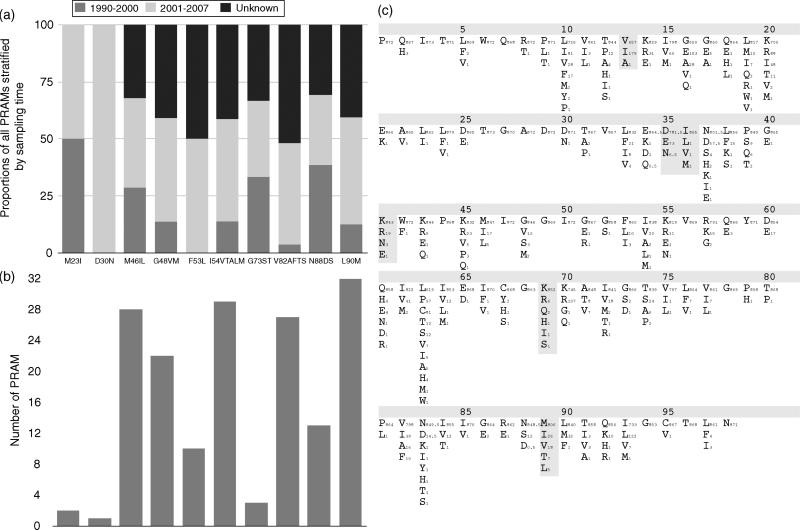
There were six differences between the consensus sequence inferred from our dataset and the subtype B consensus sequence (I13V, E35D, M36I, R41K, H69K and L89M). No polymorphisms overlapped with the list of PRAMs; however, 87 (10%) sequences contained one or more PRAMs (median = 2, range = 1–4), with L90M encountered most frequently (3.6%), and 47 sequences (5.3% of total) harbored more than 1 PRAM, with 50 (5.7%) sequences being resistant to protease inhibitor. Among the six countries with the most sequences available, sequences from Japan had the highest proportion of any major PRAM (41.3%), followed by Thailand (18.1%), Singapore (11.1%), China (3.5%), Vietnam (2.3%) and Cambodia (1.7%). Although this hierarchy roughly reflects the length of time that protease inhibitor therapy has been available in each of these countries (Roche, Bristol-Myers Squibb and Abbott, Thailand, personal communication), a comparison between the periods 1990–2000 and 2001–2007 showed no significant difference (P > 0.05) in PRAM frequencies at any of the residue sites (Fig. 1a and 1b).
Fig. 1. Observed CRF01_AE PRAMs and polymorphisms.
(a). Proportion of PRAMs by time period, (b). Number of PRAMs by residue site and (c). CRF01_AE consensus sequence and observed residue diversity. Comparisons were made between two time periods (1990–2000 and 2001–2007). The frequencies of three of the polymorphisms increased significantly from 1990–2000 to 2001–2007, residues: 16 (11.8–32.2%), 37 (3.0–11.3%) and 63 (22.8–35.4%) (P < 0.05), which corresponded to an increase in protease inhibitors in the region (Roche, Bristol-Myers Squibb and Abbott, Thailand, personal communication). Sites where CRF01_AE consensus sequence differed from subtype B consensus are marked with filled grey boxes and the subscripts represent the number of sequences observed with these nonconsensus CRF01_AE amino acids. Protease inhibitors were first introduced into each country was as follows: Thailand and Singapore, 1996; Japan, 1997; China, 1998; Vietnam and Cambodia, 2000 (Roche, Bristol-Myers Squibb and Abbott, Thailand, personal communication).
Other than PRAM sites, 5% of residues in the consensus pro sequence constructed from the CRF01_AE dataset contained a residue differing from the subtype B consensus sequence, that is, polymorphisms (grey shading, Fig. 1c). Additionally, we evaluated which polymorphisms previously associated with PRAMs in subtype B virus (http://hivdb.stanford.edu/, May 2008) were detected in the consensus sequence of the CRF01_AE virus from the dataset, and this was found to be the case for polymorphisms at codons 10 (OR = 3.4), 20 (OR = 5.2), and 62 (OR = 8.5). In order to evaluate the amino acid evolution at all sites, a phylogeny-based DEPS method was used to account for uneven substitution rates between residues and shared descent. Sites 13(I), 16(E), 19(M), 33(F), 38(S), 54(I), 64(I), 76(F), 82(F), 84(I), 88(S), 90(M) and 93(IL) were preferentially evolving towards the residues indicated in parentheses (DEPS Bayes Factor >100).
To investigate the potential contribution of HLA haplo-type to the genetic differences between subtypes, we analyzed the sequences in reference to the prevalence of particular HLA haplotypes in Thai and Chinese populations, as they provided the greatest number of sequences. There are six amino acid differences in pro between the consensus sequences of CRF01_AE and subtype B, which we define here as polymorphisms. Five of the six residues (13, 35, 36, 41 and 69) that differed between the consensus sequences of the CRF01_AE dataset and subtype B were associated with specific epitopes recognized by the HLA class I alleles present in these populations. At these five residues, 30 HLA-polymorphism associations were found, distributed among 11 (37%) HLA-A, 17 (56%) HLA-B and 2 (7%) HLA-C haplo-types. Of these associations, 24 (80%) demonstrated that the subtype B consensus amino acid would be favored during CTL escape, and 6 (20%) demonstrated that the CRF01_AE consensus amino acid would be favored. The four greatest decreases in binding affinity (i.e., the estimated CTL escape variant in the study population) were demonstrated between alleles in HLA-B* and the pro epitope in the amino acid positions 34–42, and the next two greatest decreases were between HLA-A*68 and positions 30–38. When protein structures of the subtype B consensus and the CRF01_AE dataset consensus pro sequences were modeled and compared, the six differing residues (polymorphisms) between them were not within the active site of the protease enzyme. Overall, the protease structures were almost completely identical between the subtype B and CRF01_AE dataset consensus sequences, except for some discrepancies observed at positions 35 and 36. No substantial differences in hydro-phobicity or chemical classifications were found between these polymorphisms.
Discussion
In this study, polymorphisms in the CRF01_AE protease gene were common, with the M36I polymorphism being the most frequent. M36I is a common nonsubtype B polymorphism in the absence of drug pressure and also has a higher replication capacity than subtype B wild type virus [15–18]. Thus, the presence of this polymorphism could provide a replicative advantage and suggests that 36I is most likely the natural genetic background. Polymorphisms in residues as positions 10, 20 and 62 were more likely to occur in association with major PRAMs that are often found in patients who have been treated with protease inhibitors [19–22]. In our dataset, these residues had positive correlations with one another and with major PRAMs at positions 30, 82 and 90, which are associated with resistance to nelfinavir [23]. Taken together, this pattern of covariation is likely explained by nelfinavir exposure.
Significant conservation of residues was also observed (dN/dS = 0.242), reflecting necessary constraints on genetic diversity for the conservation of protease function. Furthermore, the polymorphic changes between the consensus sequences of subtype B and CRF01_AE identified in this study were located outside the active site of protease (not shown) and did not alter their biochemical properties. As nonactive site mutations do not directly affect protease–substrate interaction [24], we theorize that the identified polymorphisms might emerge to evade CTL responses while maintaining functional catalytic activity.
Overall, 30 associations were observed between polymorphisms and specific HLA haplotypes. This result supports previous studies that identified the importance of HLA class I alleles, particularly HLA-B*, in driving HIV-1 evolution [4,5,7,25]. Recent data in subtype C virus revealed that minor differences in the amino acid sequence of an HLA-recognized epitope might affect CTL recognition leading to different clinical outcomes [6]. Of the polymorphisms identified, I13 V, E35D, M36I, R41K and H69K were associated with known epitopes recognized by HLA class I alleles predominantly found in this population. I13 V and E35D often appear during nelfinavir treatment [26]. Interestingly, R41K and H69K have not previously been reported to be associated with any currently available protease inhibitors, and the predicted HLA binding affinities at the epitope located at amino acid positions 34–42 decreases 19–40 fold when E35D, M36I and R41K are present. Given that CTL epitopes are HLA-class restricted and that sequence changes of viral escape pathways are characteristic of an HLA haplotype, the HLA haplotypes belonging to the study population most likely selected for the observed polymorphisms in CRF01_AE pro.
A number of limitations should be acknowledged. First, all sequences were downloaded from a database not designed for epidemiologic studies. Second, there are neither empirically determined linkages between the viral sequences and host HLA haplotypes, nor report of the specific antiretrovirals used by the individuals from whom these sequences were derived. Thus, future study of sequence data derived from HLA typed population is needed to confirm the current observations. Third, the reverse transcriptase gene was not studied and, as no significant difference in the protease binding site was observed between our models of protein structure, allostearic inhibition could not be assessed without an invivo assay. Fourth, a small proportion from non-Asian countries was included, though the HLA-specific and drug-specific mutations did not differ across the sequences from Asian and non-Asian sources. Ultimately, an in-vivo study that contains individual-level HIV-1 CRF01_AE pro sequences, HLA haplotype, and history of antiretroviral use is necessary to more completely delineate all the associations presented here.
In summary, this study provides the largest analysis of a curated dataset of CRF01_AE pro sequences and HLA haplotype performed to date. Polymorphisms in the protease gene were common with the M36I polymorphism being the most frequent, and it most likely represents the natural genotype of CRF01_AE, whilst the R41K and H69K polymorphisms are more likely attributable to CTL recognition of known epitopes in populations with particular HLA haplotypes. Additionally, as pro-tease function most likely requires certain biochemical properties of amino acid residues in the enzyme and the polymorphisms we observed in subtype B and the CRF01_AE consensus sequences were biochemically indistinguishable, this suggests that the residues we identified are critical for protease structure and function. Taken together, these data strongly suggests that HIV-1 CRF01_AE adapts effectively to antiretroviral and immunologic pressures in a population.
Table 2.
Hydrophobicity and chemical classifications of six different residues between subtype AE consensus and subtype B consensus sequences.
| Residue | Subtype B | Hydro-phobicity* | Chemical classifications | Subtype AE | Hydro-phobicity* | Chemical classifications |
|---|---|---|---|---|---|---|
| 13 | I | 1 | Aliphatic group | V | 1 | Aliphatic group |
| 35 | E | 4 | Negative charge | D | 4 | Negative charge |
| 36 | M | 2 | Sulphur containing group | I | 1 | Aliphatic group |
| 41 | R | 5 | Positive charge | K | 4 | Positive charge |
| 69 | H | 4 | Positive charge | K | 4 | Positive charge |
| 89 | L | 1 | Aliphatic group | M | 2 | Sulphur containing group |
Hydrophobicity 1 to 5 = from the highest hydrophobicity to the least hydrophobicity.
Acknowledgement
The authors would like to thank the University of California, San Diego Center for AIDS Research (UCSD CFAR) for support; and Rajendra Singh for his protease structure alignment. This work was supported by grants AI69432, MH083552, MH62512, AI077304, AI36214, AI047745, AI74621, AI43638, AI47745, AI57167 from the National Institutes of Health; the California HIV/AIDS Research Program RN07-SD-702; IS02-SD-701 from the University of California University wide AIDS Research Program; and by a University of California, San Diego Center for AIDS Research/NIAID Developmental Award to S.L.K.P (AI36214).
W.M., D.M.B., J.P.S., A.P., S.R.M., M.M.P., D.D.R., S.K.P. and D.M.S. contributed collectively to the analysis of the data, preparation, writing and review of the manuscript. W.M. and S.K.P. performed the statistical analysis, with input from the other authors. All authors have read and approved the final manuscript.
Footnotes
Conflicts: D.M.S. has received research support from Pfizer.
All other authors report no conflict of interest.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 2.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 4.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, et al. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahlenstiel G, Roomp K, Daumer M, Nattermann J, Vogel M, Rockstroh JK, et al. Selective pressures of HLA genotypes and antiviral therapy on human immunodeficiency virus type 1 sequence mutation at a population level. Clin Vaccine Immunol. 2007;14:1266–1273. doi: 10.1128/CVI.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngumbela KC, Day CL, Mncube Z, Nair K, Ramduth D, Thobakgale C, et al. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res Hum Retroviruses. 2008;24:72–82. doi: 10.1089/aid.2007.0124. [DOI] [PubMed] [Google Scholar]
- 7.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, et al. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 9.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 10.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 11.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G: > A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 14.Pillai SK, Kosakovsky Pond SL, Woelk CH, Richman DD, Smith DM. Codon volatility does not reflect selective pressure on the HIV-1 genome. Virology. 2005;336:137–143. doi: 10.1016/j.virol.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Grossman Z, Vardinon N, Chemtob D, Alkan ML, Bentwich Z, Burke M, et al. Genotypic variation of HIV-1 reverse transcriptase and protease: comparative analysis of clade C and clade B. AIDS. 2001;15:1453–1460. doi: 10.1097/00002030-200108170-00001. [DOI] [PubMed] [Google Scholar]
- 16.Holguin A, Alvarez A, Soriano V. High prevalence of HIV-1 subtype G and natural polymorphisms at the protease gene among HIV-infected immigrants in Madrid. AIDS. 2002;16:1163–1170. doi: 10.1097/00002030-200205240-00010. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Yue J, Wu S, Yan Y. Polymorphisms and drug resistance analysis of HIV-1 CRF01_AE strains circulating in Fujian Province, China. Arch Virol. 2007;152:1799–1805. doi: 10.1007/s00705-007-1019-9. [DOI] [PubMed] [Google Scholar]
- 18.Holguin A, Sune C, Hamy F, Soriano V, Klimkait T. Natural polymorphisms in the protease gene modulate the replicative capacity of non-B HIV-1 variants in the absence of drug pressure. J Clin Virol. 2006;36:264–271. doi: 10.1016/j.jcv.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Ives KJ, Jacobsen H, Galpin SA, Garaev MM, Dorrell L, Mous J, et al. Emergence of resistant variants of HIV in vivo during monotherapy with the proteinase inhibitor saquinavir. J Antimicrob Chemother. 1997;39:771–779. doi: 10.1093/jac/39.6.771. [DOI] [PubMed] [Google Scholar]
- 20.Condra JH, Holder DJ, Schleif WA, Blahy OM, Danovich RM, Gabryelski LJ, et al. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ariyoshi K, Matsuda M, Miura H, Tateishi S, Yamada K, Sugiura W. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J Acquir Immune Defic Syndr. 2003;33:336–342. doi: 10.1097/00126334-200307010-00007. [DOI] [PubMed] [Google Scholar]
- 22.Marcelin AG, Flandre P, de Mendoza C, Roquebert B, Peytavin G, Valer L, et al. Clinical validation of saquinavir/ritonavir genotypic resistance score in protease-inhibitor-experienced patients. Antivir Ther. 2007;12:247–252. [PubMed] [Google Scholar]
- 23.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 24.Erickson JW, Burt SK. Structural mechanisms of HIV drug resistance. Annu Rev Pharmacol Toxicol. 1996;36:545–571. doi: 10.1146/annurev.pa.36.040196.002553. [DOI] [PubMed] [Google Scholar]
- 25.John M, Moore CB, James IR, Mallal SA. Interactive selective pressures of HLA-restricted immune responses and antiretroviral drugs on HIV-1. Antivir Ther. 2005;10:551–555. [PubMed] [Google Scholar]
- 26.Shafer RW, Hsu P, Patick AK, Craig C, Brendel V. Identification of biased amino acid substitution patterns in human immunodeficiency virus type 1 isolates from patients treated with protease inhibitors. J Virol. 1999;73:6197–6202. doi: 10.1128/jvi.73.7.6197-6202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]