Abstract
Calcineurin, the conserved Ca2+/calmodulin-regulated protein phosphatase, mediates diverse aspects of Ca2+-dependent signaling. We show that substrates bind calcineurin with varying strengths and examine the impact of this affinity on signaling. We altered the calcineurin-docking site, or PxIxIT motif, in Crz1, the calcineurin-regulated transcription factor in S. cerevisiae, to decrease (Crz1PVIAVN) or increase (Crz1PVIVIT) its affinity for calcineurin. As a result, the Ca2+-dependent dephosphorylation and activation of Crz1PVIAVN are decreased, whereas Crz1PVIVIT is constitutively dephosphorylated and hyperactive. Surprisingly, the physiological consequences of altering calcineurin-Crz1 affinity depend on the growth conditions. Crz1PVIVIT improves yeast growth under several environmental stress conditions but causes a growth defect during alkaline stress, most likely by titrating calcineurin away from other substrates or regulators. Thus, calcineurin-substrate affinity determines the Ca2+ concentration dependence and output of signaling in vivo as well as the balance between different branches of calcineurin signaling in an overall biological response.
INTRODUCTION
The components of intracellular signaling cascades are often organized into stable complexes to ensure signaling efficiency and specificity. However, regulatory enzymes typically modulate many different aspects of cell physiology, and in doing so must form transient, reversible associations with multiple target proteins. To identify critical aspects of such enzyme-substrate interactions during signaling, we studied calcineurin, a major effector of Ca2+-activated cell signaling, which regulates a diverse array of cellular proteins.
Calcineurin, or PP2B, is a conserved serine-threonine protein phosphatase and the target of the immuno-suppressant drugs cyclosporin A and FK506 (Friedman and Weissman, 1991; Liu et al., 1991). Calcineurin is composed of a catalytic A subunit (CN A) and a regulatory B subunit (CN B). Interaction of Ca2+-bound calmodulin with CN A and Ca2+ binding to CN B are required for enzyme activation (Klee et al., 1998). In mammalian cells, a critical calcineurin substrate is the nuclear factor of activated T cells (NFAT), a conserved family of transcription factors that play important roles in the function and development of the immune, cardiovascular, and nervous systems (Feske et al., 2003; Hogan et al., 2003). Dephosphorylation of NFAT by calcineurin causes its accumulation in the nucleus and subsequent activation of gene expression (Hogan et al. [2003] and references therein). Besides NFAT, calcineurin dephosphorylates a number of other proteins to regulate cellular processes including synaptic function, ion homeostasis, apoptosis, cell motility, and endocytosis (Aramburu et al., 2000; Czirjak and Enyedi, 2006; Wang et al., 2005). It is not clear how these distinct calcineurin-dependent events are coordinated in vivo. In some cell types, interaction with endogenous inhibitors and scaffolding proteins may spatially restrict or target calcineurin activity (Hilioti and Cunningham, 2003; Dell’-Acqua et al., 2002; Sun et al., 1998). Overall, however, little is known about mechanisms that govern calcineurin substrate selection or signaling specificity in vivo.
A key feature of NFAT regulation is the targeting of calcineurin to a short conserved sequence in NFAT family members termed the PxIxIT motif (Aramburu et al., 1998). A PxIxIT docking site also mediates calcineurin interaction with the TRESK K+ channel substrate (Czirjak and Enyedi, 2006). A high-affinity calcineurin-binding peptide, PVIVIT peptide, derived by selection from peptide libraries comprising PxIxIT motif variations, inhibits NFAT activation in vivo and in vitro by interfering with calcineurin-NFAT docking (Aramburu et al., 1999). Furthermore, mutations in the NFAT recognition sequence that decrease binding to calcineurin impair signaling in vivo, and replacement of the native sequence by the high-affinity PVIVIT sequence causes a partial constitutive activation of NFAT in unstimulated cells (Aramburu et al., 1998, 1999). Thus, the strength of the calcineurin-NFAT interaction is tuned to the signaling requirements of the cell, and by extension, calcineurin-substrate interaction strength may in general determine the nature of calcineurin-dependent responses in vivo.
Here, we specifically address the contribution of calcineurin-substrate recognition to global aspects of calcineurin signaling, using the yeast Saccharomyces cerevisiae. These studies are tractable in yeast because of the relative simplicity of signaling in this single-celled organism and because we have previously described several calcineurin substrates and have identified their sites of interaction with the enzyme (Stathopoulos-Gerontides et al., 1999; Boustany and Cyert, 2002; Heath et al., 2004; Bultynck et al., 2006; Tabuchi et al., 2006).
Yeast calcineurin is dispensable for growth under standard laboratory conditions. In fact, under these conditions the low [Ca2+] in the yeast cytoplasm supports little calcineurin activity. However, when cells are exposed to environmental stress, i.e., high salinity, alkaline stress, cell wall damage, ER stress, or heat, intracellular Ca2+ rises and calcineurin becomes active and promotes cell survival (Cyert [2003] and references therein; Garcia et al., 2004). A key downstream target of yeast calcineurin is the transcription factor Crz1. As for NFAT, Crz1 localizes to the nucleus upon dephosphorylation where it activates transcription of a variety of genes required for adaptation to stress (Stathopoulos-Gerontides et al., 1999; Yoshimoto et al., 2002). A conserved calcineurin-dependent response element (CDRE) in the promoters of these genes mediates Crz1 binding and transactivation (Stathopoulos and Cyert, 1997). In addition to Crz1, calcineurin dephosphorylates several other yeast proteins, and studies of these proteins indicate a broader role for calcineurin in mediating cellular stress responses. The calcineurin substrate, Hph1, localizes to the endoplasmic reticulum, acts independently of Crz1, and promotes cell growth under conditions of high salt, alkaline pH, and cell wall stress through a pathway which may impact protein trafficking (Heath et al., 2004). Slm1 and Slm2 are also calcineurin substrates and are plasma membrane-localized proteins required for actin cytoskeleton polarization and heat stress-induced endocytosis of nutrient permeases (Audhya et al., 2004; Bultynck et al., 2006; Tabuchi et al., 2006). Crz1, Hph1, Slm1, and Slm2 all contain a PxIxIT-related motif, and in each case this sequence is required for CN A binding and dephosphorylation (Table 1) (Boustany and Cyert, 2002; Bultynck et al., 2006; Heath et al., 2004).
Table 1.
Binding Affinities of PxIxIT Peptides for Yeast Calcineurin
| Substrate | PxIxIT Motif | Ki (µM) |
|---|---|---|
| Crz1 | APVTPIISIQEFNE | 15 |
| Slm1 | QDQVPNIYIQTPIN | 40 |
| Slm2 | QNRVPEFYIENVDS | 20 |
| Hph1 | SSRLPVIAVNDNPV | 250 |
| Consensus peptide | GPHPVIVITGPHEE | 2 |
Here, we explore the impact of calcineurin-substrate targeting on multiple aspects of signaling. We examine four distinct substrates and show that each binds calcineurin with different strength. This interaction is determined in part by the specific sequence of the PxIxIT docking site, which is unique in each substrate. These studies demonstrate that substrate-calcineurin docking is a key determinant of signaling output and underscore the importance of maintaining low to moderate enzyme-substrate affinities in vivo. Finally, we provide insight into one mechanism that governs substrate selection by calcineurin in vivo and allows integration of distinct calcineurin-dependent reactions to determine the physiological response to signaling.
RESULTS
CN A Binds Its Substrates with a Wide Range of Affinities
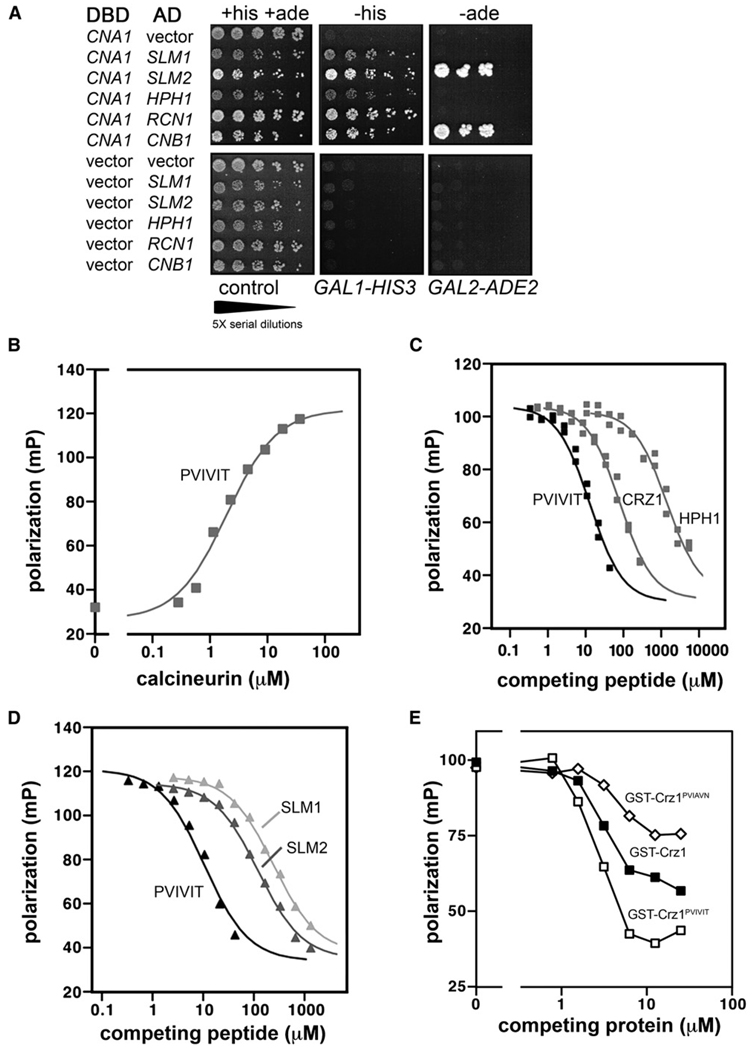
We compared interactions of Cna1, which is one of two homologous CN A subunits in yeast, with its known interacting partners, using the two-hybrid system. The PJ69-4A yeast strain contains two reporter genes: GAL1 promoter-HIS3 and GAL2 promoter-ADE2, which display low and high stringency for activation, respectively (James, 2001). Cna1 physically interacts with its substrates Hph1, Slm1, and Slm2; the Rcn1 regulator; and the B subunit Cnb1, as all of these combinations resulted in growth on medium lacking histidine (Figure 1A). However, only the interactions of Cna1 with Slm2 and Cnb1 are sufficiently strong to activate GAL2 promoter-ADE2 and allow growth on medium lacking adenine. Furthermore, the interaction of Cna1 with the substrate Slm2 promoted considerably stronger growth on media lacking histidine than did its interaction with Slm1 or Hph1 (Figure 1A). These results suggest that Cna1 interacts with its substrates with varying affinities. Crz1 was not tested in these experiments, as expression of the Crz1-Gal4AD (activation domain) fusion is toxic to yeast cells.
Figure 1. Yeast Calcineurin Substrates Have a Wide Range of Affinities for CN A.
(A) Two-hybrid assays assessing the interactions between yeast substrates and Cna1. Serial dilutions of yeast strains were spotted on medium lacking tryptophan and leucine to select for both Gal4-fusion plasmidsand on medium lacking either histidine or adenine as indicated to assay for reporter activation. DBD, Gal4-DNA binding domain fusions constructed in the plasmid pGBT9 containing the indicated ORF. AD, Gal4-activation domain fusions constructed in the plasmid pACT2 containing the indicated ORF. All plasmids were transformed into strain PJ69-4A. Cell growth was determined after incubation at 30°C for 5 days.
(B) Binding of PVIVIT peptide to purified yeast calcineurin measured by fluorescence polarization. See Experimental Procedures for details.
(C and D) Competition of PVIVIT-CN binding with nonfluorescent peptides representing the PxIxIT motifs from Crz1, Hph1, Slm2, and Slm1.
(E) Competition of PVIVIT-CN binding by purified recombinant GST-Crz1-PxIxIT proteins.
A PxIxIT motif mediates the association of Crz1, Slm1, Slm2, and Hph1 with calcineurin (Table 1). Therefore, we examined binding of yeast calcineurin to each of these motifs and to PVIVIT, the consensus PxIxIT peptide previously identified by in vitro selection for increased binding to calcineurin (Aramburu et al., 1999). Fluorescently labeled PVIVIT peptide was added to increasing concentrations of purified, active, yeast calcineurin holoenzyme, and their interaction was measured using fluorescence polarization (Li et al., 2004 and Experimental Procedures). PVIVIT bound to yeast and human calcineurin with similar affinities (Kd of ~2 µM and 0.5 µM, respectively) (Figure 1B and Li et al., 2004).
Next, peptides representing the various yeast PxIxIT sites were used to compete with yeast calcineurin-PVIVIT binding (Figures 1C and 1D, Table 1). The peptides vary in sequence; however, each contains a core motif with a proline in the first position, hydrophobic residues in the third and fifth positions, and a hydrophilic residue in the sixth position (Table 1). The PVIVIT peptide had the highest binding affinity for yeast calcineurin. Peptides encoding the calcineurin-docking sites from Crz1, Slm2, and Slm1 were of decreasing affinity (Ki of 15 µM, 20 µM, and 40 µM, respectively), whereas a peptide representing the substrate Hph1 was of considerably lower affinity (Ki of 250 µM). These results are in general agreement with the yeast two-hybrid experiments (Figure 1A) and suggest that targeting of substrates to yeast Cna1, at least via the PxIxIT motif, occurs with a wide range of affinities.
Changing the Calcineurin-Crz1 Interaction via PxIxIT Motif Mutations
As a tool to investigate the biology of calcineurin-substrate interactions, we substituted altered PxIxIT motifs into Crz1, a calcineurin substrate whose biological activity can be reliably assessed in vivo. We replaced the native Crz1 motif, PIISIQ, either with the high-affinity core sequence PVIVIT to create Crz1PVIVIT or with the low-affinity Hph1 sequence PVIAVN to create Crz1PVIAVN. Purified recombinant GST-Crz1 proteins (GST-Crz1, GST-Crz1PVIVIT, and GST-Crz1PVIAVN) were tested in the fluorescence polarization assay for their ability to compete with PVIVIT peptide for binding to calcineurin. All three proteins competed in the binding assay, with GST-Crz1PVIVIT most effective and GST-Crz1PVIAVN least effective (Figure 1E). This activity was due to CRZ1-encoded residues, as control experiments with GST revealed no appreciable interaction with fluorescent PVIVIT peptide in the concentration range used and no competition with calcineurin-PVIVIT binding (data not shown).
Competition by GST-Crz1 and GST-Crz1PVIAVN proteins differed from competition by the synthetic peptides in not achieving complete displacement of fluorescent PVIVIT. We have not investigated whether this difference at high protein concentrations reflects a technical limitation of the assay or an intrinsic property of the GST-Crz1 proteins. In either case, there are detectable differences in interaction at the PxIxIT-binding site at physiological concentrations. This fact encouraged us to proceed with biological studies.
Consequences of CN Binding-Affinity Changes on Crz1 Activity In Vivo
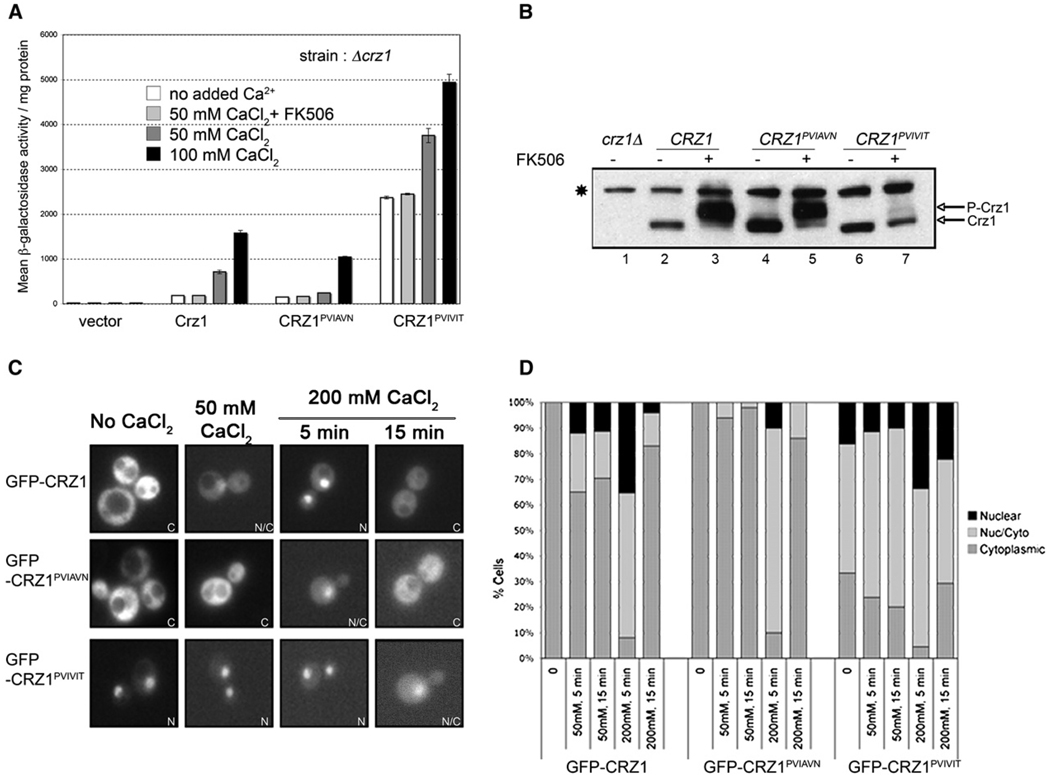
To determine the consequences of mutating the PxIxIT motif on Crz1 activity, Crz1, Crz1PVIVIT, and Crz1PVIAVN were expressed from low-copy number (centromeric) plasmids, in a strain lacking genomic Crz1 (crz1 Δ) that contains a reporter gene comprised of four tandem repeats of the Crz1 DNA binding site (4×-CDRE-lacZ) (Stathopoulos and Cyert, 1997). We assessed Crz1-dependent transcription in these cells by measuring β-galactosidase activity before and after exposure to CaCl2 (Figure 2A). In these and subsequent experiments, extracellular addition of 50–200 mM CaCl2 was used to activate calcineurin signaling. This addition results in a rapid, transient rise in intracellular Ca2+ followed by a sustained plateau and has only a modest effect on yeast cell growth (Miseta et al., 1999). A range of CaCl2 concentrations was used to achieve different levels of calcineurin activation. Similarly, in vivo, exposure of yeast to different environmental stress conditions results in Ca2+ signals of differing amplitude (Viladevall et al., 2004; Denis and Cyert, 2002).
Figure 2. PxIxIT Motif Mutations in Crz1 Alter Transcriptional Activity, In Vivo Phosphorylation States, and Nuclear Localization.
(A) β-galactosidase activity of crzlΔ 4×-CDRE-lacZ strains expressing different Crz1 variants, as noted. All plasmids were constructed in the vector pRS315. CaCl2 was added to the cell culture at the concentrations indicated, 1 hr before harvesting. FK506 was added 1 hr prior to CaCl2, where indicated. “No added Ca2+” refers to cells grown in standard media, which contains 700 µM CaCl2. Error bars indicate standard deviation (see Experimental Procedures for details).
(B) Immunoblot analysis of cell extracts from crz1Δ cells expressing vector or the indicated HA-tagged CRZ1 constructs, using anti-HA monoclonal antibody. Cells were grown either in the presence of FK506 (lanes 3,5, and 7) or solvent (lanes 1,2,4, and 6). CaCl2 (50 mM) was added to the cultures for 1 hr prior to sample preparation. A nonspecific band that is recognized by the anti-HA antibody but is independent of Crz1 is marked with an asterisk (*).
(C) Representative fluorescent images of crz1 Δ strain expressing GFP-Crz1, GFP-Crz1PVIAVN, or GFP-Crz1PVIVIT from pUG36-based plasmids. Cells were visualized within 5–15 min of CaCl2 addition as indicated. “No CaCl2” refers to cells grown in standard media, which contains 700 µM CaCl2. The panels represent examples of the localization category as indicated: N, nuclear; N/C, nuclear/cytoplasmic; and C, cytoplasmic.
(D) Graphical representation of percentage of cells scored as nuclear, nuclear/cytoplasmic, and cytoplasmic from fluorescent images. The data represent the same conditions shown in (C), i.e., no added CaCl2, and 5 or 15 min after addition of 50 mM or 200 mM CaCl2. At least 200 total cells were scored for each condition.
Cells lacking Crz1 had no detectable CDRE-lacZ activity (Figure 2A, “vector”). Cells expressing Crz1 displayed low basal CDRE-lacZ activity, which reflects the low level of calcineurin activity in cells grown in standard media (Stathopoulos and Cyert, 1997). Addition of CaCl2 caused an increase in CDRE-lacZ expression whose magnitude was concentration dependent (e.g., 3.8-fold increase for 50 mM CaCl2 and 8.5-fold increase for 100 mM CaCl2, Figure 2A). In cells expressing the low-affinity Crz1PVIAVN protein, both basal CDRE-lacZ activity and Ca2+-induced CDRE-lacZ activity were reduced. When exposed to Ca2+, these cells exhibited substantially decreased Crz1 activation as compared to wild-type (~3.0-fold less than wild-type Crz1 in 50 mM CaCl2 and 1.5-fold less in 100 mM CaCl2, Figure 2A). Cells expressing the high affinity Crz1PVIVIT protein exhibited a surprisingly high level of basal CDRE-lacZ activity (~13-fold higher than wild-type, Figure 2A). Upon treatment with Ca2+, this level was further increased (1.5- to 2.0-fold in 50 mM CaCl2 and 100 mM CaCl2, respectively, Figure 2A). The Ca2+-induced activity of all three Crz1 proteins was severely reduced by the calcineurin inhibitor, FK506, and was thus calcineurin dependent (Figure 2A). These results show that modest differences in the affinity of the Crz1 PxIxIT motif for calcineurin cause corresponding changes in both basal and Ca2+-induced Crz1-dependent transcription. Furthermore, the amount of Crz1 activity elicited by a given amount of Ca2+ changed with affinity, showing that, in vivo, the Ca2+ concentration dependence of Crz1 is determined in part by its affinity for calcineurin.
Changes in the PxIxIT Motif Affect Crz1 Phosphorylation and Nuclear Localization
CDRE-lacZ activity should directly reflect the extent of Crz1 dephosphorylation by calcineurin. Therefore, we examined each Crz1 protein by immunoblotting, as phosphorylation decreases Crz1 electrophoretic mobility (Stathopoulos-Gerontides et al., 1999) (Figure 2B). Wildtype Crz1 was largely dephosphorylated in extracts of cells containing active calcineurin and was fully phosphorylated in extracts of FK506-treated cells (Figure 2B, lanes 2 and 3). Crz1PVIAVN showed a similar FK506-dependent decrease in its electrophoretic mobility, although some slower-migrating forms were detected even in the absence of FK506 (Figure 2B, lanes 4 and 5). In contrast, Crz1PVIVIT was predominantly in the hypophosphorylated, faster-migrating form, both in the presence and absence of FK506 (Figure 2B, lanes 6 and 7). Only a small fraction of the protein appeared to be phosphorylated in extracts of FK506-treated cells.
Dephosphorylation of Crz1 by calcineurin causes its translocation to the nucleus (Stathopoulos-Gerontides et al., 1999; Boustany and Cyert, 2002; Polizotto and Cyert, 2001). Therefore, we examined the localization of GFP-tagged Crz1, Crz1PVIAVN, and Crz1PVIVIT (Figures 2C and 2D). As previously observed, Crz1 was predominantly cytoplasmic under standard growth conditions and rapidly translocated to the nucleus after addition of CaCl2. Nuclear localization peaked 5 min after treatment with 200 mM CaCl2, when 92% of cells displayed complete or partial nuclear localization. Fifteen minutes after Ca2+ addition, rephosphorylation of Crz1 resulted in its relocalization to the cytoplasm, and the population of cells that displayed full or partial nuclear localization was decreased to 18%. Exposure to 50 mM CaCl2 resulted in a significantly weaker response; 5 min after Ca2+ addition, 65% of cells still displayed cytosolic localization of Crz1.
When cells expressing Crz1PVIAVN were treated with CaCl2, nuclear localization was observed in fewer cells. For example, in response to 50 mM CaCl2, 0% of cells expressing Crz1PVIAVN displayed complete nuclear localization, compared to 12% of cells expressing wild-type Crz1 (Figure 2D). Thus, decreased Crz1PVIAVN activity correlated with decreased nuclear localization. Conversely, Crz1PVIVIT displayed nuclear localization even in cells grown without Ca2+ supplementation. Almost 70% of the cells showed complete or partial nuclear localization of Crz1PVIVIT when grown in standard media (Figure 2D). Upon addition of CaCl2, the percentage of cells in which Crz1PVIVIT was localized to the nucleus increased and then decreased again to steady-state levels (Figure 2D).
Thus, the different transcriptional activities of the Crz1 variants reflect their dephosphorylation by calcineurin and subsequent nuclear localization. We conclude that, compared to wild-type Crz1, calcineurin-dependent dephosphorylation and nuclear localization of Crz1PVIAVN is decreased and that of Crz1PVIVIT is greatly increased.
Calcineurin A Residues NIR (366–368) Mediate Interaction with the PxIxIT Motif
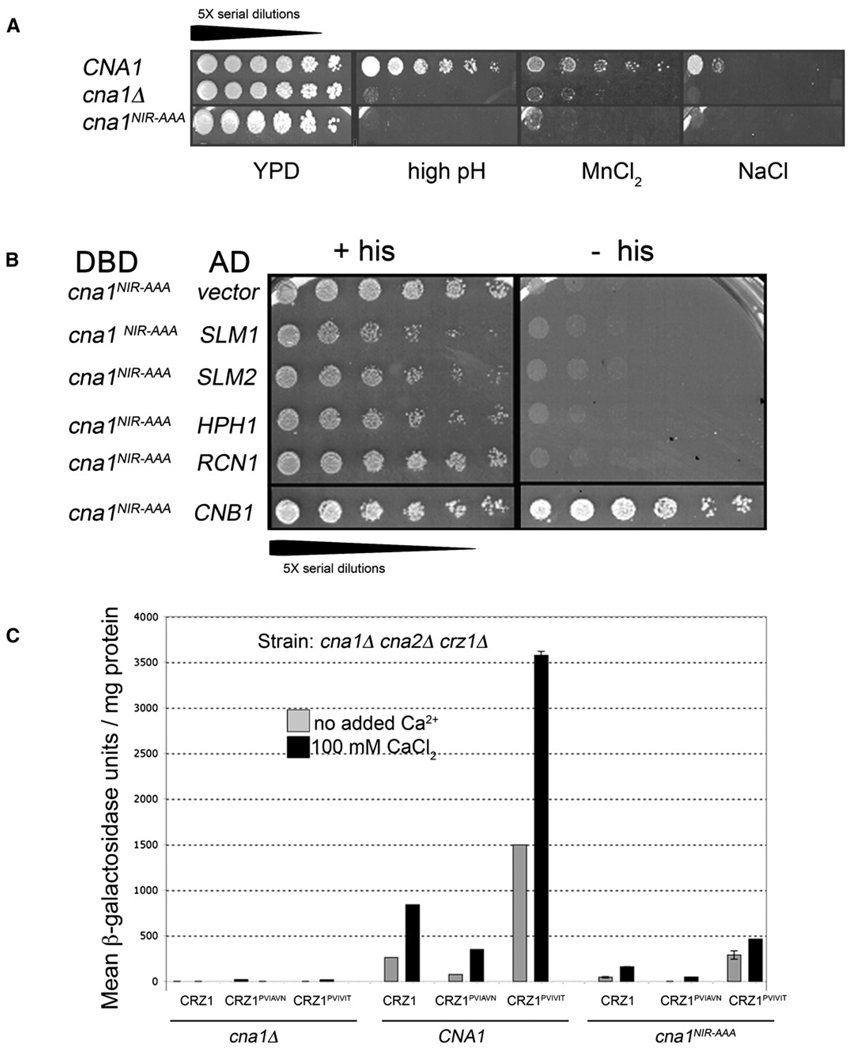
To explore further the contribution of calcineurin targeting to the distinct activities displayed by the Crz1 variants, we constructed an allele of CN A predicted to be defective in substrate interaction. In human CN A, M290 can be cross-linked to the PVIVIT peptide. Replacement of three residues that are adjacent on the protein surface to M290 (residues 330–332 in human CN Aα) with alanine (NIR-AAA), severely compromises binding to the PVIVIT peptide in vitro and dephosphorylation of NFAT in cell extracts (Li et al., 2004). We mutated the corresponding, conserved residues in CNA1 (cna1NIR-AAA, amino acids N366, I367, and R368 changed to alanine). These mutations had little effect on catalytic activity, as the bacterially expressed mutant enzyme displayed 80% of wild-type activity in vitro (see Figure S1 in the Supplemental Data available with this article online). In contrast, Cna1NIR-AAA was nonfunctional in vivo (Figure 3A), despite being expressed at levels comparable to Cna1 (data not shown). Yeast cells expressing Cna1NIR-AAA as the only CN A sub-unit displayed sensitivities to salt stress, high pH, and increased concentrations of MnCl2 that were almost as severe as those of cells lacking CN A (Figure 3A). In yeast two-hybrid assays, the interaction of Cna1NIR-AAA with CN B (Cnb1) was equivalent to that of Cna1 (Figure 3B and data not shown), but Cna1NIR-AAA failed to interact with calcineurin substrates Hph1, Slm1, or Slm2 or with the regulator Rcn1 (Figure 3B). These findings underscore the evolutionary and structural conservation of CN A-substrate interaction and highlight its importance for in vivo function.
Figure 3. Cna1NIR-AAA Is Nonfunctional Due to a Defect in Substrate Interaction, and Cells Expressing cna1NIR-AAA Show Reduced Activity of Crz1-PxIxIT Proteins.
(A) Serial dilutions of yeast strains lacking CN A (cna1 Δ cna2 Δ) and transformed with a plasmid expressing CNA1, cna1NIR-AAA, or vector were spotted on the following growth media: YPD, YPD-pH 8.0 (high pH), YPD containing 4 mM MnCl2, and YPD containing 500 mM NaCl. Growth was visualized after incubation at 30°C for 5 days (high pH) or 3 days (other media).
(B) Two-hybrid assay assessing the interactions of Cna1NIR-AAA with its substrates Hph1, Slm1, and Slm2; the Rcn1 regulator; and the B subunit Cnb1. Serial dilutions of yeast strains were spotted on medium lacking tryptophan and leucine to select for both Gal4-fusion plasmids and on medium lacking histidine, as indicated, to assay for reporter activation. DBD, Gal4-DNA binding domain fusion of Cna1NIR-AAA. AD, Gal4-activation domain fusions constructed in the plasmid pACT2 containing the indicated ORF. All plasmids were transformed into strain PJ69-4A. Cell growth was determined after 5 days incubation at 30°C.
(C) β-galactosidase activity of a cna1 Δ cna2 Δ crz1 Δ 4×-CDRE-lacZ strain transformed with plasmids expressing CNA1, cna1NIR-AAA, or vector and cotransformed with plasmids expressing Crz1-PxIxIT proteins as indicated. CaCl2 (100 mM) was added 1 hr prior to sampling, where indicated. Error bars indicate standard deviation (see Experimental Procedures for details).
In cells containing Cna1NIR-AAA, the CDRE-lacZ activity promoted by Crz1, Crz1PVIAVN, or Crz1PVIVIT was greatly reduced (~8- to 10-fold) relative to that of Cna1-expressing cells, confirming that the PxIxIT-CN A interaction is critical for their activity (Figure 3C). However, we also noted that the relative activities of Crz1, Crz1PVIAVN, and Crz1PVIVIT were the same in both strains, with Crz1PVIAVN showing the lowest activity and Crz1PVIVIT showing the highest activity (Figure 3C). Thus, residues in the PxIxIT sequence apparently still influence the weak interaction of Cna1NIR-AAA with Crz1.
Hyperactivation of Crz1PVIVIT Requires Ca2+ and CN A
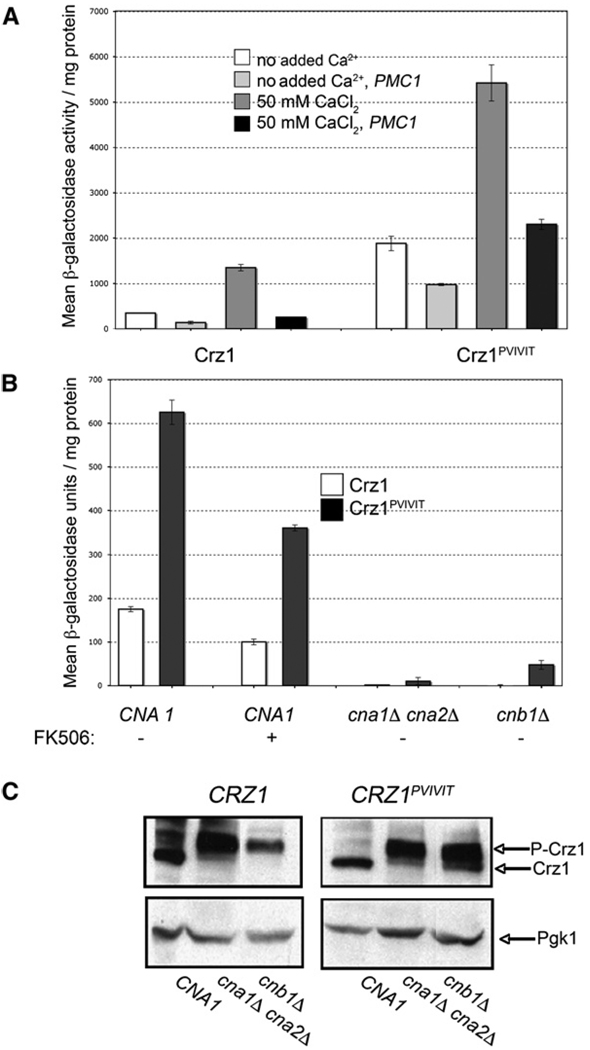
The consistent hyperactivation of Crz1PVIVIT observed, especially in cells grown in the absence of added Ca2+, led us to investigate the Crz1PVIVIT-CN A interaction in more detail. First, we examined the Ca2+ dependence of Crz1PVIVIT activation by measuring CDRE-lacZ expression in cells whose cytosolic Ca2+ concentration was lowered by overexpression of Pmc1, a vacuolar Ca2+-ATPase (Cunningham and Fink, 1994). Both the basal and Ca2+-stimulated activity of Crz1 and Crz1PVIVIT decreased under these conditions (Figure 4A), establishing that activation of each protein was Ca2+ dependent. Similar results were also observed for Crz1PVIAVN (data not shown).
Figure 4. Hyperactivation of Crz1PVIVIT Requires Ca2+ and Calcineurin Activity.
(A) β-galactosidase activity with or without CaC12 addition was determined for cells containing 4×-CDRE-lacZ and expressing Crz1 or Crz1PVIVIT, either alone or cotransformed with the multicopy plasmid pKC60 expressing PMC1. Error bars indicate standard deviation (see Experimental Procedures for details).
(B) β-galactosidase activity was determined from strains with the indicated genotype that also contained 4×-CDRE-lacZ and were transformed with plasmids expressing either CRZ1 (white bars) or Crz1PVIVIT (black bars). Cells were grown in selective media to retain the respective plasmids. Error bars indicate standard deviation (see Experimental Procedures for details).
(C) Immunoblot analysis of strains, with the indicated genotype, transformed with plasmids expressing either HA-tagged Crz1 or Crz1PVIVIT, using anti-HA antibody. Pgk1 is used as a loading control.
Next, we examined the calcineurin dependence of Crz1PVIVIT basal activity. We reasoned that increased affinity of Crz1PVIVIT for calcineurin could cause its hyperactivation in a calcineurin-dependent manner (i.e., through a decrease in the Km for the dephosphorylation reaction) and in a calcineurin-independent manner (i.e., by preventing association of Crz1PVIVIT with protein kinases). First, we tested the effect of FK506 on basal CDRE-lacZ activity and saw a 40% reduction in activity in cells expressing either Crz1 or Crz1PVIVIT (Figure 4B). This is consistent with the basal activity of these proteins being calcineurin dependent but also suggests that FK506-treated cells retain residual calcineurin activity. By comparison, in cells completely devoid of calcineurin activity and CN A polypeptide (cna1Δ cna2Δ) both Crz1 and Crz1PVIVIT are fully phosphorylated and CDRE-lacZ activity is barely detectable (≤1% of basal activity in WT cells) (Figures 4B and 4C). These findings demonstrate that in vivo, both Crz1 and Crz1PVIVIT can be phosphorylated and fully inactivated by endogenous protein kinases and that the basal activity of Crz1PVIVIT is dependent on CN A. Finally, we measured the activity of Crz1 and Crz1PVIVIT in cells lacking CN B (cnb1Δ). In these cells, CN A is present but should have little or no activity due to the absence of the regulatory subunit (Perrino et al., 1995; Cyert and Thorner, 1992). As expected, the basal CDRE-lacZ activity in cnb1Δ cells expressing Crz1 was undetectable and the protein was hyperphosphorylated in extracts of these cells (Figures 4B and 4C). Likewise, basal CDRE-lacZ activity was substantially reduced in cnb1Δ cells expressing Crz1PVIVIT (Figure 4B), confirming that the high basal activity of Crz1PVIVIT requires calcineurin activity.
Surprisingly, a low level of basal CDRE-lacZ activity was consistently observed in cnb1Δ cells expressing Crz1PVIVIT (~10% of that observed for this protein in wild-type cells), and Crz1PVIVIT was partially dephosphorylated in these cell extracts (Figure 4C). These findings may reflect the presence of residual calcineurin activity in cnb1Δ cells. Alternatively, enhanced binding of CN A to Crz1PVIVIT in these cells may interfere with the ability of kinases to phosphorylate Crz1, because two of three Crz1 kinases identified, Hrr25 and PKA, physically associate with the transcription factor (Kafadar and Cyert, 2004; Kafadar et al., 2003). Such an effect would also contribute to the hyperactivity of Crz1PVIVIT.
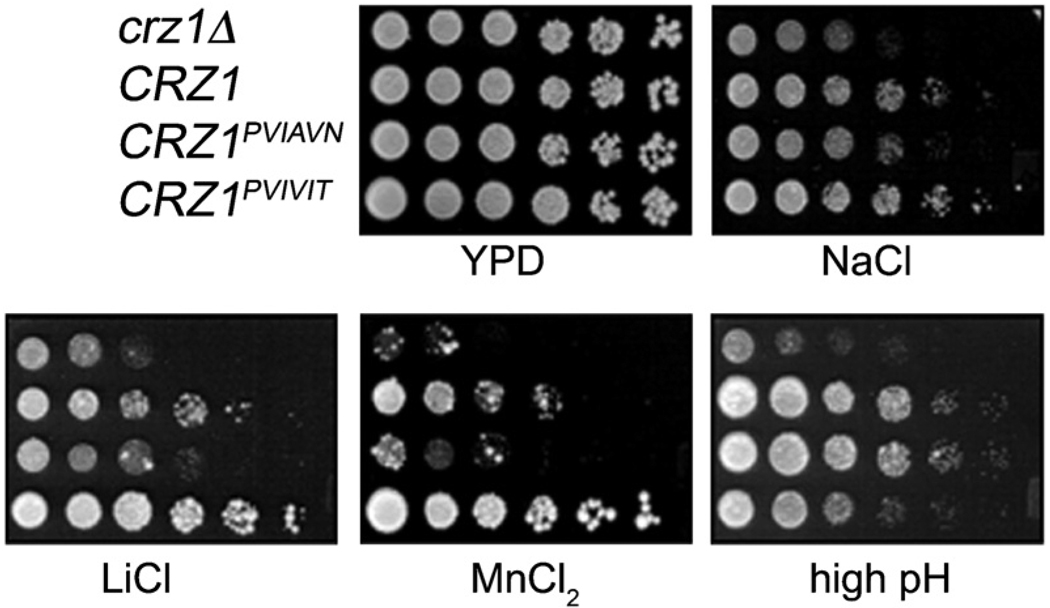
Consequences of PxIxIT Motif Mutations on Resistance to Extracellular Stresses
To assess their physiological functions, cells expressing Crz1, Crz1PVIAVN, or Crz1PVIVIT were assayed for growth under a variety of stress conditions (Figure 5). Cells lacking Crz1 (crz1Δ) showed a growth defect in the presence of high concentrations of Na+, Li+, and Mn2+ and high pH (Figure 5) that was complemented by expression of Crz1. Cells expressing Crz1PVIAVN grew less well than those expressing Crz1 on media containing high levels of Na+, Li+, and Mn2+, as well as the cell wall disruptant Congo red (Figure 5 and data not shown). Conversely, the growth of cells expressing Crz1PVIVIT was more robust than that of cells expressing Crz1 under these same conditions. Thus, in the presence of Na+, Li+, Mn2+, and Congo red, the amount of growth promoted by each Crz1 protein mirrored its level of transcriptional activity.
Figure 5. Cells Expressing Crz1, Crz1PVIAVN, or Crz1PVIVIT Display Different Growth Properties under Environmental Stress Conditions.
Five-fold serial dilutions of crz1 Δ yeast cells transformed with a plasmid expressing Crz1, Crz1PVIAVN, Crz1PVIVIT, or vector were spotted on the following media: YPD, rich medium; NaCl, YPD containing 800 mM NaCl; LiCl, YPD containing 300 mM LiCl; MnCl2, YPD containing 4 mM MnCl2; and high pH, YPD containing 100 mM Tris-HCl (pH 8.4). Growth was visualized after incubation at 30°C for 5 days (high pH) or 3 days (other media).
In contrast, in high pH medium, a different pattern of growth was displayed. Under these conditions, the growth of cells expressing Crz1 or Crz1PVIAVN was indistinguishable, whereas cells expressing Crz1PVIVIT exhibited a significant growth defect relative to cells expressing Crz1 (Figure 5). Thus, in vivo, the consequences of changing Crz1-calcineurin affinity varied depending on the environmental conditions to which cells were exposed.
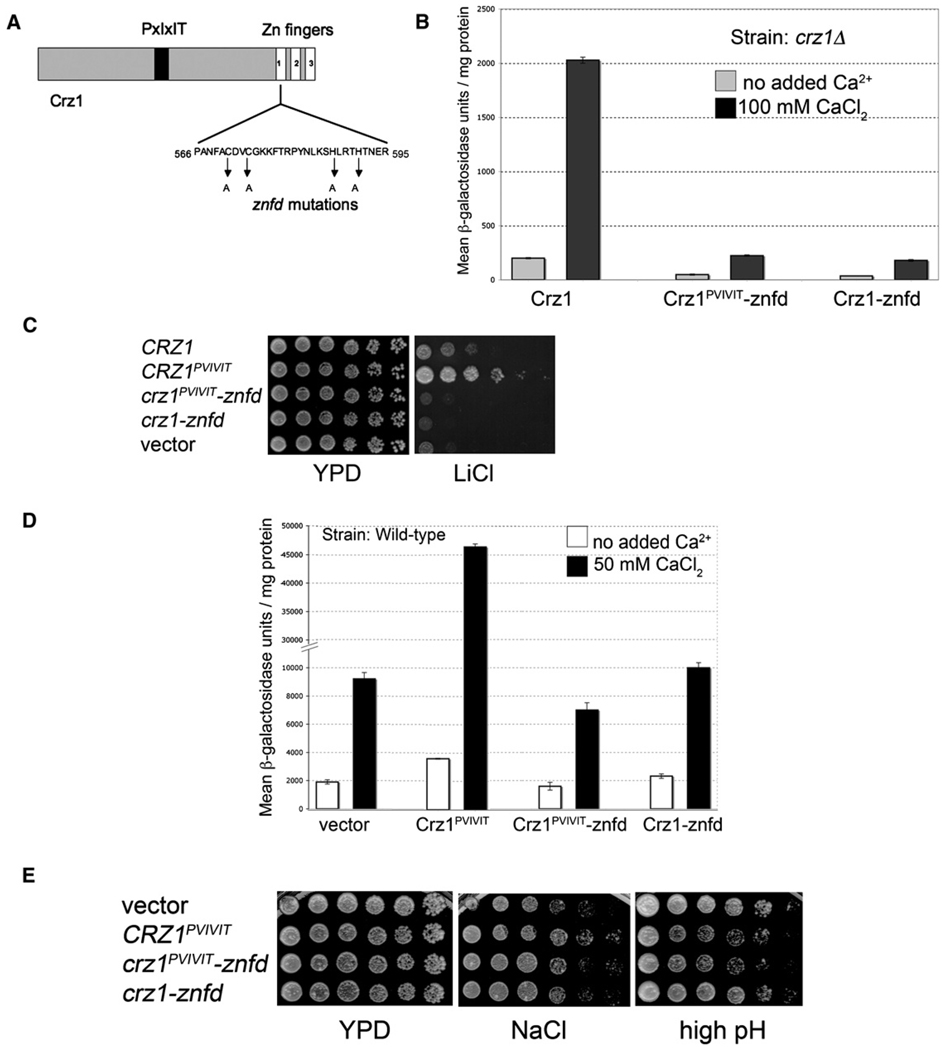
Mutations in the Zinc Finger Domain of Crz1 Elucidate One Mechanism Underlying High pH Sensitivity of CRZ1PVIVIT Cells
Crz1PVIVIT directed higher levels of transcription, suggesting that the alkaline growth defect of cells expressing this protein might result from increased expression of one or more Crz1-dependent gene products. Alternatively, the increased affinity of Crz1PVIVIT for CN A might preclude calcineurin’s interaction with other substrates or regulators whose activity is required for growth in high pH medium.
Nonfunctional Crz1 proteins were used to separate the effects of Crz1 binding to calcineurin from its effects on gene expression. We introduced mutations into the first zinc finger domain in Crz1 that should abrogate DNA binding and thus interfere with its ability to drive transcription (Figure 6A). Two such mutant alleles were constructed, crz1-znfd (for zinc-finger defective) and crz1PVIVIT-znfd. These mutants exhibited little CDRE-lacZ activity and were defective for growth under a variety of stress conditions (Figures 6B and 6C and data not shown). Both mutant proteins were expressed at levels similar to wildtype Crz1 (data not shown).
Figure 6. Mutations in the Crz1 Zinc Finger Domain Separate Transcriptional Activity from CN Binding.
(A) Schematic diagram of Crz1 sequence indicating location of zinc finger domain (-znfd) mutations.
(B) -znfd mutations reduce Crz1PVIVIT transcriptional activity. β-galactosidase activity of crz1 Δ 4×-CDRE-lacZ strain transformed with plasmids expressing Crz1, Crz1PVIVIT-znfd, or Crz1-znfd. CaCl2 (100 mM) was added 2 hr prior to sample preparation where indicated. Error bars indicate standard deviation (see Experimental Procedures for details).
(C) -znfd mutations reduce Crz1 function. Five-fold serial dilutions of crz1 Δ yeast cells expressing different Crz1 variants from plasmids were spotted on solid growth media, i.e., YPD or YPD containing 300 mM LiCl. Growth was visualized after incubation at 30°C for 3 days.
(D) Dominant effects of CRZ1 alleles on transcriptional activity. β-galactosidase activity of CRZ1 4×-CDRE-lacZ yeast strain (wild-type) expressing various CRZ1 alleles as shown. CaCl2 (50 mM) was added 1 hr prior to sample preparation, where indicated. Error bars indicate standard deviation (see Experimental Procedures for details).
(E) Dominant effects of CRZ1 alleles on growth under high pH. Three-fold serial dilutions of wild-type yeast cells expressing various CRZ1 alleles (described in [D]) were spotted on YPD, YPD plus 800 mM NaCl, or YPD-pH 8.4 plates. Growth was visualized after incubation at 30°C for 10 days (high pH) or 3 days (other media).
Expression of Crz1PVIVIT-znfd in wild-type cells resulted in reduced CDRE-lacZ activity (Figure 6D). Thus, Crz1PVIVIT-znfd dominantly interferes with Crz1 function. This is likely due to titration of CN A, as expression of Crz1-znfd showed no such effect. In contrast, Crz1PVIVIT expression significantly increased CDRE-lacZ activity.
The effects of these mutant alleles on wild-type cell growth were examined under conditions of environmental stress. In growth medium with elevated Li+, Na+, or Mn2+, expression of Crz1PVIVIT, but not Crz1PVIVIT-znfd or Crz1-znfd, improved growth relative to wild-type cells (Figure 6E and data not shown), confirming that Crz1PVIVIT promotes growth under these conditions by increasing Crz1-dependent transcription. Expression of either Crz1PVIVIT or Crz1PVIVIT-znfd in wild-type cells, but not expression of Crz1-znfd, resulted in decreased growth on high pH medium (Figure 6E). However, as described above, Crz1PVIVIT and Crz1PVIVIT-znfd have opposing effects on CDRE-lacZ expression in these cells (Figure 6D). Thus, expression of Crz1PVIVIT is not detrimental under these conditions because of increased Crz1-dependent transcription. Rather, we conclude that Crz1PVIVIT disrupts calcineurin interaction with substrates other than Crz1 and that de-phosphorylation of non-Crz1 substrates is the principal means by which calcineurin improves growth during alkaline stress.
DISCUSSION
Our studies demonstrate that calcineurin binds to its natural substrates with varying affinities and that the PxIxIT motif is a major determinant of binding affinity in several yeast substrates. The conclusions are supported by two-hybrid studies, in vitro peptide binding data, and experiments in which variant PxIxIT motifs were grafted into Crz1. Our findings do not, however, preclude the existence in some substrates of additional sites of contact with the enzyme, such as those proposed for the NFAT transcription factors (Liu et al., 1999, 2001; Park et al., 2000; Martinez-Martinez et al., 2006), or alternative mechanisms for calcineurin-substrate interaction (Aramburu et al., 1999).
A second important conclusion is that calcineurinsubstrate affinity determines the Ca2+ concentration dependence and output of signaling. We mutated the PxIxIT motif of the transcription factor Crz1 to increase or decrease its affinity for calcineurin and found that the Ca2+ dependence and activity of Crz1 in vivo changed in parallel with its affinity. Indeed, mutations that cause a substantial loss of protein-protein interaction lead to impaired signaling in many other signal transduction pathways. For the HOG response in yeast, in particular, a detailed correlation has been shown between decreased proteinprotein interaction of the osmosensor/scaffold protein Sho1 with the MAPKK Pbs2 and decreased magnitude of the signal produced (Marles et al., 2004). Our calcineurin-Crz1 experiments extend such observations to enzyme-substrate interactions and further document that increased protein-protein interaction leads to enhanced biological signaling.
A third, and unexpected, finding is that increasing the calcineurin-Crz1 affinity above the native level results in impaired physiological activation of a second pathway, the pathway controlling growth during alkaline stress. Cells expressing Crz1PVIVIT grew better than cells expressing Crz1 on media containing high levels of Na+, Li+, Mn2+, or Congo red, suggesting that the extent of Crz1 activation is the major calcineurin-dependent event that limits growth of wild-type cells under these conditions. However, the same Crz1PVIVIT-expressing cells grew considerably less well at high pH. This effect was independent of Crz1-dependent transcription, indicating that it is an effect on another calcineurin substrate rather than an effect of enhanced expression of a Crz1-regulated gene. In fact, calcineurin mutants show a significantly greater growth defect than crz1Δ cells at high pH (Heath et al., 2004), confirming that responses to alkaline pH are mediated by calcineurin substrates other than Crz1. Although this apparent partitioning of calcineurin among its cellular substrates was produced artificially by expressing Crz1PVIVIT, the results demonstrate that availability of calcineurin is limiting in cells and that binding affinity and/or kinetics can directly impact substrate selection. In vivo, such effects may be particularly important in response to brief activation of calcineurin or to activation of a limited fraction of the enzyme. Exposure of yeast cells to increased extracellular Ca2+, alkaline pH, or high salinity elicits Ca2+ transients of differing kinetics and magnitude (Denis and Cyert, 2002; Miseta et al., 1999; Viladevall et al., 2004), and each condition may cause dephosphorylation of a distinct set of calcineurin substrates, determined in part by their affinity for the phosphatase. For mammalian cells, the amplitude and frequency of oscillatory Ca2+ signals are known to specify differential activation of downstream targets (Dolmetsch et al., 1997, 1998).
How can these findings be rationalized? In the simplest model, calcineurin activity directed to a substrate is proportional to the amount of calcineurin-substrate complex. CN A is present at ~14,000 copies per cell (Ghaemmaghami et al., 2003), which, distributed in a volume ≥20 fl (Woldringh et al., 1993), translates to a concentration <~1 µM. Much of the enzyme is likely engaged with substrates and/or regulators, and therefore the cellular concentration of uncomplexed calcineurin will be substantially lower. Crz1 is present at ~1100 copies per cell (Ghaemmaghami et al., 2003) or <0.1 µM. Thus, concentrations of uncomplexed calcineurin and Crz1 are less than the Kd measured here for binding of the Crz1 PxIxIT peptide to calcineurin. Accordingly, an increase in affinity will result in more calcineurin-substrate complex, and a decrease in affinity will result in less complex, causing corresponding changes in dephosphorylation. A realistic quantitative model would, of course, require experimental data on the fraction of calcineurin activated, the kinetics of calcineurin binding and dissociation, the rates of dephosphorylation of specific substrate residues by bound calcineurin, and the corresponding parameters for the relevant kinases.
Protein-protein interactions underlie signaling specificity in cells (Pawson and Nash, 2003; Remenyi et al., 2006). Here, we show that for the transient weak interactions of calcineurin with its substrates, the strength of interaction has further crucial implications for biological signaling. Because signaling proteins commonly engage in transient interactions with several different partner proteins during cellular responses, the principles illustrated by calcineurin are likely to be fundamental and conserved properties of many signaling pathways.
EXPERIMENTAL PROCEDURES
Yeast strains, media, general methods, and all plasmids used in this study are described in the Supplemental Data. For two-hybrid analysis, Gal4-activation domain fusions (in pACT2) and Gal4-DNA binding domain fusions (in pGBT9) were expressed in strain PJ69-4A (James, 2001). Growth was scored by dilution plating.
To determine CDRE-lacZ activity, cells were grown to mid-log phase, in media containing CaCl2 when noted. β-galactosidase activity is reported as maximum rate OD415 change/minute/mg of protein and was determined as described (Bultynck et al., 2006) using an equal amount of protein (5–500 µg) in each assay. Values represent an average of two independent extracts, each measured in triplicate. Error bars indicate the standard deviation.
For determination of Crz1 phosphorylation, 50 µg of cell extract, prepared by glass bead disruption (Kafadar et al., 2003), was separated on 4%–20% SDS polyacrylamide gels, analyzed by immunoblotting using anti-HA (16B12, Covance, CA) and anti-PGK (Molecular Probes, Invitrogen) antibodies, and visualized by ECL (Pierce, Rockford, IL). Expression of Cna1NIR-AAA was analyzed by immunoblotting with anti-Cna1 antisera (Jiang and Cyert, 1999).
Purification of Yeast Calcineurin
GST-Cna1 (residues 1–417) and Cnb1 were coexpressed in E. coli, and the heterodimer was purified using glutathione-Sepharose. GST was cleaved off with factor Xa. (See the Supplemental Data for details.)
Peptide and Protein Binding to Yeast Calcineurin
Peptides listed in Table 1 were synthesized at Tufts University Core Facility and purified by reversed phase HPLC. Fluorescence polarization assays with Oregon green-labeled PVIVIT 14-mer peptide were carried out as described (Li et al., 2004) with modifications detailed in the Supplemental Data.
Purification of GST-Tagged Crz1 Proteins
GST-Crz1 proteins were purified from E. coli strain BL21 carrying pLys using glutathione-Sepharose, eluted with buffer containing 15 mM glutathione, and dialyzed against PBS buffer with 10% glycerol. For Figure 1E, GST-Crz1 proteins were purified by cation exchange and gel filtration (see the Supplemental Data). Attempts to remove GST by protease digestion were unsuccessful and resulted in Crz1 degradation.
Fluorescence Microscopy
Yeast cells expressing GFP-Crz1 proteins were grown to mid-log phase (OD600, 0.4–0.8). CaCl2 was added to the desired concentration, and cells were immediately concentrated by centrifugation and visualized with a Nikon Eclipse E600 microscope with fluorescence optics and an HB100 mercury lamp. Fluorescein filter sets (Chroma Technology, Brattlebori, VT) were used to visualize GFP. Photographs were taken with a Hamamatsu 47420-95 digital charge-coupled device camera and QED software (QED Imaging). A total of 200–275 cells were counted for every condition to generate the data presented in Figure 3D.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kyle Cunningham for providing plasmid pKC60, and Liansen Liu and Michael Rexach for technical advice. We thank Geert Bultynck, Allyson O’Donnell, and other members of the Cyert laboratory for helpful advice and discussions. We are indebted to Patrice Wout for critical reading of the manuscript. Funding for this work was provided by NIH research grants GM-48728 to M.S.C. and AI40127 to Anjana Rao.
Footnotes
Supplemental Data
Supplemental Data include one figure, two tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/25/6/889/DC1/.
REFERENCES
- Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Rao A, Klee CB. Calcineurin: from structure to function. Curr. Top. Cell. Regul. 2000;36:237–295. doi: 10.1016/s0070-2137(01)80011-x. [DOI] [PubMed] [Google Scholar]
- Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004;23:3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boustany LM, Cyert MS. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 2002;16:608–619. doi: 10.1101/gad.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultynck G, Heath VL, Majeed AP, Galan JM, Haguenauer-Tsapis R, Cyert MS. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 2006;26:4729–4745. doi: 10.1128/MCB.01973-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 2003;311:1143–1150. doi: 10.1016/s0006-291x(03)01552-3. [DOI] [PubMed] [Google Scholar]
- Cyert MS, Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czirjak G, Enyedi P. Targeting of calcineurin to an NFAT-like docking site is required for the calcium-dependent activation of the background K+ channel, TRESK. J. Biol. Chem. 2006;281:14677–14682. doi: 10.1074/jbc.M602495200. [DOI] [PubMed] [Google Scholar]
- Dell’Acqua ML, Dodge KL, Tavalin SJ, Scott JD. Mapping the protein phosphatase-2B anchoring site on AKAP79. Binding and inhibition of phosphatase activity are mediated by residues 315–360. J. Biol. Chem. 2002;277:48796–48802. doi: 10.1074/jbc.M207833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V, Cyert MS. Internal Ca(2+) release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J. Cell Biol. 2002;156:29–34. doi: 10.1083/jcb.200111004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- Feske S, Okamura H, Hogan PG, Rao A. Ca2+/calcineurin signalling in cells of the immune system. Biochem. Biophys. Res. Commun. 2003;311:1117–1132. doi: 10.1016/j.bbrc.2003.09.174. [DOI] [PubMed] [Google Scholar]
- Friedman J, Weissman I. Two cytoplasmic candidates for immunophilin action are revealed by affinity for a new cyclophilin: one in the presence and one in the absence of CsA. Cell. 1991;66:799–806. doi: 10.1016/0092-8674(91)90123-g. [DOI] [PubMed] [Google Scholar]
- Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Heath VL, Shaw SL, Roy S, Cyert MS. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:695–704. doi: 10.1128/EC.3.3.695-704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z, Cunningham KW. The RCN family of calcineurin regulators. Biochem. Biophys. Res. Commun. 2003;311:1089–1093. doi: 10.1016/s0006-291x(03)01515-8. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- James P. Yeast two-hybrid vectors and strains. Methods Mol. Biol. 2001;177:41–84. doi: 10.1385/1-59259-210-4:041. [DOI] [PubMed] [Google Scholar]
- Jiang B, Cyert MS. Identification of a novel region critical for calcineurin function in vivo and in vitro. J. Biol. Chem. 1999;274:18543–18551. doi: 10.1074/jbc.274.26.18543. [DOI] [PubMed] [Google Scholar]
- Kafadar KA, Cyert MS. Integration of stress responses: modulation of calcineurin signaling in Saccharomyces cerevisiae by protein kinase A. Eukaryot. Cell. 2004;3:1147–1153. doi: 10.1128/EC.3.5.1147-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar KA, Zhu H, Snyder M, Cyert MS. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes Dev. 2003;17:2698–2708. doi: 10.1101/gad.1140603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- Li H, Rao A, Hogan PG. Structural delineation of the calcineurin-NFAT interaction and its parallels to PP1 targeting interactions. J. Mol. Biol. 2004;342:1659–1674. doi: 10.1016/j.jmb.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Liu J, Masuda ES, Tsuruta L, Arai N, Arai K. Two independent calcineurin-binding regions in the N-terminal domain of murine NF-ATx1 recruit calcineurin to murine NF-ATx1. J. Immunol. 1999;162:4755–4761. [PubMed] [Google Scholar]
- Liu J, Arai K, Arai N. Inhibition of NFATx activation by an oligopeptide: disrupting the interaction of NFATx with calcineurin. J. Immunol. 2001;167:2677–2687. doi: 10.4049/jimmunol.167.5.2677. [DOI] [PubMed] [Google Scholar]
- Marles JA, Dahesh S, Haynes J, Andrews BJ, Davidson AR. Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol. Cell. 2004;14:813–823. doi: 10.1016/j.molcel.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez S, Rodriguez A, Lopez-Maderuelo MD, Ortega-Perez I, Vazquez J, Redondo JM. Blockade of NFAT activation by the second calcineurin binding site. J. Biol. Chem. 2006;281:6227–6235. doi: 10.1074/jbc.M513885200. [DOI] [PubMed] [Google Scholar]
- Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell DM. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 1999;451:132–136. doi: 10.1016/s0014-5793(99)00519-0. [DOI] [PubMed] [Google Scholar]
- Park S, Uesugi M, Verdine GL. A second calcineurin binding site on the NFAT regulatory domain. Proc. Natl. Acad. Sci. USA. 2000;97:7130–7135. doi: 10.1073/pnas.97.13.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Perrino BA, Ng LY, Soderling TR. Calcium regulation of calcineurin phosphatase activity by its B subunit and calmodulin. Role of the autoinhibitory domain. J. Biol. Chem. 1995;270:7012. doi: 10.1074/jbc.270.12.7012. [DOI] [PubMed] [Google Scholar]
- Polizotto RS, Cyert MS. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J. Cell Biol. 2001;154:951–960. doi: 10.1083/jcb.200104078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999;13:798–803. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Youn HD, Loh C, Stolow M, He W, Liu JO. Cabin 1, a negative regulator for calcineurin signaling in T lymphocytes. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L, Serrano R, Ruiz A, Domenech G, Giraldo J, Barcelo A, Arino J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:43614–43624. doi: 10.1074/jbc.M403606200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shibasaki F, Mizuno K. Calcium signal-induced cofilin dephosphorylation is mediated by Slingshot via calcineurin. J. Biol. Chem. 2005;280:12683–12689. doi: 10.1074/jbc.M411494200. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Huls PG, Vischer NO. Volume growth of daughter and parent cells during the cell cycle of Saccharomyces cerevisiae a/alpha as determined by image cytometry. J. Bacteriol. 1993;175:3174–3181. doi: 10.1128/jb.175.10.3174-3181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.