Abstract
Canonical and noncanonical nuclear factor κB (NF-κB) signaling are the two basic pathways responsible for the release of NF-κB dimers from their inhibitors. Enhanced NF-κB signaling leads to inflammatory and proliferative diseases; thus, inhibitory pathways that limit its activity are critical. Whereas multiple negative feedback mechanisms control canonical NF-κB signaling, none has been identified for the noncanonical pathway. Here, we describe a mechanism of negative feedback control of noncanonical NF-κB signaling that attenuated the stabilization of NF-κB–inducing kinase (NIK), the central regulatory kinase of the non-canonical pathway, induced by B cell–activating factor receptor (BAFF-R) and lymphotoxin β receptor (LTβR). Inhibitor of κB (IκB) kinase α (IKKα) was previously thought to lie downstream of NIK in the non-canonical NF-κB pathway; we showed that phosphorylation of NIK by IKKα destabilized NIK. In the absence of IKKα-mediated negative feedback, the abundance of NIK increased after receptor ligation. A form of NIK with mutations in the IKKα-targeted serine residues was more stable than wild-type NIK and resulted in increased noncanonical NF-κB signaling. Thus, in addition to the regulation of the basal abundance of NIK in unstimulated cells by a complex containing tumor necrosis factor receptor–associated factor (TRAF) and cellular inhibitor of apoptosis (cIAP) proteins, IKKα-dependent destabilization of NIK prevents the uncontrolled activity of the noncanonical NF-κB pathway after receptor ligation.
INTRODUCTION
The regulation of gene transcription by dimers of nuclear factor κB (NF-κB) proteins is important for various immune processes and for the modulation of cell death and survival (1). In resting cells, NF-κB dimers are sequestered in the cytosol by inhibitory proteins and are released through the actions of two basic pathways termed canonical and noncanonical NF-κB signaling (2). These two pathways lead to the release of different NF-κB heterodimers with vastly different kinetics and perform nonredundant physiological functions. Whereas canonical NF-κB signaling leads to the release of NF-κB dimers within minutes in response to many stimuli, noncanonical signaling is activated by a select group of receptors, such as B cell–activating factor receptor (BAFF-R), lymphotoxin β receptor (LTβR), and receptor activator of NF-κB (RANK) on a limited set of cells, including B cells, fibroblasts, and macrophages (3–5). Although the pathways are regulated distinctly, they both play nonredundant physiological roles in B cell survival, lymphoid organogenesis, and osteoclast differentiation (6).
Whereas the regulation of canonical NF-κB signaling has been studied extensively, the architecture and regulation of noncanonical NF-κB signaling are only beginning to be understood (7). The central event that leads to noncanonical NF-κB signaling is the stabilization of NF-κB–inducing kinase (NIK), which is normally degraded to an undetectable amount in resting cells, but which accumulates after stimulation of the appropriate receptors (8). Accumulated NIK phosphorylates and activates inhibitor of κB kinase α (IKKα), which in turn phosphorylates the C terminus of p100, an NF-κB precursor protein (9, 10). The C terminus of p100 sequesters its N-terminal domain, known as p52. Upon phosphorylation by IKKα, the C terminus of p100 is degraded, which enables the release of the p52-RelB NF-κB dimer (11). How NIK is constitutively degraded in unstimulated cells and subsequently released upon receptor ligation is beginning to be understood (12, 13). NIK is thought to be recruited by tumor necrosis factor (TNF) receptor (TNFR)–associated factor 3 (TRAF3) to a complex composed of TRAF2, TRAF3, and cellular inhibitor of apoptosis 1 (cIAP1) and cIAP2, which, in resting cells, constitutively ubiquitinate NIK, leading to its degradation. Receptor ligation results in the degradation of TRAF3, which enables the release of NIK from the control of the TRAF-cIAP complex, and, consequently, accumulation of NIK.
Whereas the TRAF-cIAP model provides substantial insight into how noncanonical NF-κB signaling is inhibited in an unstimulated context, it is not known whether or how noncanonical NF-κB signaling is inhibited after receptor stimulation. The need for an improved understanding of the regulation of poststimulatory noncanonical NF-κB signaling has gained increased importance because conditions that simulate the long-term activation of noncanonical NF-κB signaling lead to substantial proliferative and autoimmune pathology. For example, deletion of genes that encode components of the TRAF-cIAP complex or amplifications of NIK, leading to constitutive NIK stability, are frequently the key oncogenic events in multiple myeloma and B cell lymphomas (14–16). Oncogenic viruses such as Epstein-Barr virus and human T cell leukemia virus have proteins that mimic receptor activation of the pathway (17, 18). Furthermore, substantially increased concentrations of BAFF, which primarily activates the noncanonical NF-κB pathway, are often seen in the plasma of patients with autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, and Sjogren’s syndrome (19–21). Mouse models of autoimmune diseases have confirmed clinical findings that the increased activity of noncanonical NF-κB signaling is a critical driver of inflammatory pathology (22, 23).
Given the substantial contribution that long-term activation of non-canonical NF-κB signaling makes to proliferative and inflammatory diseases, it is critical to understand how signaling through this pathway is inhibited after stimulation. As is the case for canonical NF-κB signaling, defects in negative feedback mechanisms ultimately may be found to be the root cause of disease pathogenesis in certain situations (24). Here, we report a previously uncharacterized role for IKKα in the regulation of the stability of NIK and describe an IKKα-mediated mechanism of negative feedback within noncanonical NF-κB signaling that serves to attenuate signal-induced accumulation of NIK. We showed that a deficiency in IKKα or a defect in the interaction between NIK and IKKα led to substantially higher amounts of NIK protein in multiple cell types after receptor ligation than when IKKα was present and able to interact with NIK. The mechanism by which IKKα served to destabilize NIK after receptor ligation was through IKKα-mediated phosphorylation of particular residues in the C terminus of NIK. We found that mutation of these residues, thus eliminating IKKα-mediated negative feedback, led to the enhanced stability of NIK and the increased activity of noncanonical NF-κB signaling. Therefore, although NIK is regulated by the previously described TRAF-cIAP complex during unstimulated conditions, our findings provide evidence that IKKα-mediated phosphorylation of NIK prevents uncontrolled activity of the noncanonical NF-κB pathway after receptor ligation.
RESULTS
IKKα inhibits NIK
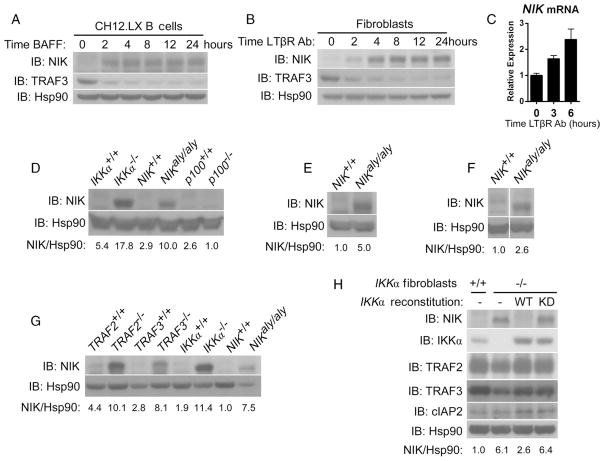
According to the current model for the regulation of noncanonical NF-κB signaling, NIK is not only constitutively synthesized but also constitutively targeted for degradation, which is mediated by the TRAF-cIAP complex in unstimulated cells (8, 12, 13). Because stimulation of the appropriate receptors releases NIK from the control of the TRAF-cIAP complex, through the degradation of TRAF3, it would be expected that the abundance of NIK would increase over time. However, we found in both B cells and fibroblasts that the abundance of NIK protein did not continually increase after ligation of BAFFR or LTβR, but instead remained constant even though TRAF3 protein was not resynthesized and the abundance of NIK messenger RNA (mRNA) increased slightly (Fig. 1, A to C). Given this discrepancy, we hypothesized that the current model for the regulation of NIK was incomplete and that a negative feedback mechanism existed that prevented a continual increase in the abundance of NIK after receptor ligation. We hypothesized that this putative negative feedback mechanism would not be functional if noncanonical NF-κB signaling were disrupted through the deletion of downstream mediators of the pathway. Accordingly, we assessed the stability of NIK in unstimulated fibroblasts derived from mice deficient in IKKα or p100 and from alymphoplasia (aly) mice, which carry a single amino acid mutation in NIK that ablates the interaction between NIK and IKKα and thus prevents downstream noncanonical NF-κB signaling (3). We detected NIK protein in fibroblasts from IKKα−/− and NIKaly/aly mice, but not from p100−/− mice (Fig. 1D). In addition to fibroblasts, freshly isolated splenic B cells and RANK-stimulated, primary bone marrow macrophages from aly mice contained substantially more NIK protein than did the same cells from wild-type mice, which suggested that the proposed regulatory mechanism was not cell-specific (Fig. 1, E and F).
Fig. 1.
IKKα inhibits NIK. (A and B) Long-term receptor ligation leads to the accumulation of a constant amount of NIK, in the absence of the resynthesis of TRAF3, as assayed by Western blotting in CH12.LX B cells treated with hBAFF (100 ng/ml) (A) or in murine fibroblasts treated with an agonistic antibody against LTβR (2 μg/ml) (B). (C) The abundance of NIK mRNA was assayed by quantitative polymerase chain reaction (PCR) assay in murine fibroblasts and did not decrease after treatment with agonistic antibody against LTβR (2 μg/ml). (D) NIK is basally stable in untreated fibroblasts from IKKα−/− and NIKaly/aly but not wild-type or p100−/− fibroblasts as assessed by Western blotting. Values reported for the quantitation of bands here (in numbers beneath the blots) and in all other cases are relative arbitrary units as described in Materials and Methods. (E and F) Enhanced stability of NIK in freshly isolated splenic B cells (E) and primary bone marrow macrophages treated with recombinant mRANK (100 ng/ml) (F) from NIKaly/aly compared to that in cells isolated from wild-type mice. (G) Enhanced basal stability of NIK from IKKα−/− and NIKaly/aly fibroblasts is comparable to that observed in TRAF2−/− and TRAF3−/− fibroblasts as assayed by Western blotting. (H) Enhanced basal stability of NIK observed in IKKα−/− fibroblasts is dependent on the kinase activity of IKKα as evidenced by retroviral reconstitution of IKKα−/− fibroblasts with pBABE-Empty Vector (−), pBABE-IKKα-WT (WT), or pBABE-IKKα-K44A (KD), the kinase-defective mutant of IKKα.
To better understand the relative effect of this previously uncharacterized mechanism of negative regulation on the stability of NIK compared to that of the TRAF-cIAP complex, we compared the basal stability of NIK in IKKα−/− and NIKaly/aly cells with that in fibroblasts deficient in TRAF2 or TRAF3 and we noted comparable amounts of NIK among these cells (Fig. 1G). We then attempted to determine the role of the kinase activity of IKKα in regulating the stability of NIK by retrovirally reconstituting IKKα−/− fibroblasts with wild-type IKKα or a kinase-defective mutant of IKKα (25). Reconstitution of cells with wild-type IKKα resulted in a return of the basal abundance of NIK protein to undetectable amounts, whereas reconstitution with kinase-defective IKKα did not, indicating a role for the kinase activity of IKKα in the inhibitory mechanism. In all cases, the abundances of TRAF2, TRAF3, and cIAPs were roughly equivalent, which indicated that perturbation of the amounts of the constituents of the TRAF-cIAP complex was not responsible for IKKα-mediated modulation of NIK stability (Fig. 1H).
TRAF-cIAP–mediated control of NIK is preserved in IKKα-deficient and aly cells
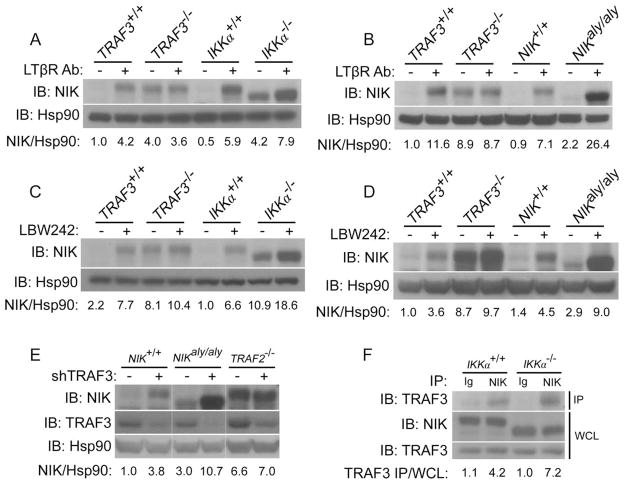
We initially considered the possibility that the enhanced stability of NIK in IKKα−/− and aly cells might be due to a previously unappreciated role for IKKα in the TRAF-cIAP–mediated degradation of NIK. For example, an interaction between NIK and functional IKKα may be required for the successful degradation of NIK by the TRAF-cIAP complex. If this were the case, one would expect that the abundance of NIK in IKKα-deficient cells or in aly cells would no longer be under the control of the TRAF-cIAP complex; thus, no increase in the abundance of NIK would be expected to be seen under stimuli that inhibit the activity of the TRAF-cIAP complex. For example, in TRAF3-deficient fibroblasts in which NIK cannot interact with the TRAF-cIAP complex, no increase in the abundance of NIK was observed under conditions of receptor stimulation or treatment with LBW242, an inhibitor of the cIAPs (Fig. 2, A and C). However, the abundance of NIK in IKKα-deficient cells and aly cells continued to be responsive to receptor stimulation and inhibition of cIAP (Fig. 2, A to D). That the abundance of NIK in IKKα-deficient cells and aly cells continued to be modulated by the TRAF-cIAP complex suggested that TRAF-cIAP–mediated degradation of NIK was not dependent on the presence of IKKα or on the interaction between NIK and IKKα.
Fig. 2.
TRAF-cIAP–mediated control of the stability of NIK is preserved in IKKα-deficient cells and aly cells. (A and B) Western blotting analysis revealed that the abundance of NIK continued to be responsive to LTβR stimulation with 4-hour treatment of IKKα−/− fibroblasts (A) and NIKaly/aly fibroblasts (B) with an agonistic antibody against LTβR. (C and D) Western blotting analysis showed that the abundance of NIK continued to be responsive to inhibition of cIAPs with 4-hour treatment of IKKα−/− fibroblasts (C) and NIKaly/aly fibroblasts (D) with LBW242 (1 μM), a small-molecule inhibitor of cIAP1 and cIAP2. (E) Knockdown of TRAF3 in both wild-type and NIKaly/aly fibroblasts results in the enhanced stability of NIK as evidenced by retroviral reconstitution of wild-type, NIKaly/aly, and TRAF2−/− fibroblasts with control shRNA (−) or with shRNA specific for TRAF3 (+). (F) NIK in IKKα−/− fibroblasts continues to interact with TRAF3 as evidenced by coimmunoprecipitation of endogenous TRAF3 with NIK with control immunoglobulin (Ig) or antibody against NIK (NIK) from extracts of wild-type or IKKα−/− fibroblasts. The abundance of NIK in these cells was equilibrated with the proteasome inhibitor MG132.
To further confirm this point, we used short hairpin RNA (shRNA) to knock down TRAF3 in wild-type and NIKaly/aly cells. As a control, we also performed knockdown of TRAF3 in TRAF2-deficient fibroblasts in which NIK should not be under the control of the TRAF-cIAP complex because of the absence of TRAF2, which normally recruits the cIAPs to the complex. As expected, knockdown of TRAF3 in TRAF2-deficient fibroblasts did not result in an increase in the abundance of NIK; however, in both wild-type and NIKaly/aly cells, knockdown of TRAF3 resulted in the enhanced stability of NIK (Fig. 2E). In addition to these functional and genetic approaches, we determined whether the interaction between NIK and TRAF3 was preserved in IKKα-deficient cells, because TRAF3 is the only component of the TRAF-cIAP complex that is thought to directly interact with NIK. Indeed, coimmunoprecipitation of endogenous TRAF3 with NIK was comparable in IKKα+/+ and IKKα−/− fibroblasts (Fig. 2F). Together, these data suggest that NIK continued to interact with TRAF3 in IKKα-deficient cells or aly cells and continued to be subject to inhibition by the TRAF-cIAP complex.
IKKα phosphorylates NIK on specific C-terminal residues
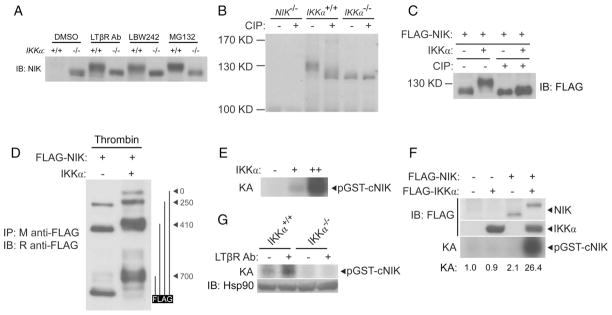
In the course of our experiments, we noted that the species of NIK protein that accumulated in response to various stimuli in IKKα-deficient cells migrated substantially further on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) than did NIK protein from wild-type fibroblasts (Fig. 3A). A difference in the migration of NIK was also observed in extracts from NIKaly/aly and NIK+/+ cells (Fig. 2, B and D). The difference in the migration of NIK isolated from extracts of IKKα+/+ or IKKα−/− fibroblasts was eliminated upon treatment with calf intestinal phosphatase (CIP), suggesting that the difference in migration might be due to the IKKα-dependent phosphorylation of NIK (Fig. 3B). Furthermore, this phenomenon was recapitulated in experiments with human embryonic kidney (HEK) 293T cells in which cotransfection with plasmids encoding IKKα and NIK resulted in NIK protein that exhibited a substantially slower migration on SDS-PAGE gels than occurred with NIK from cells transfected with NIK-encoding plasmid alone (Fig. 3C). Again, this migratory difference was eliminated by treatment of the extracts with CIP (Fig. 3C). An increase in the abundance of IKKα in transfected HEK 293T cells did not result in the destabilization of NIK, which was likely due to the high abundance of these proteins in the transiently transfected cells. However, IKKα did destabilize NIK in fibroblasts when a retroviral expression system was used that resulted in the abundance of the ectopically expressed proteins more closely matching that of their endogenous counterparts (fig. S1). To determine the general location of the phosphorylated residues in NIK, we performed partial thrombin digestions of NIK protein alone or in the presence of IKKα and found that phosphorylation occurred in the C-terminal 247 amino acid residues of NIK (Fig. 3D). We then generated a glutathione S-transferase (GST)-fusion protein containing these C-terminal 247 residues of NIK (GST-cNIK) and found that purified IKKα phosphorylated this substrate in an in vitro kinase assay (Fig. 3E).
Fig. 3.
IKKα phosphorylates NIK. (A) NIK from IKKα−/− fibroblasts migrates further on SDS-PAGE gels than does NIK from IKKα+/+ fibroblasts regardless of the stimulus used to trigger its accumulation. Fibroblasts were treated for 4 hours with the indicated stimuli and the abundance of NIK was roughly equilibrated by loading different amounts of extract to facilitate migration analysis. (B) Differential migration of NIK from IKKα+/+ and IKKα−/− fibroblasts is due to phosphorylation. The accumulation of NIK was triggered with the proteasome inhibitor MG132, after which NIK was immunopurified and subjected to treatment with CIP. (C) IKKα leads to the phosphorylation of NIK in transfected HEK 293T cells. HEK 293T cells were cotransfected with plasmid encoding FLAG-tagged NIK alone or with plasmid encoding IKKα. Extracts were subjected to immunoprecipitation with agarose conjugated to antibody against FLAG. Samples were split, and half were subjected to treatment with CIP. (D) IKKα-dependent phosphorylation of NIK is retained in the C-terminal 247 amino acid residues of NIK. An additional thrombin cleavage site at residue 700 of NIK was generated by means of a T702G/D703R mutation of the protein. HEK 293T cells were cotransfected with plasmid encoding NIK, FLAG-tagged at the C terminus, with or without plasmid encoding IKKα. FLAG-NIK was immunopurified, subjected to partial digestion with thrombin, and analyzed by Western blotting with polyclonal rabbit antibody against FLAG. Potential thrombin cleavage products are shown on the right. (E) IKKα phosphorylates the C terminus of NIK. In vitro kinase assay (KA) of the GST-cNIK(700–942) substrate with increasing amounts of immunopurified IKKα from HEK 293T cells. (F) Addition of NIK substantially enhances the IKKα-dependent phosphorylation of the C terminus of NIK. HEK 293T cells were transfected as indicated; lysates were immunopurified with M2-conjugated agarose and subjected to kinase assay (KA). (G) Endogenous IKKα phosphorylates the C terminus of NIK in a signal-dependent manner. IKKα+/+ or IKKα−/− fibroblasts were treated for 3 hours with an agonistic antibody against LTβR (2 μg/ml). Lysates were immunoprecipitated with an antibody against IKKα and subjected to KA.
The primary role of NIK in noncanonical NF-κB signaling is thought to be the phosphorylation of IKKα, which stimulates its kinase activity (9). Activated IKKα was thought to then phosphorylate the C terminus of p100, targeting it for subsequent degradation (10). However, in addition to the phosphorylation of p100, stimulation of the kinase activity of IKKα by NIK might also serve to enhance IKKα-mediated phosphorylation of the C terminus of NIK. Indeed, we reduced the amount of IKKα used in our kinase assays such that its kinase activity on the GST-cNIK substrate was undetectable; however, inclusion of NIK in these reactions resulted in enhanced IKKα-mediated phosphorylation of GST-cNIK (Fig. 3F). Additionally, stimulation of LTβR on fibroblasts resulted in the enhanced phosphorylation of the GST-cNIK substrate by endogenous IKKα (Fig. 3G). Together, these data suggest the existence of a previously uncharacterized kinase loop operating within the noncanonical NF-κB pathway in which receptor stimulation leads to stabilization of NIK, which enables NIK-mediated stimulation of the kinase activity of IKKα, which in turn phosphorylates the C terminus of NIK. IKKα-mediated phosphorylation of NIK may lead to its degradation, similar to IKKα-induced degradation of p100, thus explaining the enhanced stability of NIK in IKKα-deficient cells or aly cells (10). To confirm this hypothesis, we needed to identify the precise residues in the C terminus of NIK that were phosphorylated by IKKα.
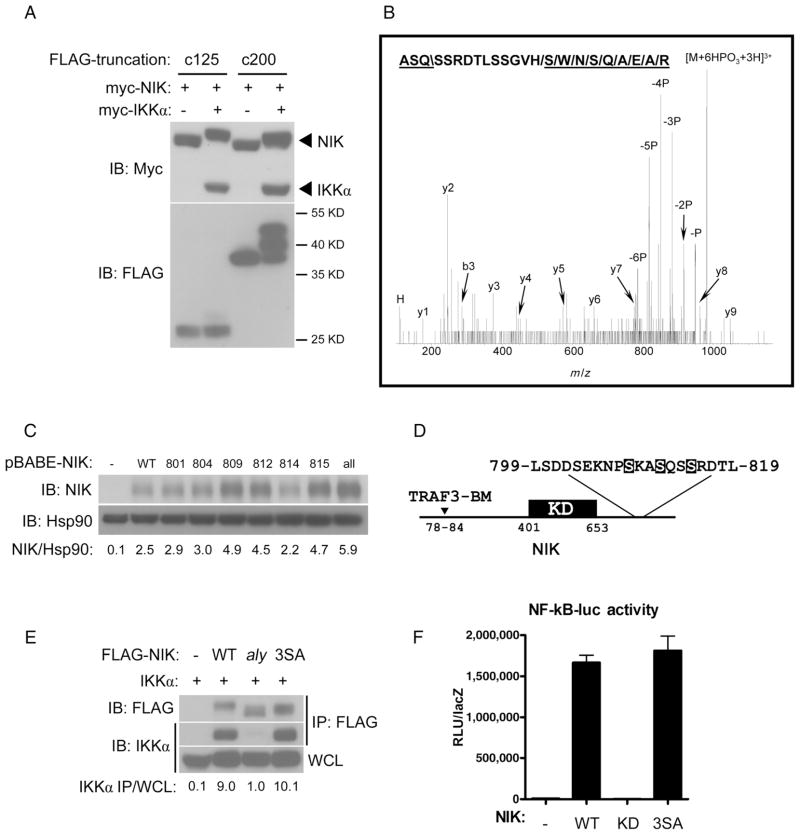
To further define the site in the C terminus of NIK that was phosphorylated by IKKα, we used a truncation strategy and generated two truncations in NIK that spanned the C-terminal 125 (residues 817 to 942) and 200 (residues 742 to 942) amino acid residues. Transfection of HEK 293T cells with plasmids encoding these truncated variants of NIK with plasmid encoding IKKα resulted in a shift in the apparent molecular mass on an SDS-PAGE gel of the 200-residue fragment but not of the 125-residue fragment even though both truncated proteins interacted with IKKα, which suggested that the IKKα target serines were within residues 742 to 817 of NIK (Fig. 4A and fig. S2). We then purified NIK from HEK 293T cells transfected with plasmids encoding NIK and IKKα and subjected it to analysis by mass spectrometry (MS), which identified a multiphosphorylated peptide of NIK that spanned amino acid residues 810 to 827 (Fig. 4B). Although we were unable to obtain sufficient tandem MS (MS/MS) data to assign the phosphorylation sites to particular residues, the analysis suggested that the IKKα-target residues were likely positioned toward the C-terminal end of the 742 to 817 region identified by our truncation analysis.
Fig. 4.
IKKα phosphorylates NIK on Ser809, Ser812, and Ser815. (A) Truncation analysis implicates amino acid residues 742 to 817 of NIK as encompassing the region that contains IKKα target residues. HEK 293T cells were cotransfected with plasmids encoding FLAG-tagged truncations of NIK spanning the C-terminal 200 residues (c200, residues 742 to 942) or 125 residues (c125, residues 817 to 942) with or without plasmid encoding IKKα. In all cases, NIK was included to stimulate the kinase activity of IKKα. (B) MS/MS spectrum of 981.1 (3+). Matching band y-ions, and one immonium ion have been marked. “P” indicates peaks resulting from neutral loss of phosphoric acid. The backbone fragmentation necessary to generate the b- (“\”) and y-ions (“/”) have been marked in the peptide sequence spanning residues 810 to 827 of NIK. (C) Ser→Ala mutations at residues Ser809, Ser812, or Ser815 lead to enhanced stability of NIK. NIK−/− fibroblasts were retrovirally reconstituted with pBABE-empty (−), Ser→Ala point mutations of NIK at given residues (8××), or combined mutation of all six serines (all). (D) Diagram of NIK indicating its TRAF3-binding motif (TRAF3-BM), kinase domain (KD), and expansion of the area harboring IKKα target serine residues (highlighted). (E) Combined Ser→Ala mutation of NIK at residues Ser809, Ser812, and Ser815 generates a mutant protein, henceforth referred to as NIK-3SA, that interacts with IKKα. HEK 293T cells were transfected with plasmid encoding IKKα in combination with either empty vector (−), FLAG-tagged wild-type NIK (NIK-WT), NIK-aly, or NIK-3SA. Lysates subjected to immunoprecipitation with antibody against FLAG were analyzed by Western blotting for IKKα. (F) The NIK-3SA mutant retains kinase activity as assessed by transfecting HEK 293T cells with a 2xκB-luciferase reporter, CMV-βgal, and either pCDNA3-emtpy (−), NIK-WT (WT), the NIK KK430/431AA kinase-defective mutant (KD), or NIK-3SA (3SA). Mean raw light units (RLU) normalized to lacZ activity are shown for triplicate samples. Abbreviations for the amino acid residues are as follows: A, Ala; D, Asp; E, Glu; G, Gly; H, His; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; and W, Trp.
Because IKKα is a serine and threonine kinase, we generated individual Ser→Ala mutations for each of the six serine residues between residues 800 to 817 of NIK and reconstituted NIK-deficient fibroblasts individually with these mutant forms of NIK. Although other residues may be phosphorylated by IKKα in this region, Ser→Ala mutations at residues 809, 812, and 815 resulted in mutant NIK proteins that had substantially enhanced basal stability compared to that of wild-type NIK (Fig. 4C). Additionally, IKKα displayed in vitro kinase activity on Ser809, Ser812, and Ser815 (fig. S3). These three residues of NIK together conform to a tandem IKKα consensus sequence, defined by SxxS/T, where x is any amino acid residue, and are positioned C-terminal to the kinase domain of NIK and far from its N-terminal TRAF3-binding motif (Fig. 4D) (26, 27). That Ser→Ala mutants of NIK at residues 809, 812, and 815 displayed enhanced stability would be expected if phosphorylation of these residues by IKKα promoted the degradation of NIK, which would explain the increased stability of NIK in IKKα-deficient cells and aly cells. Alternatively, Ser→Ala substitutions at these residues might disrupt the interaction between NIK and IKKα, recapitulating the aly phenotype, or affect the kinase activity of NIK. However, the affinity of IKKα for NIK and the kinase activity of NIK were preserved for the NIK mutant proteins that harbored the combined S809A/S812A/S815A mutation, henceforth referred to as NIK-3SA (Fig. 4, E and F). This suggested that residues Ser809, Ser812, and Ser815 were required for IKKα-mediated negative feedback regulation of NIK stability. Finally, we wished to reconfirm our findings that the binding of the TRAF-cIAP complex and the degradation of NIK were not affected by IKKα-mediated feedback phosphorylation of NIK. Consistent with this, we found that TRAF3 coimmunoprecipitated comparably with wild-type NIK and NIK-3SA in the presence of IKKα (fig. S4A). Furthermore, the degradation of NIK that was observed in HEK 293T cells cotransfected with plasmids encoding all of the components of the TRAF-cIAP complex and IKKα was not affected by the presence of NIK-3SA (fig. S4B).
Negative feedback controls the stability of NIK and regulates noncanonical NF-κB signaling
To determine the functional importance of IKKα-dependent phosphorylation of NIK, we reconstituted NIK-deficient fibroblasts with wild-type NIK, the NIK-3SA mutant, or, for comparison, the aly mutant of NIK. As expected, the reconstituted aly mutant of NIK was substantially more stable than wild-type NIK because it cannot interact with IKKα, recapitulating the results observed in NIKaly/aly cells (Fig. 5A). However, the NIK-3SA mutant, which interacted as well as wild-type NIK with IKKα, was as stable as NIK-aly, suggesting that the NIK residues Ser809, Ser812, and Ser815 are target residues for IKKα that are critical for the enhanced stability of NIK observed in aly fibroblasts (Fig. 5A). Treatment of reconstituted cells with the proteasome inhibitor MG132 resulted in equalization of the abundance of all NIK species, suggesting that differences in the abundance of NIK were not due to differential expression and that the differential stabilities of wild-type NIK and NIK-3SA were proteasome-dependent (Fig. 5B). Wild-type NIK, NIK-aly, and NIK-3SA were also equivalently stable in IKKα-deficient cells, confirming that the differential stability of these NIK proteins was dependent on IKKα-mediated negative feedback (Fig. 5C).
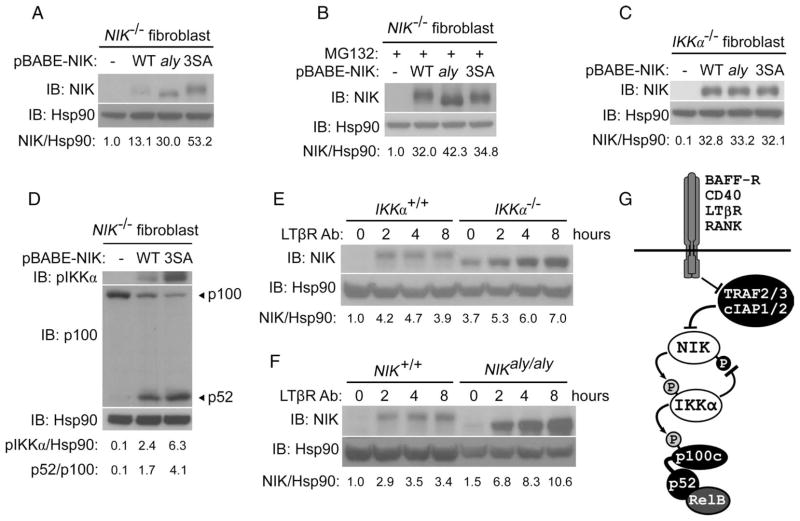
Fig. 5.
Negative feedback controls the stability of NIK and regulates noncanonical NF-κB signaling. (A) Mutation of Ser809, Ser812, and Ser815 to alanine leads to substantially enhanced NIK stability, comparable to that of the NIK aly mutant. Western blotting of untreated NIK−/− fibroblasts retrovirally reconstituted with pBABE empty (−), NIK-WT, NIK-aly, or the NIK-3SA mutant. (B) Differential stability of NIK mutants seen in (A) was not due to differences in gene expression because the abundance of NIK was equalized upon treatment of cells for 4 hours with the proteasome inhibitor MG132. (C) Differential stability of NIK mutants seen in (A) was dependent on IKKα feedback as evidenced by their equivalent abundance when retrovirally reconstituted into IKKα−/− fibroblasts. (D) Elimination of IKKα-dependent feedback results in enhanced noncanonical NF-κB signaling. NIK−/− fibroblasts were retrovirally reconstituted with pBABE-empty (−), NIK-WT, or NIK-3SA (3SA). Extracts were analyzed by Western blotting with an antibody against pIKKα and with an antibody against the N terminus of p100 that recognizes both p100 and its processed form, p52. (E and F) Elimination of IKKα-dependent feedback results in unabated accumulation of NIK during long-term stimulation of receptor. Western blotting analysis of IKKα+/+ and IKKα−/− fibroblasts (E) or NIK+/+ and NIKaly/aly fibroblasts (F) treated for the indicated times with an agonistic antibody against LTβR (2 μg/ml). (G) A refined model of noncanonical NF-κB signaling that includes the role of IKKα-dependent negative feedback on the stability of NIK. As shown, NIK stability is inhibited by the previously studied TRAF-cIAP complex under unstimulated conditions, whereas IKKα-dependent phosphorylation, described here, controls the abundance of NIK after receptor stimulation.
The enhanced stability of NIK-3SA compared to that of wild-type NIK in retrovirally reconstituted NIK-deficient cells resulted in enhanced noncanonical NF-κB signaling, as evidenced by the detection of substantially greater amounts of phosphorylated IKKα (pIKKα) and processing of p100 to p52. This demonstrates the importance of IKKα-dependent phosphorylation of NIK to the inhibition of noncanonical NF-κB signaling (Fig. 5D). These data establish a previously uncharacterized role for IKKα in modulating the stability of NIK and show the existence of a negative feedback loop that prevents the excessive accumulation of NIK after receptor stimulation. This is best illustrated by comparing the kinetics of NIK accumulation in wild-type and IKKα-deficient or aly fibroblasts. Whereas the accumulation of NIK in wild-type cells was attenuated after receptor ligation, resulting in little or changes to the amount of NIK over time, activation of noncanonical signaling in IKKα-deficient and aly cells led to continuous accumulation of NIK far beyond that observed in wild-type cells (Fig. 5, E and F).
DISCUSSION
Although the posttranslational stability of NIK is the central regulatory element of the noncanonical NF-κB pathway, the molecular mechanism by which the degradation of NIK is controlled in different contexts is only just beginning to be understood (8). Constitutive degradation of NIK in unstimulated cells is mediated by the TRAF-cIAP complex; however, little is known about control of NIK after receptor stimulation (12, 13). In this report, we describe a previously uncharacterized negative feedback mechanism within noncanonical signaling that controls the abundance of NIK after receptor ligation. Negative feedback occurred through IKKα-mediated phosphorylation of serine residues in the C terminus of NIK that promoted its destabilization. Consequently, receptor-induced stabilization of NIK and the resultant activation of the kinase activity of IKKα ultimately fed back to prevent the uncontrolled accumulation of NIK and enhanced noncanonical NF-κB signaling after receptor stimulation. IKKα has traditionally been viewed as a downstream target of the kinase activity of NIK within noncanonical NF-κB signaling, a component of the pathway that is activated by NIK and that transmits that signal forward through the phosphorylation of p100 (9, 10). However, our study demonstrates that IKKα plays a critical role in the regulation of noncanonical NF-κB signaling by additionally acting as an upstream kinase of NIK and in so doing, controlling the stability of NIK (Fig. 5G).
How the stability of NIK is controlled and the effect of its dysregulation in vivo have been of substantial interest. To date, all such studies have focused on the roles of TRAF2 and TRAF3 in controlling the degradation of NIK. For example, in the past several years various groups have generated mice that lack TRAF2 or TRAF3 in combination with p100 or NIK, that have conditional deletion of TRAF2 and TRAF3 in various cell types, and that transgenically express NIK mutants that cannot interact with TRAF3 (13, 28–31). We addressed the possibility that IKKα-mediated phosphorylation of NIK might somehow be required for activity of the TRAF-cIAP complex; however, our studies suggested that this was not the case because the abundance of NIK in IKKα−/− cells and aly cells continued to be responsive to stimuli that modulated the activity of the TRAF-cIAP complex. Furthermore, the affinity of NIK for TRAF3, the only component of the TRAF-cIAP complex thought to interact with NIK, did not change in the presence or absence of IKKα and was not affected by inclusion of the 3SA mutation in NIK. These data are in accordance with findings that the serine residues in the C terminus of NIK that are phosphorylated by IKKα are far from the N-terminal TRAF3-binding motif of NIK. Finally, the degradation of NIK in cells in which all of the components of the TRAF-cIAP complex were ectopically coexpressed was not affected by the presence of the 3SA mutation. Therefore, it appears that the abundance of NIK is controlled by two separate mechanisms: the previously studied TRAF-cIAP complex, which is responsible for the degradation of NIK in unstimulated contexts, and the previously uncharacterized IKKα-dependent mechanism that we describe here, which is responsible for the inhibition of NIK after receptor stimulation.
That the differential stabilities of reconstituted wild-type NIK and NIK-3SA were eliminated upon inhibition of the proteasome suggests that proteasomal degradation may be the ultimate mechanism by which feedback phosphorylation of NIK results in its degradation (Fig. 5B). Proteins degraded by the proteasome are frequently targeted for destruction by ubiquitination (32). However, we were unable to detect any enhancement in the extent of ubiquitination of NIK in HEK 293T cells cotransfected with plasmids encoding IKKα and NIK (fig. S5). Although ubiquitination-independent degradation by the proteasome has been described for a number of proteins, it is also possible that the necessary endogenous cofactors required for mediating phosphorylation-dependent ubiquitination of NIK are not found in sufficient abundance in HEK 293T cells to exert detectable ubiquitination of NIK (33). If ubiquitination is ultimately found to be the molecular mechanism by which phosphorylated NIK is degraded, it will be of interest to determine whether the lysine residues in NIK that are targeted by ubiquitin are different from those targeted by the TRAF-cIAP complex.
In the case of p100, whose ubiquitination and proteasomal processing is stimulated by IKKα-mediated phosphorylation, ubiquitination is mediated by recruitment of the SCFβ-TrCP ubiquitin ligase complex (10). We investigated whether a similar mechanism might be operational for NIK, but we found no enhancement in the recruitment of β-TrCP to NIK in comparison to that to NIK-3SA in the presence or absence of IKKα. Furthermore, the phosphorylation site that we identified bears no homology to the well-described β-TrCP consensus phosphodegron (34). Further studies will be required to elaborate the molecular constituents that stimulate the degradation of NIK in response to IKKα-dependent phosphorylation of the C-terminal serine residues that we identified.
In fibroblasts derived from IKKα-deficient mice or aly mice, the basal stability of NIK in the absence of stimulation was also increased, to an extent comparable to that seen in fibroblasts deficient in components of the TRAF-cIAP complex. Therefore, it is possible that, for some cell types, both the TRAF-cIAP complex and the IKKα-dependent phosphorylation system are nonredundantly required to completely eliminate all of the NIK protein that is synthesized during the unstimulated state. This is consistent with the observation that IKKα-mediated phosphorylation of NIK in fibroblasts appeared to occur even in the absence of receptor ligation, as evidenced by the difference in the apparent molecular mass of NIK species from IKKα+/+ and IKKα−/− fibroblasts even under conditions in which just the proteasome was inhibited (Fig. 3A) and by the observation of the phosphorylation of NIK by endogenous IKKα in unstimulated fibroblasts (Fig. 3G).
In addition to its role in noncanonical NF-κB signaling, NIK can enhance canonical NF-κB signaling in certain cellular contexts through the direct activation of the IKK complex (35–37). For example, NIK stabilized in fibroblasts because of a deficiency in TRAF3 or because of pretreatment with agonists of the LTβR results in the substantial enhancement of TNFα-induced activation of the IKK complex (35). Activation of canonical NF-κB signaling by NIK is dependent on functional IKKα (35–37). Consequently, it may be that in the absence of IKKα-dependent destabilization of NIK, canonical NF-κB signaling may also be enhanced.
Enhanced noncanonical signaling is now recognized to cause various hematological cancers and autoimmune pathology. Thus far, this is thought to occur through deletion of components of the TRAF-cIAP complex, genetic amplifications of NIK, or long-term ligand stimulation of cells by increased concentrations of BAFF in the plasma (14–16, 19–23). Notably, defective negative feedback in canonical NF-κB signaling leads to malignancies (24). As such, determining the in vivo sequelae of defective negative feedback of noncanonical NF-κB signaling in various cell types will be of important interest for future studies.
MATERIALS AND METHODS
Antibodies and reagents
Antibodies against the hemagglutinin (HA) tag, heat shock protein of 90 kD (Hsp90), TRAF2, TRAF3, and cIAP2 were from Santa Cruz Biotechnology; antibodies against NIK, IKKα, pIKKα, and p100 were from Cell Signaling Technology; antibody against the FLAG tag (M2) and agarose conjugated to M2 were from Sigma; rabbit antibody against FLAG was from Covance; and antibody against LTβR was from Alexis Biochemicals. Recombinant human BAFF (hBAFF) and murine RANK (mRANK) proteins were obtained from R&D Systems. Calf intestinal alkaline phosphatase was obtained from New England Biotechnology. Thrombin was obtained from Amersham. MG132 was obtained from American Peptide, and LBW242 was a generous gift from Novartis Pharmaceuticals. Bone marrow macrophages were isolated as described previously from femurs and tibia, whereas B cells were isolated from spleens as described previously (38, 39).
Cell culture and transfections
HEK 293T cells and fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech) supplemented with 5% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Bone marrow–derived macrophages were cultured as above except that the medium contained 10% FCS and 2% of the supernatant of the macrophage colony–stimulating factor (M-CSF)-producing L949 cell line. CH12.LX B cells were cultured in RPMI (Mediatech) supplemented with 10% FCS, penicillin, streptomycin, and 50 μM β-mercaptoethanol. All retroviral transduction experiments were performed with the ψ-A Maloney murine leukemia virus in conjunction with pBABE-puro alone or a pBABE-puro–derived construct, as described previously (12). Transient transfections of HEK 293T cells were performed with the Fugene 6 reagent (Roche).
Immunoprecipitations
Cells were lysed in modified radioimmunoprecipitation assay (RIPA) buffer containing 0.5% (v/v) NP-40 and 0.1% (w/v) sodium deoxycholate supplemented with protease inhibitor tablets (Roche). Extracts were then incubated for 2 hours with the indicated antibodies at 4°C and for 1 hour with Protein G beads (Amersham), or for 2 hours with beads conjugated to M2 antibodies, or in the case of immunoprecipitation of endogenous proteins with TrueBlot antibody against rabbit immunoglobulin G (IgG) (eBiosciences). Subsequently, beads were washed three times with lysis buffer, resolved by SDS-PAGE, and analyzed by Western blotting with the indicated antibodies. In the case of immunoprecipitation of endogenous proteins, TrueBlot horseradish peroxidase–conjugated secondary antibody was used.
Protease, phosphatase, and in vitro kinase reactions
To assay the activities of proteases and phosphatases, cellular extracts were subjected to immunoprecipitation as described earlier followed by treatment with 10 U of thrombin (Amersham) in phosphate-buffered saline (PBS) or with 10 U of calf intestinal phosphatase (NEB) in NEB Buffer 3, respectively. In the case of thrombin digestions, the initial immunoprecipitation was performed with M2 beads and Western blotting was performed with polyclonal rabbit antibody against the FLAG tag. In vitro kinase reactions were performed as described previously (40).
Knockdown by shRNA
Cells were infected with retroviruses generated with pSIREN-puro vectors (Clontech) containing control shRNA or TRAF3-specific shRNA that targeted nucleotides 314 to 333 of mouse TRAF3. Cells were then selected by culturing in medium containing puromycin (3 μg/ml, Sigma).
Purification of NIK and mass spectrometry
HEK 293T cells were cotransfected with plasmids encoding 2xFLAG-NIK and IKKα; 48 hours after transfection, NIK was immunoprecipitated as described above with M2-conjugated beads and proteins were eluted with protein solubilization buffer. Removal of SDS and tryptic digestion in 30% dimethyl sulfoxide (DMSO) were performed as described previously (41). The tryptic peptides were fractionated manually by electrostatic repulsion–hydrophilic interaction chromatography (ERLIC) (42). In short, the peptides were loaded onto microcolumns packed with PolyWAX LP (Nest Group, Inc) and eluted by a step gradient starting at a high percentage of acetonitrile and ending at a high concentration of ammonium formate in 20% acetonitrile, 5% formic acid. Highly phosphorylated peptides were eluted late in the gradient at high salt concentrations. The fractions that eluted with 250 mM ammonium formate, 5% formic acid, and 20% acetonitrile were dried, resuspended in 5% formic acid, loaded onto a microcolumn packed with 20 μm R3 Poros material (Applied Biosystems), and eluted directly into a glass emitter (Proxeon Biosystems) (43). The peptide mixture was analyzed with a QqTOF mass spectrometer (Applied Biosystems/Sciex QSTAR Pulsar XL) and the peptide sequence was assigned manually.
Image quantitation and experimental design
Quantitation of all images was performed with ImageJ software and relative arbitrary units are shown. Briefly, the intensity of the band of interest was normalized to its corresponding control band. For each panel, all values were divided by an arbitrary number to obtain relative arbitrary units. All experiments were performed at least three times and the data shown are representative.
Supplementary Material
www.sciencesignaling.org/cgi/content/full/3/123/ra41/DC1
Fig. S1. Ectopic expression of IKKα results in destabilization of ectopically expressed NIK.
Fig. S2. Truncated mutants of NIK continue to interact with IKKα.
Fig. S3. IKKα phosphorylates Ser809, Ser812, and Ser815 of NIK.
Fig. S4. NIK-3SA does not affect the interaction between wild-type NIK and TRAF3 or its degradation by the TRAF-cIAP complex.
Fig. S5. Ubiquitination of NIK is not enhanced by the presence of IKKα in HEK 293T cells.
Acknowledgments
We thank L. Zawel and Novartis Pharmaceuticals for providing LBW242; Amgen for providing Nik−/− mice; W.-C. Yeh and T. W. Mak for providing TRAF2−/− mouse embryo fibroblasts (MEFs), and M. Karin for IKKα−/− MEFs.
Funding: Part of this work was supported by NIH research grants T32 GM008042 (to the UCLA Medical Scientist Training Program supporting B.R.), F30 HL095309 (to B.R.), AI33068 and CA69381 (to C.F.W.), and R01 AI069120 and R01 GM078607 (to G.C.).
Footnotes
Author contributions: B.R., B.Z., A.J.Y., T.S., and P.W.D performed all experiments; B.R., A.J.Y., C.F.W., J.A.L., and G.C. designed experiments and analyzed data; B.R. and G.C. wrote the paper.
Competing interests: None to report.
REFERENCES AND NOTES
- 1.Baeuerle PA, Baltimore D. NF-κB: Ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Matsushima A, Kaisho T, Rennert PD, Nakano H, Kurosawa K, Uchida D, Takeda K, Akira S, Matsumoto M. Essential role of nuclear factor (NF)-κB–inducing kinase and inhibitor of κB (IκB) kinase α in NF-κB activation through lymphotoxin β receptor, but not through tumor necrosis factor receptor I. J Exp Med. 2001;193:631–636. doi: 10.1084/jem.193.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 5.Novack DV, Yin L, Hagen-Stapleton A, Schreiber RD, Goeddel DV, Ross FP, Teitelbaum SL. The IκB function of NF-κB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 7.Scheidereit C. IκB kinase complexes: Gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 8.Qing G, Qu Z, Xiao G. Stabilization of basally translated NF-κB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-κB2 p100. J Biol Chem. 2005;280:40578–40582. doi: 10.1074/jbc.M508776200. [DOI] [PubMed] [Google Scholar]
- 9.Senftleben U, Cao Y, Xiao G, Greten FR, Krähn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 10.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J Biol Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 11.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 12.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh W, Mak TW, Korneluk RG, Cheng G. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DAA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD, Jr, Kuehl WM, Staudt LM. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkinson PGP, Coope HJ, Rowe M, Ley SC. Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-κB2 p100 to p52. J Biol Chem. 2003;278:51134–51142. doi: 10.1074/jbc.M304771200. [DOI] [PubMed] [Google Scholar]
- 18.Xiao G, Cvijic ME, Fong A, Harhaj EW, Uhlik MT, Waterfield M, Sun SC. Retroviral oncoprotein Tax induces processing of NF-κB2/p100 in T cells: Evidence for the involvement of IKKα. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Roschke V, Baker KP, Wang Z, Alarcón GS, Fessler BJ, Bastian H, Kimberly RP, Zhou T. Cutting edge: A role for B lymphocyte stimulator in systemic lupus erythematosus. J Immunol. 2001;166:6–10. doi: 10.4049/jimmunol.166.1.6. [DOI] [PubMed] [Google Scholar]
- 20.Cheema GS, Roschke V, Hilbert DM, Stohl W. Elevated serum B lymphocyte stimulator levels in patients with systemic immune-based rheumatic diseases. Arthritis Rheum. 2001;44:1313–1319. doi: 10.1002/1529-0131(200106)44:6<1313::AID-ART223>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Groom J, Kalled SL, Cutler AH, Olson C, Woodcock SA, Schneider P, Tschopp J, Cachero TG, Batten M, Wheway J, Mauri D, Cavill D, Gordon TP, Mackay CR, Mackay F. Association of BAFF/BLyS overexpression and altered B cell differentiation with Sjögrens syndrome. J Clin Invest. 2002;109:59–68. doi: 10.1172/JCI14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aya K, Alhawagri M, Hagen-Stapleton A, Kitaura H, Kanagawa O, Novack DV. NF-κB–inducing kinase controls lymphocyte and osteoclast activities in inflammatory arthritis. J Clin Invest. 2005;115:1848–1854. doi: 10.1172/JCI23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enzler T, Bonizzi G, Silverman GJ, Otero DC, Widhopf GF, Anzelon-Mills A, Rickert RC, Karin M. Alternative and classical NF-κB signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–415. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S, Mori S, Ishikawa Y, Okamoto K, Tobinai K, Nakagama H, Nakahata T, Yoshino T, Kobayashi Y, Ogawa S. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 25.Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y. IKKα shields 14-3-3σ, a G2/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol Cell. 2007;27:214–227. doi: 10.1016/j.molcel.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 26.Chariot A. The NF-κB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 28.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G. Rescue of TRAF3-null mice by p100 NF-κB deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27:253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki Y, Calado DP, Derudder E, Zhang B, Shimizu Y, Mackay F, Nishikawa S, Rajewsky K, Schmidt-Supprian M. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc Natl Acad Sci USA. 2008;105:10883–10888. doi: 10.1073/pnas.0805186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 33.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Fuchs SY, Spiegelman VS, Kumar KGS. The many faces of β-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 35.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-κB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci USA. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.OMahony A, Lin X, Geleziunas R, Greene WC. Activation of the heterodimeric ’ IκB kinase α (IKKα)-IKKβ complex is directional: IKKα regulates IKKβ under both basal and stimulated conditions. Mol Cell Biol. 2000;20:1170–1178. doi: 10.1128/mcb.20.4.1170-1178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-κB activation pathways by NF-κB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- 39.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G. Rescue of TRAF3-null mice by p100 NF-κB deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 41.Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ytterberg J, Ogorzalek-Loo R, Boontheug P, Loo J. 55th ASMS Conference on Mass Spectrometry and Allied Topics; Indianapolis, IN. June 2007. [Google Scholar]
- 43.Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencesignaling.org/cgi/content/full/3/123/ra41/DC1
Fig. S1. Ectopic expression of IKKα results in destabilization of ectopically expressed NIK.
Fig. S2. Truncated mutants of NIK continue to interact with IKKα.
Fig. S3. IKKα phosphorylates Ser809, Ser812, and Ser815 of NIK.
Fig. S4. NIK-3SA does not affect the interaction between wild-type NIK and TRAF3 or its degradation by the TRAF-cIAP complex.
Fig. S5. Ubiquitination of NIK is not enhanced by the presence of IKKα in HEK 293T cells.