Abstract
Editing of mRNA transcribed from the mitochondrial cryptogenes ND8 (G1), ND9 (G2), G3, G4, ND3 (G5), RPS12 (G6) was investigated in Leishmania mexicana amazonensis, strain LV78, by amplification of the cDNA, cloning and sequencing. For each of these genes, extensively and partially edited transcripts were found to be relatively abundant compared to the respective pre-edited molecules. Moreover, the editing patterns observed in a majority of transcripts of each gene were consistent among themselves which allowed for inferring consensus editing sequences. The open reading frames contained in the consensus sequences were predicted to encode polypeptides that were highly similar to their counterparts in other species of Trypanosomatidae. Several kinetoplast DNA minicircles from this species available in the public domain were found to contain genes for guide RNAs which mediate editing of some of the mRNAs. The results indicate that the investigated strain of L. m. amazonensis has preserved its full editing capacity in spite of the long-term maintenance in culture. This property differs drastically from the other Leishmania species which lost some or all of the G1–G5 mRNA editing ability in culture.
1. Introduction
Mitochondrial genes in trypanosomatid protists are encoded by maxicircles which together with more abundant minicircles are catenated in the form of a kinetoplast DNA (kDNA) network [1–3]. The genes localized in maxicircles include two ribosomal RNA genes (12S and 9S rRNA) and eighteen protein-coding genes that represent ribosomal protein S12 (RPS12) and subunits of mitochondrial respiratory complexes I, III, IV and V [4]. There are no tRNA genes, and all tRNAs participating in mitochondrial translation are imported from the cytoplasm [5]. The most striking feature of the kinetoplast-mitochondrial genetic system is the process of RNA editing whereby messenger RNAs of several protein-coding genes are modified by specific insertions and (less frequently) deletions of uridylate residues [6]. Positions of the insertion/deletion sites and numbers of the U’s involved are defined by sequences of guide RNAs (gRNA) that are encoded mainly in the kDNA minicircles [7–9]. Although each minicircle class encodes only 1 to 3 different gRNAs, a single kDNA network contains dozens or hundreds of minicircle classes, thereby providing the required multitude of gRNAs [10]. Editing information is passed on from the guide RNAs to the messengers by interaction of the two RNA molecules mediated by complex enzymatic machinery (reviewed in [11–15]). Besides the 20S core “editosome” or “L-complex” [16–18], several additional macromolecular complexes have been identified including the complexes involved in gRNA biogenesis [19, 20] and mRNA polyadenylation [21].
Editing results in conversion of a pre-edited transcript with a defective reading frame into a mature mRNA [22–25]. Translation of the edited apocytochrome b (Cyb) mRNA was demonstrated [26].
The magnitude of RNA editing varies from a few inserted U-residues, such as those needed to repair a −1 frameshift in the COII mRNA [6,27], to hundreds of residues, as exemplified by ND7 or A6 transcripts that contain no recognizable ORF prior to editing [22,23,25]. Such extensively edited (pan-edited) mRNAs are processed with involvement of multiple gRNAs each of which being responsible only for a short ‘editing block’ (30–50 nt) of mRNA sequence [28–30]. Editing begins at the 3′ end of a pre-edited transcript by formation of a 10–15 nt ‘anchor’ duplex between the first gRNA and the mRNA and then expands towards the 5′ end until the entire gRNA is base-paired with the mRNA [7]. This is followed by annealing of the second gRNA which displaces the first gRNA from the overlap region and then extends the edited sequence further upstream, and so on until the entire mRNA is edited [31].
Such a complex process can be vulnerable to deleterious mutations and random genetic drift [32]. Indeed, only the indispensable mRNAs types, such as mRNA encoding subunits of cytochrome c oxidase, cytochrome bc1, and F1F0 ATPase or RPS12, are invariably edited in the investigated trypanosomatid species with functional oxidative phosphorylation suggesting the presence of a strong selection pressure to maintain intactness of the editing system [29,33]. This is not the case with respect to the five short cryptogenes, earlier referred to as G-rich regions G1–G5 (also called CR1–CR5) [29,33]. These are thought to encode subunits of respiratory Complex I (NADH dehydrogenase), although sequence-based identification was only possible for G1 (=ND8) [34], G2 (=ND9) [35] and, with lower confidence, G5(=ND3) [36]. In line with the uncertain function, and even the very existence, of Complex I in cultured Leishmania cells [37], it was found that productive editing of the G1–G5 mRNA is disrupted in the old laboratory strain of L. tarentolae [29,38]. It was hypothesized that in the absence of selection to maintain the full repertoire of guide RNAs, including those necessary for editing of G1-G5 mRNAs, the minicircles that encode these gRNAs had been lost in culture [32]. This view was supported by finding that in a more recently isolated strain of the same species the editing was almost intact [28].
Consequently, it can be thought that periodic passages of cultures through an animal or a sandfly should prevent the loss of editing at the population level. It was surprising, therefore, to find that a virulent strain of L. donovani, with a presumed ability to propagate in animal and insect hosts [39], also showed disruption of the G1–G5 mRNA editing, as well as insertional inactivating of one additional subunit of Complex I [33]. However, the other investigated mRNAs were efficiently edited in the same strain, indicating that the editing machinery in general was intact. These findings suggest that the selective loss of editing in this strain might occur relatively rapidly (since the most recent passage through the host or vector). Alternatively, the G1–G5 subunits or possibly even the entire Complex I, might be dispensable not only in culture but also in hosts, at least in the laboratory settings that might not fully recapitulate crucial steps of the natural life cycle. In this case the observed anomalies might have occurred over a longer period of time (since the initial isolation in culture in 1962).
Another question to be addressed is to what extent the observed dispensability of the G1–G5 products represents a general feature which, therefore, can also be observed in other Leishmania species, especially those that also were maintained in the laboratory for long time. In this work the editing has been investigated in L. mexicana amazonensis initially isolated in 1972. The strain (LV78) is virulent and has been used intensively for investigation of various aspects of Leishmania cell biology and pathogenesis (see e.g [40–43]). The results presented below indicate that in this organism, unlike in L. tarentolae and L. donovani, the editing of G1–G5 transcripts is intact. Such maintenance of editing indicates that there are significant, although not yet fully appreciated, differences among Leishmania species which allow for the loss of editing in some cases but prevent this in others.
2. Materials and Methods
2.1. Culture conditions, isolation of kDNA and recombinant DNA procedures
A culture of L. m. amazonensis (MPRP/BR/72/M1845), strain LV78, virulent clonal line 12–1, was provided by K.-P. Chang. Promastigotes were maintained at 26 °C in the M199 medium (Invitrogen) supplemented with 10% (v/v) heat inactivated fetal bovine serum and other components as described previously [39]. Other aspects of culture maintenance and growth of large (1–2 L) batch cultures were performed essentially as described for L. tarentolae [44]. Kinetoplast DNA was isolated by the CsCl cushion sedimentation procedure [45]. PCR amplification, cloning and sequencing were performed using standard procedures as described elsewhere [33]. DNA sequences were analyzed using Vector NTI.
2.2. Reverse transcription PCR and related procedures
Trizol (Invitrogen) extraction procedure was used to isolate total cell RNA of promastigotes. RT-PCR was performed using SuperScript™ III One-Step RT-PCR system (Invitrogen). The following oligonucleotides were used for RT-PCR of mitochondrial mRNA:
ND8 (G1), M268 (TAAATATAATAAGAAGTATATTAAT) and M269 (AACATTAACTAAATTTTTACTGCAA);
ND9 (G2), M270 (TATAATAAACTGTTTATATTAA) and M271 (AAAATTTAGTTAATGTTTAAGT);
G3, M272 (AAAATAAAATAACATGTATTAAGTA) and M273 (CTTTTTACTACAAAAATAATAGCTA);
G4, M274 (TAGATATAAAAAATATTAAATAAG) and M275 (TAATAAAAATAAATTCATATCCA);
ND3 (G5), M276 (TCACTCAAAAAAATTCACGCTT) and M277 (TATTATTTTAGTCAAGAGAAAGTTT);
RPS12 (G6): M278 (CATAAATACCTATTTAGACCTTT) and M279 (TTATTTTTATAAAAATATAAATAATTAAAC).
cDNA was purified, cloned and sequenced as described previously [33]. Guide RNA search was performed using the UWGCG program BESTFIT [46].
3. Results and Discussion
3.1. Partial sequence analysis of L. m. amazonensis LV78 maxicircle kDNA
The maxicircle DNA region encompassing two adjacent G-rich cryptogenes ND8 and ND9 and also the region containing the G3 cryptogene were amplified by PCR using flanking oligonucleotide primers chosen with the aid of the available L. tarentolae and L. donovani maxicircle sequences. Subsequently, the regions were expanded and merged into a single 9,373 bp contig by amplification of additional segments and by primer walking. The sequenced region represented more than a half of the maxicircle conserved region and included nearly the entire 12S rRNA gene (except for 60–65 nt missing at the 5′ end), followed by the genes for 9S rRNA, ND8, ND9, MURF5, ND7, COIII, Cyb, A6, MURF1 (putative ND2 [47]), G3 (a cryptogene), ND1, and flanked on the other side with the 5′ half of the COII gene. The genes were found in the same order and with the same polarity as in other investigated trypanosomatid species. The region including the G4 cryptogene (574 bp) and another region including the adjacent cryptogenes ND3 and RPS12 (876 bp) were amplified using conserved sequences in the flanks.
The determined gene sequences from L. m. amazonensis show a high degree of similarity (85–90%) to homologous genes from other Leishmania species. The maxicircle genome conservation includes positions and lengths of the pre-edited regions which therefore can be easily identified by sequence comparisons. Thus, in L. m. amazonensis, as in other investigated Leishmania species, the ND7 gene contains two short pre-edited regions: the first one at the 5′ end of the coding sequence and the second one ~170 nt downstream; the COIII and Cyb genes are 5′-edited; the A6 gene contains an extensively edited 5′-region; and transcripts of the ND1, MURF1 and MURF5 genes do not require editing (data not shown).
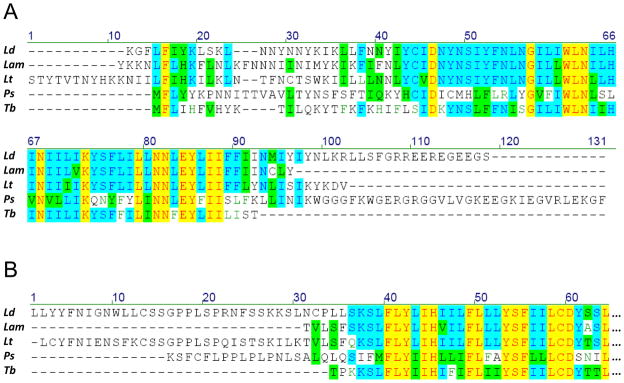
The conservation even includes some idiosyncratic features of the MURF5 and MURF1 ORFs as illustrated in Fig. 1. The in silico translated MURF5 amino acid sequences from several species (Fig. 1A) show a high degree of similarity except for the very N- and C-termini. The homology in that region is also shared by the Phytomonas serpens and the Trypanosoma brucei sequences. The conserved part, therefore, represents the functional polypeptide product. However, positions of the start and the end codons are less certain. Unlike in the other two species, none of the predicted Leishmania sequences begins with methionine. A conserved leucine residue, encoded by a TTG codon and found at alignment position 15, probably takes its place. This suggests that instead of ATG, the MURF5 initiation involves a non-canonical codon TTG in Leishmania. Earlier, TTG was proposed to serve as initiation codon in pan-edited G4 and ND3 mRNA of T. brucei [36,48]. It also frequently functions as initiation codon in mitochondria of nematodes [49].
Fig. 1.
Comparison of MURF1 and MURF5 amino acid sequences from Leishmania mexicana amazonensis (Lam) with their homologs from Leishmania donovani (Ld), Leishmania tarentolae (Lt), Phytomonas serpens (Ps), and Trypanosoma brucei (Tb). Multiple sequence alignment was generated using Vector NTI. Identical residues found in a given position in all sequences are highlighted with yellow, and those in the majority of sequences are indicated with blue, while conservative substitutions are shown with green. Dashes represent alignment gaps: (A) multiple alignment of the complete predicted MURF5 sequences; (B); alignment of the N-terminal ends of the MURF1 sequences.
The position of translation termination codon is also problematic in the MURF5 sequences. Conservation of the amino acid sequences ends at positions 93–97 which coincide with the end of the L. m. amazonensis ORF, but not in the other species (Fig. 1A). A possibility exists that in these cases the termination codon is created by polyadenylation of an in-frame TA or T that are localized in this region but this has not been verified experimentally.
Most of the predicted MURF1 N-terminal sequences, including L. m. amazonensis (Fig. 1B), also lack methionine. The exception is the P. serpens sequence where it is found at the start of the conserved region (alignment position 39). The corresponding position in the other sequences is occupied by leucine which is encoded by TTG. This again points at the potential role of this triplet as a noncanonical initiation codon in trypanosomatids.
The encoded initiation and termination codons in other L. m. amazonensis genes are either ATG (Met), as in ND1 and COII, or ATA (Ile) in ND7, although in the last case there is also a conserved in-frame ATG six codons downstream (data not shown). The initiation codons of COIII, Cyb and A6 are likely to be localized within the respective 5′ pre-edited regions and created by editing out of the encoded AG dinucleotides, as in the other Leishmania species. The encoded termination codons were canonical TAA (ND7, Cyb, A6, MURF1, ND1) or TAG (COIII). The TGA codon, as in other trypanosomatids, is predicted to code for tryptophan.
3.2. Editing of pan-edited cryptogene mRNAs
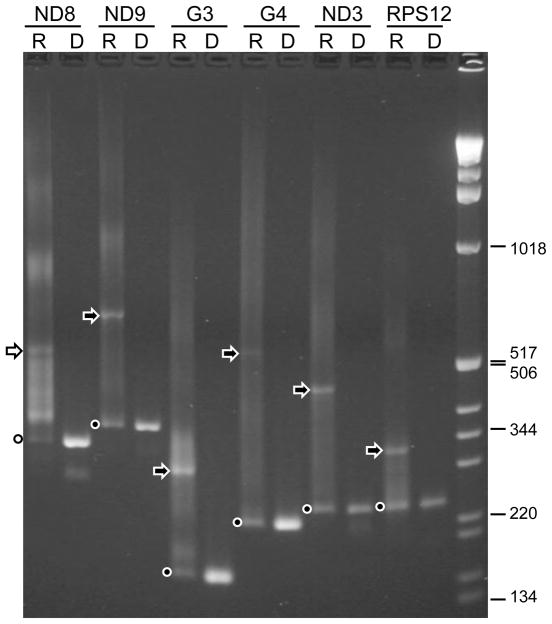
Reverse-transcription PCR was employed to investigate editing of transcripts derived from the six pan-edited cryptogenes of L. m. amazonensis. The amplification strategy was based on the presence of unedited sequences at the 5′ and 3′ ends of the transcripts that flank the pan-edited mRNA regions [46]. The flanking sequences are used to design PCR primers which, therefore, allow for amplification of all types of mRNA (pre-edited, partially edited and fully edited) transcribed from a targeted cryptogene. The flanking sequences in the L. m. amazonensis were identified by comparison with the sequences from L. tarentolae [28]. Due to the noticeable size increase (nearly two-fold) after extensive editing, cDNA products representing pan-edited transcripts can be distinguished from pre-edited products. Indeed, after gel electrophoresis of the amplified material for each cryptogene, a discrete cDNA band with the size larger than the pre-edited product was observed (Fig. 2, arrow). Moreover, judging by relative abundance of the edited cDNA products (arrows) compared to the pre-edited cDNA products (dots), it can be tentatively concluded that the edited products represent a significant fraction in the steady-state mRNA pool. These results were the first indication that productive editing takes place in each case.
Fig. 2.
Results of RT-PCR amplification of mitochondrial cryptogene transcripts from promastigotes of L. m. amazonensis after electrophoresis in a 2% (w/v) agarose gel. Each cryptogene, indicated above the panel, was analyzed in two reactions: lanes indicated with R represent RT-PCR with mitochondrial RNA as the source of material; lanes labeled with D represent standard amplification reactions using kDNA. Dots indicate the bands representing pre-edited cDNA amplification products (with sizes matching the respective genomic fragments), arrows show positions of presumptive fully edited cDNA products. Positions of 1 kb DNA Ladder bands (Invitrogen) are shown to the right.
To verify this, the amplified material was cloned in the plasmid vector and sequenced. The presence of inserted or deleted uridylates in the cDNA sequences was deduced from comparison with the corresponding gene sequences. Edited sequences of individual clones were arranged in the order reflecting the 3′ to 5′ progression of editing and used to derive the editing consensus sequences [46].
The ND8 (=G1) edited sequence was derived from the sequences of 41 cDNA clones that represented various stages of editing. Translation of the consensus yielded a 146 aa polypeptide that was 91.1% identical to the ND8 from L. tarentolae LEM125 (Fig. 3), the only other Leishmania species from which this sequence was available [28], indicating that the functional, fully edited ND8 mRNA is also present in L. m. amazonensis. The canonical initiation and termination codons are created by editing in both species.
Fig. 3.
Pan-edited mitochondrial ND8 (NADH dehydrogenase subunit 8) mRNA sequence of L. m. amazonensis. The sequence (Lam, upper lane) and its in silico translation (the lane underneath the mRNA sequence) are compared with the respective protein and mRNA sequences from L. tarentolae LEM125 (Lt). Edited regions on both mRNA are underlined. Unedited 5′ and 3′ sequences expected to be present in Lam mRNA and used as PCR primers are shown with bold italics. Encoded nucleotides preserved in the edited transcripts are shown with upper case letters; inserted uridylates are shown with low case u’s and deleted uridylates are shown with asterisks. Dashes represent alignment gaps. The exact 5′ and 3′ ends of the Lam mRNA are unknown, while the actual ends of the Lt mRNA are as shown.
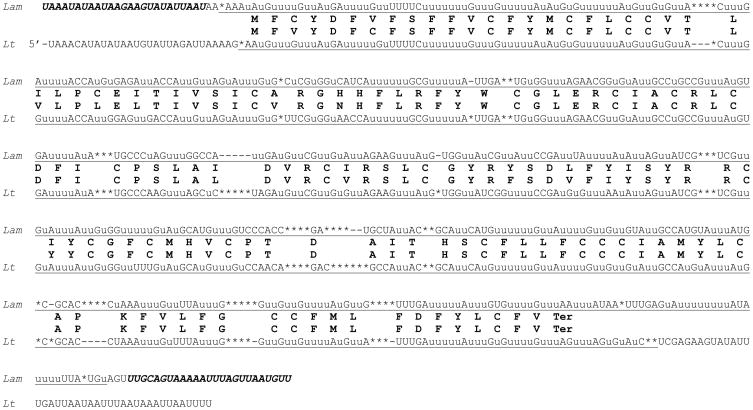
An ND9 (=G2) edited sequence (Fig. 4) was derived from the analysis of 16 cDNA clones including six clones that were consistently edited throughout the entire length. The 203 amino acid sequence derived from the editing consensus sequence is highly (84.7%) similar to the 190 aa L. tarentolae sequence except for the very C-terminal parts. It needs to be mentioned that there was a minor group of partially edited cDNA clones that deviated from the editing consensus in this region by the presence of a single additional U residue (highlighted with black in Fig. 4). Interestingly, the amino acid sequence derived from this alternative editing pattern is nearly identical to the L. tarentolae sequence. If this alternative (yet more conserved) pattern were present in the fully edited L. m. amazonensis mRNA, then the proteins from the two species would be 92.7% identical. It is uncertain if this pattern is present in at least some of the mature mRNA molecules, so that two types of proteins may be produced, and what the function of this duplicity would be. Potential diversification of protein sequences by alternative editing has been noticed earlier [36, 50]. The initiation and the conserved termination codons in the L. m. amazonensis sequence are canonical and created by editing.
Fig. 4.
Pan-edited mitochondrial ND9 (NADH dehydrogenase subunit 9) mRNA sequence of L. m. amazonensis. Most designations are as in Fig. 3. The 3′-end alternative editing pattern caused by the presence of an extra U (highlighted with black) in some Lam transcripts (see the text) is also shown. A minicircle-encoded guide RNA, gND9-V, is shown matched with the Lam edited sequence with canonical base pairs indicated with vertical lines and non-canonical G-U base pairs indicated with colons.
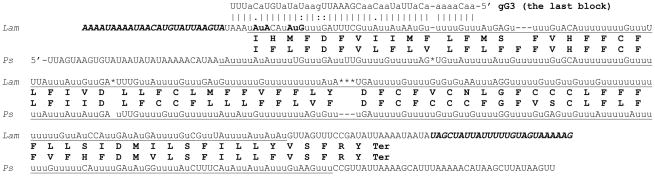
The edited G3 sequence of L. m. amazonensis (Fig. 5) was derived from the analysis of 39 cDNA clones including six clones that were edited over the entire length of the cDNA. The derived 77 aa polypeptide could not be compared with a Leishmania homolog due the absence of a productive G3 editing in the two species investigated earlier [28, 29, 33]. However, edited G3 sequences are available from P. serpens (59.0% identity) and T. brucei (40.3% identity). The initiation codon in the L. m. amazonensis sequence is either a conserved noncanonical AuA or a canonical AuG that is localized just a few nucleotides downstream. The L. m. amazonensis polypeptide has two predicted transmembrane segments and is expected to be an integral membrane protein. BLAST search has not revealed G3 homologs in other organisms and its function remains unknown.
Fig. 5.
Pan-edited mitochondrial G3 mRNA sequence of L. m. amazonensis. The Lam sequence is compared with the G3 edited sequence of Phytomonas serpens (Ps). A minicircle-encoded gRNA, mediating the last editing block, is also shown. Other designations are as in Fig. 3 and 4.
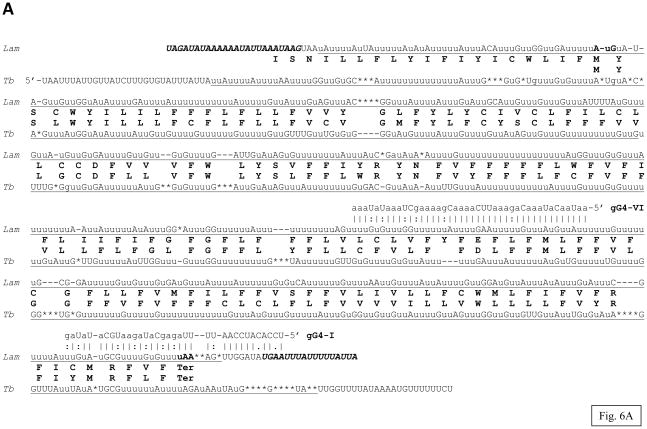
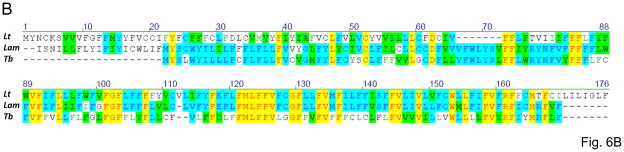
The inferred G4 edited sequence is shown in Fig. 6A. The edited consensus was bases on the analysis of 30 cDNA clones including two clones that were edited over the entire length. The encoded polypeptide is 166 amino acids long and includes six transmembrane segments but shows no identifiable homology besides the counterparts in other trypanosomatids. The previously determined sequence from L. tarentolae LEM125 is highly similar (67.3% identity) with the L. m. amazonensis sequence in the C-terminal half, however the level of identity in the N-terminal half is only 18.1% (Fig. 6B). Remarkably, the entire L. m. amazonensis sequence is rather similar to the sequence from Trypanosoma brucei (55.8% identity in the overlapping region), which lends additional credence to the L. m. amazonensis consensus. There are several alternatives for the position of the translation initiation codon in the L. m. amazonensis sequence. One possibility represents a canonical AuG that is conserved in T. brucei (Fig. 6A). However, this would leave an unusually long untranslated edited sequence upstream and that would represent a situation not typically observed in trypanosomatids. The alternatives represent several in-frame Auu (Ile) codons in the upstream edited sequence. The predicted termination codon is canonical uAA found in the same position as in T. brucei.
Fig. 6.
Pan-edited mitochondrial G4 mRNA and protein sequences of L. m. amazonensis: (A) comparison of the edited G4 mRNA and the derived amino acid sequence with the T. brucei sequences (Tb). Two minicircle-encoded gRNAs, gG4-I and gG4-IV, are also shown; (B) multiple alignment of the G4 sequences from the two aforementioned species and L. tarentolae (Lt). The remaining designations are as in Fig. 1, 3 and 4.
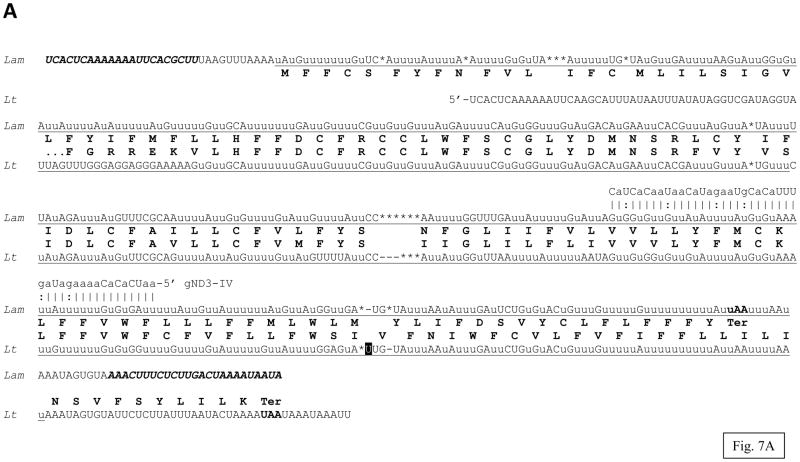
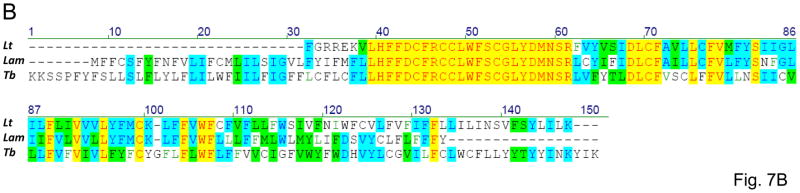
The G5 (putative ND3) edited sequence of L. m. amazonensis was derived from 15 cDNA clones including seven clones consistently edited almost over the entire length. The canonical initiation and termination codons flank the ORF found in the sequence (Fig. 7A). The 125 aa polypeptide has three transmembrane regions but no homologs could be found in non-trypanosomatid organisms. Both the edited G5 mRNA sequence and its in silico translation are highly similar to the fully edited sector of the published L. tarentolae LEM125 sequence (85.8% and 62.7% identity, respectively) [28]. However, the presence of an extra U residue (highlighted in Fig. 7A) in the L. tarentolae sequence shifts the translation frame with respect to the L. m. amazonensis sequence, and that leads to reduction of the similarity between the C-terminal parts of the two proteins. The possibility exists that in either species this difference is eliminated by a subsequent re-editing within this region or that there is a minor fraction of alternatively edited transcripts (as discussed above for ND9 mRNA) but this has not been investigated. The C-terminal, as well as the N-terminal part of the L. m. amazonensis protein shows a noticeable conservation with respect to the T. brucei sequence (fig. 7B). The N-terminal segment of the L. tarentolae protein is not available, apparently due to incomplete editing of the G5 mRNA in that strain [28].
Fig. 7.
Pan-edited mitochondrial ND3 (NADH dehydrogenase subunit 3) mRNA and protein sequences of L. m. amazonensis: (A) comparison with the edited ND3 mRNA and the derived amino acid sequences from L. tarentolae LEM125 (Lt). The unmatched U-residue in the Lt sequence which is responsible for the relative reading frame shift is highlighted with black. A minicircle-encoded gRNA, gND3-IV, is also shown; (B) multiple alignment of the ND3 amino acid sequences. Other designations are as in Fig. 1, 3 and 4.
The RPS12 (=G6) edited sequence (Fig. 8) was based on the analysis of 25 cDNA clones including 19 clones that were consistently edited almost over the entire length. The edited consensus encoded a polypeptide that was 76.1% identical to the L. tarentolae sequence and 87.5% identical to the L. donovani sequence. In the Leishmania species the reading frame starts with a conserved noncanonical Auu (Ile) codon that is created by editing and ends with an encoded conserved UAA. The conservation also includes the presence of two Domain Connection Sequences in each species [31,33].
Fig. 8.
Pan-edited mitochondrial RPS12 (ribosomal protein S12) mRNA sequence of L. m. amazonensis. The Domain Connections Sequences DCS-I and DCS-II are boxed. Other designations are as in Fig. 3.
3.3. Guide RNAs and gRNA genes
Most guide RNA genes are localized in minicircles and can be identified by finding a sequence match between a minicircle and an edited mRNA sequence [8–10, 46]. Several minicircle sequences from the L. mexicana species complex (that includes L. m. amazonensis) are available in public databases. The 0.7–0.8 kb minicircles of this species share the same basic organization with other Leishmania including a 100–130 bp conserved region that contains replication origins and a variable region that defines differences among minicircle classes, including encoding different types of gRNAs. As expected, in several minicircles a match was found between the edited sequences described above and a specific segment of the variable region. This segment, a putative gRNA gene, is localized at a distance of 240–260 bp from a CSB-3 sequence element GGGGTTGGTGTA on the opposite strand. The organization of gRNA genes in L. mexicana is, therefore, the same as in L. tarentolae and L. donovani [8,10,33].
A potential gRNA gene for editing of ND9 mRNA (Fig. 4) was localized in the minicircle class represented by the GanBank™ entries M94090, Z11551 and Z11552. The mRNA segment edited with this putative gRNA accurately corresponds to editing block V (gND9-V) of the L. tarentolae ND9 editing cascade in which a complete gRNA set has been described [28].
The gene potentially encoding a gRNA for the last editing block of the G3 mRNA is found in the minicircles M21235 and M21236 (Fig. 5). Two putative guide RNAs were identified for the G4 mRNA (Fig. 6A). The first editing block (gG4-I) is encoded by the minicircles M94088, M94089, Z11549, Z11550, Z11554, Z11556. Another guide RNA is encoded by the minicircles M94091, Z11553, Z11555. According to its position with respect to the edited sequence it represents block VI gRNA (gG4-VI). A potential gene encoding a guide RNA for editing block IV of ND3 mRNA (gND3-IV) was found in the minicircle entry M21327 (Fig. 7A).
A gene for potential block II Cyb gRNA is contained within the sequenced region of the maxicircle in the same locus as in the other species (positions 1791–1740 of the GenBank™ HM439238 entry, the complementary strand).
4. Conclusion
I have found that mRNA derived from each of the six mitochondrial pan-edited cryptogenes is productively edited in promastigotes of L. m. amazonensis, strain LV78. So far, this organism was the only investigated Leishmania that possessed the full editing capacity; in the other investigated species of this genus the editing of G1–G5 mRNA was fully or partially disrupted [28, 29, 33]. It is also noteworthy that the productive editing is observed in L. m. amazonensis in spite of the fact that this is another example of an old laboratory strain which had passed through numerous replication cycles in culture under a supposedly relaxed selection to maintain the complete minicircle and the gRNA repertoire. This lack of selection pressure was believed to be the main factor for the loss of editing in the other cases. This new finding suggests that some important, yet not fully appreciated, biochemical differences among the strains or species play an important role in maintaining intactness of the editing system or allowing for its loss in some cases. The availability of a representative set of genomic sequences from this genus will shed light in this and other questions addressing evolution and comparative biology of this group.
Acknowledgments
I am indebted to Prof. K.-P. Chang for providing the strain of L. m. amazonensis and for valuable advice and discussions. Technical assistance by C. Kim and A. Banerjee is greatly appreciated. This work was supported by the NIH grant AI070927 to D. A. M.
Abbreviations
- aa
amino acid
- CO
cytochrome c oxidase
- Cyb
apocytochrome b
- gRNA
guide RNA
- kDNA
kinetoplast DNA
- mRNA
messenger RNA
- MURF
maxicircle unassigned reading frame
- ORF
open reading frame
- RPS12
ribosomal protein S12
- rRNA
ribosomal RNA
- tRNA
transfer RNA
Footnotes
Note: Nucleotide sequence data reported in this work are available in the GenBank™ database under the accession numbers HM439238 and HM443080.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simpson L. The mitochondrial genome of kinetoplastid protozoa: Genomic organization, transcription, replication and evolution. Ann Rev Microbiol. 1987;41:363–82. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- 2.Stuart K. Kinetoplast DNA, a mitochondrial DNA with a difference. Mol Biochem Parasitol. 1983;9:93–104. doi: 10.1016/0166-6851(83)90103-2. [DOI] [PubMed] [Google Scholar]
- 3.Lukeš J, Guilbride DL, Votýpka J, et al. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson L, Wang SH, Thiemann OH, et al. U-insertion/deletion Edited Sequence Database. Nucl Acids Res. 1998;26:170–6. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfonzo JD, Soll D. Mitochondrial tRNA import--the challenge to understand has just begun. Biol Chem. 2009;390:717–22. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benne R, Van den Burg J, Brakenhoff J, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–26. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 7.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–98. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 8.Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–84. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- 9.Pollard VW, Rohrer SP, Michelotti EF, et al. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990;63:783–90. doi: 10.1016/0092-8674(90)90144-4. [DOI] [PubMed] [Google Scholar]
- 10.Simpson L. The genomic organization of guide RNA genes in kinetoplastid protozoa: Several conundrums and their solutions. Mol Biochem Parasitol. 1997;86:133–41. doi: 10.1016/s0166-6851(97)00037-6. [DOI] [PubMed] [Google Scholar]
- 11.Simpson L, Aphasizhev R, Gao G, et al. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–70. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart KD, Schnaufer A, Ernst NL, et al. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim Biophys Acta. 2008;1779:270–80. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koslowsky DJ. Complex interactions in the regulations of trypanosome mitochondrial gene expression. Trends Parasitol. 2009;25:252–5. doi: 10.1016/j.pt.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Osato D, Rogers K, Guo Q, et al. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009;15:1338–44. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aphasizhev R, Aphasizheva I, Nelson RE, et al. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–24. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carnes J, Trotter JR, Peltan A, et al. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–30. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Ge P, Hui WH, et al. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci USA. 2009;106:12306–10. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng J, Aphasizheva I, Etheridge RD, et al. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:1–12. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimi H, Čičová Z, Novotná L, et al. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–99. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etheridge RD, Aphasizheva I, Gershon PD, et al. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhat GJ, Koslowsky DJ, Feagin JE, et al. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61:885–94. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- 23.Feagin JE, Abraham J, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–22. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 24.Feagin JE, Shaw JM, Simpson L, et al. Creation of AUG initiation codons by addition of uridines within cytochrome b transcripts of kinetoplastids. Proc Natl Acad Sci USA. 1988;85:539–43. doi: 10.1073/pnas.85.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koslowsky DJ, Bhat GJ, Perrollaz AL, et al. The MURF3 gene of T. brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell. 1990;62:901–11. doi: 10.1016/0092-8674(90)90265-g. [DOI] [PubMed] [Google Scholar]
- 26.Horváth A, Berry EA, Maslov DA. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–40. doi: 10.1126/science.287.5458.1639. [DOI] [PubMed] [Google Scholar]
- 27.Shaw J, Campbell D, Simpson L. Internal frameshifts within the mitochondrial genes for cytochrome oxidase subunit II and maxicircle unidentified reading frame 3 in Leishmania tarentolae are corrected by RNA editing: Evidence for translation of the edited cytochrome oxidase subunit II mRNA. Proc Natl Acad Sci. 1989;86:6220–4. doi: 10.1073/pnas.86.16.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao GG, Kapushoc ST, Simpson AM, et al. Guide RNAs of the recently isolated LEM125 strain of Leishmania tarentolae: An unexpected complexity. RNA. 2001;7:1335–47. doi: 10.1017/s1355838201018076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiemann OH, Maslov DA, Simpson L. Disruption of RNA editing in Leishmania tarentolae by the loss of minicircle-encoded guide RNA genes. EMBO J. 1994;13:5689–700. doi: 10.1002/j.1460-2075.1994.tb06907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corell RA, Feagin JE, Riley GR, et al. Trypanosoma brucei minicircles encode multiple guide RNAs which can direct editing of extensively overlapping sequences. Nucleic Acids Res. 1993;21:4313–20. doi: 10.1093/nar/21.18.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–67. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 32.Simpson L, Thiemann OH, Savill NJ, et al. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci USA. 2000;97:6986–93. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neboháčová M, Kim CE, Simpson L, et al. RNA editing and mitochondrial activity in promastigotes and amastigotes of Leishmania donovani. Int J Parasitol. 2009;39:635–44. doi: 10.1016/j.ijpara.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souza AE, Myler PJ, Stuart K. Maxicircle CR1 transcripts of Trypanosoma brucei are edited, developmentally regulated, and encode a putative iron-sulfur protein homologous to an NADH dehydrogenase subunit. Mol Cell Biol. 1992;12:2100–7. doi: 10.1128/mcb.12.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souza AE, Shu H-H, Read LK, et al. Extensive editing of CR2 maxicircle transcripts of Trypanosoma brucei predicts a protein with homology to a subunit of NADH dehydrogenase. Mol Cell Biol. 1993;13:6832–40. doi: 10.1128/mcb.13.11.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read LK, Wilson KD, Myler PJ, et al. Editing of Trypanosoma brucei maxicircle CR5 mRNA generates variable carboxy terminal predicted protein sequences. Nucleic Acids Res. 1994;22:1489–95. doi: 10.1093/nar/22.8.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opperdoes FR, Michels PA. Complex I of Trypanosomatidae: does it exist? Trends Parasitol. 2008 doi: 10.1016/j.pt.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Maslov DA, Thiemann O, Simpson L. Editing and misediting of transcripts of the kinetoplast maxicircle G5 (ND3) cryptogene in an old laboratory strain of Leishmania tarentolae. Mol Biochem Parasitol. 1994;68:155–9. doi: 10.1016/0166-6851(94)00160-x. [DOI] [PubMed] [Google Scholar]
- 39.Goyard S, Segawa H, Gordon J, et al. An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol. 2003;130:31–42. doi: 10.1016/s0166-6851(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 40.Chang KP, Bray RS, Leaney AJ. Infection of mouse macrophages in vitro by sandfly-derived promastigotes of Leishmania mexicana amazonensis. Trans R Soc Trop Med Hyg. 1981;75:475–6. doi: 10.1016/0035-9203(81)90128-0. [DOI] [PubMed] [Google Scholar]
- 41.Lee S-Y, Lee S-T, Chang K-P. Transkinetoplastidy--A novel phenomenon involving bulk alterations of mitochondrion-kinetoplast DNA of a trypanosomatid protozoan. J Protozool. 1992;39:190–6. doi: 10.1111/j.1550-7408.1992.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 42.McGwire B, Chang KP. Genetic rescue of surface metalloproteinase (gp63)-deficiency in Leishmania amazonensis variants increases their infection of macrophages at the early phase. Mol Biochem Parasitol. 1994;66:345–7. doi: 10.1016/0166-6851(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 43.Thiakaki M, Rohousova I, Volfova V, et al. Sand fly specificity of saliva-mediated protective immunity in Leishmania amazonensis-BALB/c mouse model. Microbes Infect. 2005;7:760–6. doi: 10.1016/j.micinf.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Simpson L, Simpson AM, Blum B. RNA editing in mitochondria. In: Higgins SJ, Hames BD, editors. RNA Processing. IRL Press; 1994. pp. 69–105. [Google Scholar]
- 45.Simpson L, Berliner J. Isolation of the kinetoplast DNA of Leishmania tarentolae in the form of a network. J Protozool. 1974;21:382–93. doi: 10.1111/j.1550-7408.1974.tb03675.x. [DOI] [PubMed] [Google Scholar]
- 46.Maslov DA, Simpson L. Strategies of kinetoplastid cryptogene discovery and analysis. Methods Enzymol. 2007;424:127–39. doi: 10.1016/S0076-6879(07)24006-6. [DOI] [PubMed] [Google Scholar]
- 47.Kannan S, Burger G. Unassigned MURFI of kinetoplastids codes for NADH dehydrogenase subunit 2. BMC Genomics. 2008;9:455. doi: 10.1186/1471-2164-9-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corell RA, Myler P, Stuart K. Trypanosoma brucei mitochondrial CR4 gene encodes an extensively edited mRNA with completely edited sequence only in bloodstream forms. Mol Biochem Parasitol. 1994;64:65–74. doi: 10.1016/0166-6851(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 49.Okimoto R, Macfarlane JL, Wolstenholme DR. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucl Acids Res. 1990;18:6113–8. doi: 10.1093/nar/18.20.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochsenreiter T, Hajduk SL. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 2006;7:1128–33. doi: 10.1038/sj.embor.7400817. [DOI] [PMC free article] [PubMed] [Google Scholar]