Human induced pluripotent stem (iPS) cells derived from somatic cells of patients hold great promise for modelling human diseases. Dermal fibroblasts are frequently used for reprogramming, but require an invasive skin biopsy and a prolonged period of expansion in cell culture prior to use. Here, we report the derivation of iPS cells from multiple human blood sources including peripheral blood mononuclear cells (PBMCs) harvested by routine venipuncture. Peripheral blood-derived human iPS lines are comparable to human embryonic stem (ES) cells with respect to morphology, expression of surface antigens, activation of endogenous pluripotency genes, DNA methylation and differentiation potential. Analysis of Immunoglobulin and T-cell receptor gene rearrangement revealed that some of the PBMC iPS cells were derived from T-cells, documenting derivation of iPS cells from terminally differentiated cell types. Importantly, peripheral blood cells can be isolated with minimal risk to the donor and can be obtained in sufficient numbers to enable reprogramming without the need for prolonged expansion in culture. Reprogramming from blood cells thus represents a fast, safe and efficient way of generating patient-specific iPS cells.
Somatic cells can be induced to the pluripotent state by the enforced expression of several transcription factors including OCT4, SOX2, KLF4, MYC, NANOG and LIN28 (Takahashi et al. 2007; Yu et al. 2007; Park et al. 2008a). Human iPS cells are commonly generated from dermal fibroblasts harvested by surgical skin biopsy (Park et al. 2008b). Exposure of the dermis to ultraviolet light increases the risk for chromosomal aberrations (Ikehata et al. 2003), raising concerns for whether iPS cells will reflect the patient’s constitutional genotype. For routine clinical application, it would be desirable to reprogram cell types that are safe and can be collected non-invasively in large numbers.
Blood is a cell source that can be easily obtained from patients. Mouse B and T-cells are amenable to reprogramming by over-expressing Oct4, Sox2, Klf4 and Myc with the ectopic expression of Cepbα and p53 knockdown respectively (Hanna et al. 2008; Hong et al. 2009). iPS cell lines have also been generated from mouse bone marrow progenitor cells (Okabe et al. 2009). We have previously reprogrammed cytokine-mobilized human CD34+ peripheral blood cells to pluripotency, but such harvests are cumbersome, expensive, and time-consuming (Loh et al. 2009). Several recent studies reported the generation of iPS cells from human bone marrow and cord blood (Ye et al. 2009; Giorgetti et al. 2009; Haase et al. 2009), but bone marrow harvesting is an invasive procedure, and cord blood is available for only a minority of individuals who have their samples banked at birth. A recent study using peripheral blood from donors with myeloproliferative disorder (MPD) isolated iPS colonies that contain the JAK2-V617F mutation (Ye et al. 2009), but MPD is characterized by abnormally high numbers of circulating CD34+ cells from the bone marrow. These previous studies demonstrating successful reprogramming of blood cells into iPS cells have relied on specialized blood cell sources with high proliferative potential.
CD34+ hematopoietic stem/progenitor cells mobilized into the donor’s peripheral blood by pre-treatment with granulocyte colony stimulating factor (G-CSF) can be successfully reprogrammed to pluripotency (Loh et al. 2009). To test if we can reprogram cells from routine peripheral blood (PB) sources, we obtained CD34+ purified blood samples from a healthy 49 year-old male donor who had undergone simple apheresis without cytokine priming. We also isolated mononuclear cells (PBMCs) from the peripheral blood samples collected by venipuncture of four healthy donors (28–49 years old) using Ficoll density centrifugation.
To induce reprogramming of enriched CD34+ blood cells, we infected with lentiviruses expressing OCT4, SOX2, KLF4, MYC reprogramming factors (Figure 1A). Colonies with well-defined hESC-like morphology were first observed 21 days after transduction (Figure 1B). For reprogramming of fresh peripheral blood mononuclear cells (PBMCs), we employed two rounds of lentiviral infection (day 0 and day 8), and isolated colonies with distinct flat and compact morphology with clear-cut round edges reminiscent of hES cells after a slightly longer latency of around 35 days (Figure 1C). Interestingly, a previous study using a single round of lentiviral infection of PBMCs failed to observe iPSC colony formation (Haase et al. 2009). In a separate set of experiments, we tested the ability of retroviruses encoding the human reprogramming factors to generate iPSCs from human PBMCs, and despite low infection efficiency, observed iPSC colonies after 25–35 days (Figure 1D).
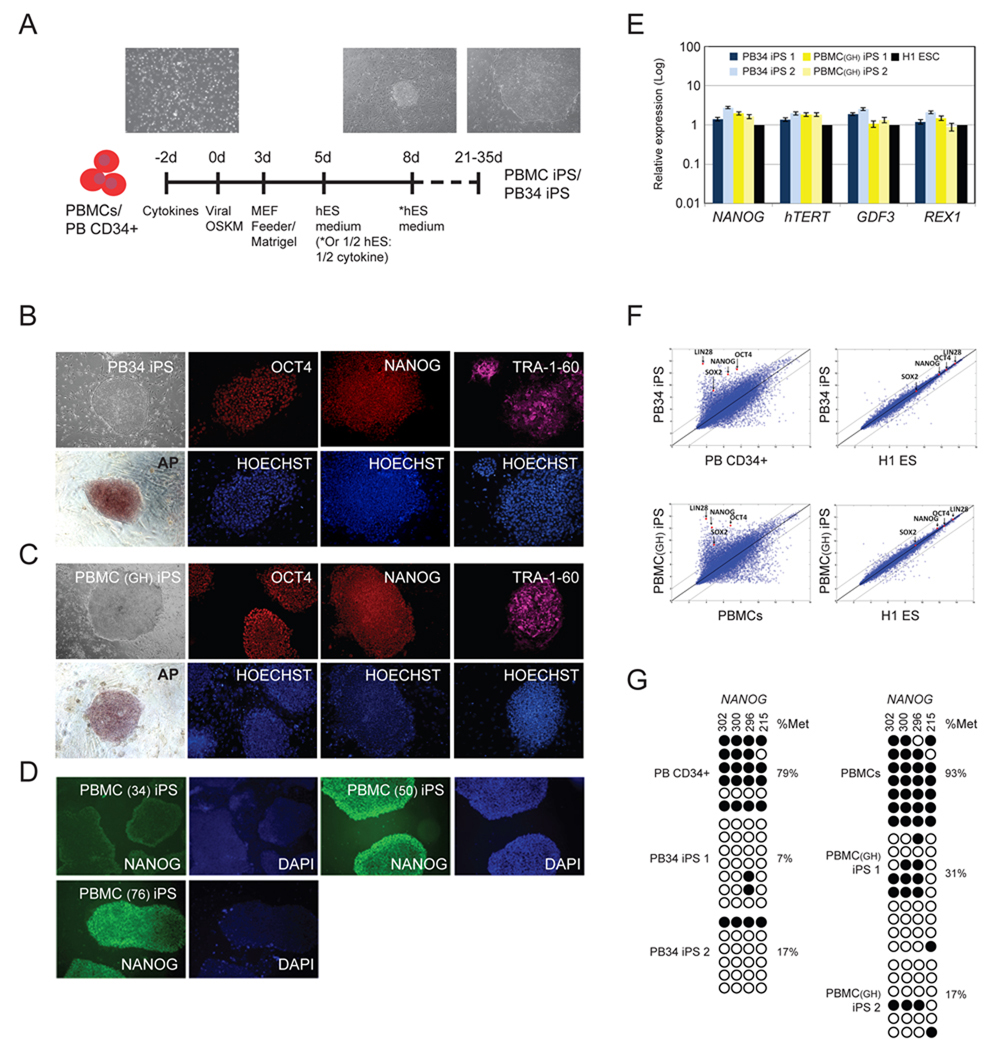
Figure 1. Reprogramming of peripheral blood cells to pluripotent iPS cells.
(A) Scheme for reprogramming human peripheral blood (PB) mononuclear cells (PBMCs) and CD34+ cells (PB CD34+). Morphology of the typical peripheral blood cells and images of hES cell-like iPS colonies are shown.
(B) Images of PB34 iPS colonies. Bright field images were acquired with a standard microscope (Nikon, Japan) with a 10x objective. Immunohistochemistry of PB derived iPS cell colonies expressing markers for OCT4, NANOG, Tra-1-60 and alkaline phosphatase (AP). Hoechst staining indicates the total cell content per field. Fibroblasts surrounding human iPS colonies serve as internal negative controls for immunohistochemistry staining. Images were acquired with a standard microscope (Nikon, Japan) with a 10x objective.
(C) Images of PBMC (Donor GH) iPS colonies. Bright field images were acquired with a standard microscope (Nikon, Japan) with a 10x objective. Immunohistochemistry of PB derived iPS cell colonies expressing markers for OCT4, NANOG, Tra-1-60 and alkaline phosphatase (AP). Hoechst staining indicates the total cell content per field.
(D) Images of PBMC (Donor 34, 50, 76) iPS colonies. Bright field images were acquired with a standard microscope (Nikon, Japan) with a 10x objective. Immunohistochemistry of PB derived iPS cell colonies expressing marker for NANOG. DAPI staining indicates the total cell content per field.
(E) Quantitative reverse transcription–PCR analyses for the expression of ES cell-marker genes NANOG, hTERT, GDF3 and REX1 in PB CD34+ and PBMCs derived iPS cells and human H1 ES cells. Individual PCR reactions were normalized against β-ACTIN and plotted (Log10 scale) relative to the expression level in the H1 ES cells, which was set to 1.
(F) Scatter plots comparing PB34 iPS and PBMC iPS cells global gene expression profiles to parental (Left) and H1 human ES cells (Right). The black lines indicate the linear equivalent and two-fold changes in gene expression levels between the paired cell types. Positions of pluripotency genes Oct4, Sox2, Nanog and Lin28 in scatter plots are indicated.
(G) Bisulfite genomic sequencing of the NANOG promoters reveals demethylation in the iPS cell lines. Each horizontal row of circles represents an individual sequencing reaction for a given amplicon. Open and filled circles represent unmethylated and methylated CpGs dinucleotides, respectively. Percentage of methylation is indicated for each cell line. For further characterization of the peripheral blood-derived iPSC clones see also Figures S1 and Table S1.
Using immunohistochemistry and flow cytometry, we analysed the iPS cell lines for expression of markers shared with hES cells. Consistent with their hES cell-like morphology, both PB34 iPS and PBMC iPS cells stained positive for Tra-1-81, NANOG, OCT4, Tra-1-60, SSEA4, and alkaline phosphatase (AP) staining (Chan et al. 2009) (Figure 1B–D, S1A-C). We routinely observed a reprogramming efficiency of 0.002% for PB CD34+ cells (Table S1), comparable to prior experience with primary fibroblasts, mobilized PBMCs, and cord blood cell reprogramming (Takahashi et al. 2007, Park et al. 2008a, Loh et al. 2009 and Haase et al. 2009). For PBMCs, we obtained hES-like colonies at the lower efficiency of 0.0008-0.001% (Table S1).
We further characterized the PB34 iPS and PBMC iPS cell lines for properties specific to hES cells. Efficient transgene silencing is essential for the derivation of pluripotent iPS cell lines (Brambrink et al. 2008). qRT-PCR using primers specific for endogenous and total transcripts of the reprogramming factors confirmed that OCT4, SOX2, KLF4 and MYC transgenes were efficiently silenced in the blood-derived iPS cells (Figure S1D). Additional analysis using quantitative PCR revealed the activation of pluripotency markers NANOG, hTERT, REX1 and GDF3 to a level similar to the expression in H1 hES cells (Figure 1E).
We next performed global gene expression analysis of the peripheral blood-derived iPS cells comparing it to hES, fibroblast iPS and somatic parental cells. Clustering analysis revealed a high degree of similarity among the reprogrammed iPS cells (dH1F-iPS, PBMC iPS1, PB34 iPS1, PB34 iPS2), which clustered together with the H1 and H9 ES cells and were distant from the parental somatic cells, as determined by a Euclidean distance metric (Figure S1E). Analysis of scatter plots similarly shows a tighter correlation among reprogrammed iPS cells (PB34 iPS, PBMC iPS) and human ES cells (H1 ES) than between differentiated parental cells and their reprogrammed derivatives (Figure 1F). Consistent with the activation of endogenous pluripotency-associated gene expression, reprogramming of the blood cells was accompanied by the demethylation of CpG dinucleotides at the NANOG promoters (Figure 1G). Moreover, cytogenetic analysis showed normal karyotypes for the iPS lines (Figure S1F).
Next, we evaluated the developmental potential of the iPS cell lines by in vitro embryoid body differentiation, hematopoietic colony forming assays, and in vivo teratoma induction. The iPS cells readily formed embryoid bodies upon induction (Figure S2A). qRT–PCR of the differentiated cells showed strong suppression of the pluripotency genes and activation of lineage-specific genes representing the three germ layers (Figure S2B, S2C). Hematopoietic differentiation of iPS cell lines resulted in erythroid, myeloid and granulocytic colony formation (Figure 2A, 2B). Interestingly, all PB CD34+ derived iPS lines we tested show greater hematopoietic colony forming activity than PBMC iPS. (Figure 2A).
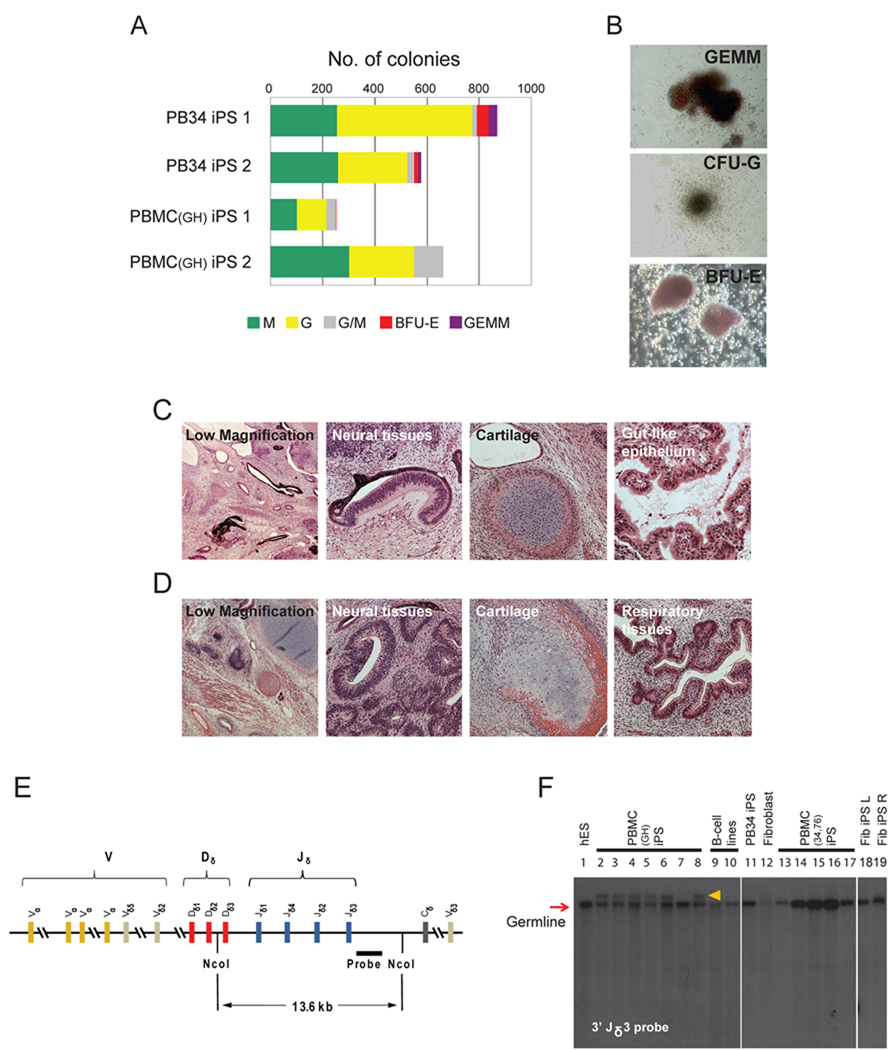
Figure 2. Pluripotency and V(D)J re-arrangement of peripheral blood-derived iPS cells.
(A) Embryoid bodies derived from PB34 and PBMC iPS cells yield hematopoetic colonies in semisolid methylcellulose media: burst forming unit-erythroid (BFU-E), colony forming unit-granulocyte (CFU-G), colony forming unit-macrophage (CFU-M), colony forming unit-granulocyte, macrophage (CFU-GM) and colony forming unit-granulocyte, erythroid, macrophage (CFU-GEMM). Total number of each type of colony was counted.
(B) Representative images of various types of hematopoietic colonies. Images were acquired with a standard microscope (Nikon, Japan) with a 20x objective.
(C, D) Hematoxylin and eosin staining of teratomas derived from immunodeficient mice injected with PB34 iPS (C) and PBMC iPS (D) cells show tissues representing all three embryonic germ layers.
(E) Genomic DNA from peripheral blood-derived iPS lines grown was digested with Nco I and analyzed for V(D)J rearrangements at the TCRδ (T-cell receptor Delta) locus by Southern blotting using a 3’Jδ3 probe.
(F) TCRδ V(D)J recombination of blood-derived iPS cell clones. Lanes 2–8 and lanes 13–17 are PBMC iPS lines. B-cell lines on lanes 8 and 9 showed no rearrangement. TCRδ rearrangement was observed for some PBMCs derived iPS lines (lanes 2–6 and 8). Lanes 1, 11, 12, 18, 19 are H1 hES cells, PB34 iPS cells, fibroblast cells, fibroblast derived iPS using retrovirus and lentivirus, respectively. The red arrow indicates expected size of the germline band. Orange arrow indicates re-arranged bands.
For further information on the pluripotency, V(D)J rearrangement and fingerprint analysis performed on the peripheral blood-derived iPSC clones see also Figures S2 and Table S2.
The most rigorous test for pluripotency of human ES cells is the formation of teratomas in immunodeficient mouse hosts (Lensch et al. 2007). Upon subcutaneous injection into immunodeficient Rag2−/− γc−/− mice, the iPS cell lines generated well-differentiated cystic teratomas representing all three embryonic germ layers (Figure 2C, 2D). DNA fingerprinting analysis verified that these cells were indeed derived from the parental blood cells, and not a result of contamination from existing hES or iPS cell lines (Table S2). The iPS clones have been propagated for at least 20 passages as of this submission.
As peripheral blood mononuclear cells consist of both myeloid and lymphoid elements (Figure S2D), we were interested in determining the lineage of origin of the reprogrammed cells. We tested the iPS clones for the presence of functionally rearranged immunoglobulin and T cell receptor genes using probes specific for IgH, TCRδ and TCRβ2. Among 12 independent clones from 4 separate individuals, we failed to detect IgH recombination, indicating that none of our lines arose from B lymphocytes (Figure S2E). As reported for the mouse, reprogramming human B lymphocytes may require additional factors like CEBPα (Hanna et al. 2008). Next, we analyzed the iPS lines for TCRδ and TCRβ2 recombination (Figure 2E, 2F, S2F). No PBMC iPS lines demonstrated TCRβ2 recombination, whereas 7 of 8 PBMC iPS lines isolated from a single donor sample exhibited rearrangement of the TCRδ locus, indicative of derivation from cells of the T lineage (Figure 2F). In contrast, PBMC iPS lines from donors 34 and 76 lacked rearrangement of IgH, TCRδ and TCRβ2, indicating derivation from non-lymphoid lineages (Figure 2F, S2E, S2F).
Isolation of iPS cells from T lymphocytes represents definitive proof that even terminally differentiated human cells are susceptible to reprogramming to pluripotency. Distinct protocols of cytokine stimulation and viral infection of the PBMC cells may pre-dispose to derivation from lymphoid versus non-lymphoid hematopoietic cells from peripheral blood sources, as can pre-selection of lymphoid target cells prior to reprogramming (Hong et al. 2009). PBMCs from donor GH were grown in medium containing IL-3, which is known to stimulate the growth of subsets of CD4+ T cells (Mueller et al. 1994) (Figure S2D). In contrast, PBMCs from donor 34 and 76 were cultured in medium promoting expansion of dendritic cells, and yielded iPS cells with germ line IgH and TCR alleles. For applications in regenerative medicine, iPS cells containing antibody or T cell receptor gene rearrangement may be undesirable (Serwold et al. 2007).
In conclusion, we have successfully reprogrammed cells from peripheral blood sources including samples obtained through routine venipuncture. Our study provides a strategy for the reliable generation of induced pluripotent stem cells from peripheral blood mononuclear cells. Although the per-cell derivation efficiency is low, peripheral blood is an accessible source of a large number of primary cells (easily 105-106), thus enabling reliable iPS cell isolation from only a few milliliters of whole blood. Future application of viral and transgene-free reprogramming or protein transduction (Kaji et al. 2009; Woltjen et al. 2009; Yu et al. 2009; Kim et al. 2009; Zhou et al. 2009) to peripheral blood reprogramming will greatly facilitate the development of efficient and safe ways of generating patient-specific pluripotent stem cells.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by grants from the National Institutes of Health (NIH) and the Howard Hughes Medical Institute to G.Q.D. J.J.C. is supported by SysCODE (Systems-based Consortium for Organ Design & Engineering), NIH grant # RL1DE019021. Y.H.L. is supported by the overseas fellowship from Agency of Science, Technology and Research (A*Star), and the Institute of Medical Biology, Singapore. We are grateful to Ann M. Mullally, Anupama Narla, Benjamin L. Ebert, Lars UW Müller, Axel Schambach and Stelios Andreadis for technical assistances. We acknowledge Sabine Loewer for helpful discussions and critical comments on the manuscript. While this manuscript was under revision, a manuscript appeared online demonstrating isolation of iPS cells from peripheral blood, including a single line that showed evidence for both TCRδ and TCRβ2 rearrangement by PCR (Kunisato et al. 2010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Information includes Supplemental Experimental Procedures, two figures, and two tables and can be found with this article online
Conflict-of-interest disclosure: GQD is a member of the Scientific Advisory Board of iPierian, Inc. MG and SI are employed by iPierian, Inc, a biotechnology company using iPS cells for drug discovery.
REFERENCES
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodríguez-Pizà I, Vassena R, Raya A, Boué S, Barrero MJ, Corbella BA, et al. Cell Stem Cell. 2009;5:353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, Zweigerdt R, Gruh I, Meyer J, Wagner S, et al. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata H, Masuda T, Sakata H, Ono T. Environ Mol Mutagen. 2003;41:280–292. doi: 10.1002/em.10153. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisato A, Wakatsuki M, Shinba H, Ota T, Ishida I, Nagao K. Stem Cells Dev. 2010 doi: 10.1089/scd.2009.0149. Published online. [DOI] [PubMed] [Google Scholar]
- Lensch MW, Schlaeger TM, Zon LI, Daley GQ. Cell Stem Cell. 2007;1:253–258. doi: 10.1016/j.stem.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DL, Chen ZM, Schwartz RH, Gorman DM, Kennedy MK. The Journal of Immunology. 1994;153:3014–3027. [PubMed] [Google Scholar]
- Okabe M, Otsu M, Ahn DH, Kobayashi T, Morita Y, Wakiyama Y, Onodera M, Eto K, Ema H, Nakauchi H. Blood. 2009;114:1764–1767. doi: 10.1182/blood-2009-02-203695. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Nature. 2008a;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Nat Protoc. 2008b;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Serwold T, Hochedlinger K, Inlay MA, Jaenisch R, Weissman IL. J Immunol. 2007;15:928–938. doi: 10.4049/jimmunol.179.2.928. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.