Abstract
Exchanging and sharing scientific results are essential for researchers in the field of computational modelling. BioModels.net defines agreed-upon standards for model curation. A fundamental one, MIRIAM (Minimum Information Requested in the Annotation of Models), standardises the annotation and curation process of quantitative models in biology. To support this standard, MIRIAM Resources maintains a set of standard data types for annotating models, and provides services for manipulating these annotations. Furthermore, BioModels.net creates controlled vocabularies, such as SBO (Systems Biology Ontology) which strictly indexes, defines and links terms used in Systems Biology. Finally, BioModels Database provides a free, centralised, publicly accessible database for storing, searching and retrieving curated and annotated computational models. Each resource provides a web interface to submit, search, retrieve and display its data. In addition, the BioModels.net team provides a set of Web Services which allows the community to programmatically access the resources. A user is then able to perform remote queries, such as retrieving a model and resolving all its MIRIAM Annotations, as well as getting the details about the associated SBO terms. These web services use established standards. Communications rely on SOAP (Simple Object Access Protocol) messages and the available queries are described in a WSDL (Web Services Description Language) file. Several libraries are provided in order to simplify the development of client software. BioModels.net Web Services make one step further for the researchers to simulate and understand the entirety of a biological system, by allowing them to retrieve biological models in their own tool, combine queries in workflows and efficiently analyse models.
Keywords: BioModels.net, Systems Biology, modelling, Web Services, annotation, ontology
INTRODUCTION
In the past decades, the biological data recorded in computer databases has been growing enormously, thanks to the advances of molecular and cellular biology. This paved the way to Systems Biology approaches, which often use computational modelling as an important tool to make sense of large amount of disparate information. For conveniently exchanging and reusing models, the community is increasingly relying on (i) standards for biological quantitative models curation, (ii) agreed-upon vocabularies for annotating models and (iii) a publicly available database of trusted and annotated computational models. The continuous effort of BioModels.net (http://www.biomodels.net) is contributing in these three key areas with the projects BioModels Database, MIRIAM Resources and the Systems Biology Ontology (SBO).
MIRIAM (the Minimum Information Requested in the Annotation of Models) [1] was created for standardising the curation and annotation processes of biological quantitative models. In order to identify each model element and to keep consistent interoperability of the annotations in models, MIRIAM requires the use of standard uniform resource identifiers (URIs). MIRIAM resources [2] has been developed for cataloguing and maintaining these URIs, as well as providing peripheral services for using them. In order to identify and append semantics to the processes and entities used by quantitative models, the SBO [3] has been created, a set of controlled vocabularies tailored for various problems encountered in Systems Biology. BioModels Database [4] is a free platform for storing, searching, browsing and retrieving published and curated biological quantitative models. Diverse services are provided for accessing, using and analysing these models.
MIRIAM Resources, SBO and BioModels Database provide a broad range of tools and services for biological modelling. An easy way to access them is via an interface displayed in a Web browser. Here we describe an alternative way to use these resources: via the BioModels.net Web Services. These kinds of services have several advantages. First, they do not require any installation or maintenance from the user: the code is hosted on our side and updated by our team. Second, as the computing tasks are executed on our servers, they deliver, in general, faster results than running them on a traditional desktop. Third, they can be accessed from everywhere (even from networks protected by firewalls). Finally, they only rely on an Internet connection, which most scientists have access to nowadays.
Although each of these services is independent, they can be combined in workflows in order to achieve more complex tasks (for example, by using the Taverna workbench [5]). These services provide a broader range of functions than the ones already available via the Web interface and allow more powerful interaction with the data, like batch processing of large datasets. More importantly, these services can be considered as a complete toolkit which allows other software to programmatically access the data and services of the BioModels.net project. This makes BioModels.net Web Services a unique solution for the community to access, analyse and understand biological models.
BIOMODELS WEB SERVICES
BioModels Database is a public resource of curated and published quantitative models in biology. Some of these models come from direct submission by publication authors, as several publishers (for example: Nature Publishing Group, Public Library of Science or BioMed Central) advise deposition in BioModels Database as part of the paper submission process. Some others are created by our team of curators, based on peer reviewed publications. A complete Web application allows users to browse the models, analyse them (by launching simulations online) and export the models in various formats, such as SBML [6] or CellML [7]. It is also possible to extract and save sub-models.
In addition, BioModels Database provides a programming interface via Web Services. These allow the search, download and analysis of models. To date, these services can be queried from two different endpoints: http://www.ebi.ac.uk/biomodels-main/services/BioModelsWebServices and http://biomodels.caltech.edu/services/BioModelsWebServices. Moreover, the WSDL can be accessed at: http://www.ebi.ac.uk/biomodels-main/services/BioModelsWebServices?wsdl and http://biomodels.caltech.edu/services/BioModelsWebServices?wsdl respectively.
Every model in BioModels Database is uniquely identified. A submission identifier is assigned to any model during the submission process (this identifier is composed of the string ‘MODEL’ followed by ten digits, for example ‘MODEL7984093336’). Once a model is published, a BioModels identifier is created (composed of the string ‘BIOMD’ followed by ten digits, for example ‘BIOMD0000000216’). Both identifiers are unique and will never be re-assigned to a different model. Therefore users who want to download a specific model, and know its identifier, can use the method getModelSBMLById(). For instance, in the publication about dynamical modeling of syncytial mitotic cycles in Drosophila embryos [8], a model is quoted with the submission identifier MODEL7984093336; by using getModelSBMLById(‘MODEL7984093336’), one can retrieve the serialised string of the model's; SBML file. Users can use either submission identifiers or BioModels identifiers to retrieve models. When the model's; identifier is unknown, users can search models by providing keywords, such as the model's; name (with getModelsIdByName()), the author's; name (with getModelsIdByPerson()) or some publication information (with getModelsIdByPublication()). However, this might return more than one model. Alternatively, as models in BioModels Database are cross-referenced with some external resources, a user can launch a search by giving the name or identifier of an external entity used to annotate the model. For example, getModelsIdByGO(‘mitotic cell cycle’) or getModelsIdByGOId(‘GO:0000278’) will return models related with ‘mitotic cell cycle’.
Since the launch of BioModels Database, the size of the submitted models has been increasing rapidly. If one is only interested in some specific parts of a model, the method getSubModelSBML( ) can help extracting these parts, and assembling them into a valid and coherent sub-model. It is necessary to know the identifiers of the parts which will be extracted prior to call this method. This can be done by downloading the complete model first by the way introduced above.
BioModels Database, so far, is the only model repository which provides a complete set of Web Services for accessing and using its content. For more information and documentation about these services, please refer to: http://www.ebi.ac.uk/biomodels-main/webservices. For bug reports or feature requests, please use the SourceForge tracker at: http://sourceforge.net/projects/biomodels/.
MIRIAM WEB SERVICES
MIRIAM Resources fulfills several needs. It catalogues consistent ensemble of datasets available online (that we call data types), supplies a URI for identifying them, and lists the online physical locations where the data can be accessed (that we call resources). It also provides a way to generate and resolve unique and perennial identifiers (in a form of a URI, that we call MIRIAM URI) for datasets; that is a specific piece of knowledge belonging to a data type. This allows the community to unambiguously identify entities by using a single URI, which is one of the requirements of the MIRIAM Standard. This also shields the user from the possibly numerous and likely variable resources distributing the datasets. In order to support the usage of these URIs by modellers and model users, MIRIAM Resources provides several resolution and conversion services. It is important to emphasise that MIRIAM Resources is not designed to be a end-user utility, but rather a tool used by other software via application-to-application communications. Therefore, the programmatic access to MIRIAM URIs is one of the most important part of MIRIAM Resources.
A wide range of services are available. These include methods for retrieving the information stored about a given data type, like its definition (getDataTypeDef( )), the list of its synonyms (getDataTypeSynonyms( )) or the regular expression matchings the dataset identifiers (getDataTypePattern( )). There are also methods for generating a MIRIAM URI from a data type name and the identifier of a dataset (getURI( )) and resolving the physical locations where knowledge about a dataset can accessed given its MIRIAM URI (getLocations( )).
A library for the Java programming language is also available in order to handle all the communication processes involved when invoking the Web Services. This makes the consumption of the services very easy. For example, it can allow the developer of a tool that creates and edits models to handle MIRIAM Annotations making the usage of MIRIAM URIs transparent for the modeller.
These services can be queried via the following endpoint: http://www.ebi.ac.uk/miriamws/main/MiriamWebServices and the WSDL can be accessed at: http://www.ebi.ac.uk/miriamws/main/MiriamWebServices?wsdl. Moreover, all the documentation about these services is accessible from: http://www.ebi.ac.uk/miriam/main/mdb?section=ws. It includes the full description of all the available queries (listing their parameters and the type of result they return), as well as examples of usage of the library. For bug reports or feature requests, please use the SourceForge tracker at: http://sourceforge.net/projects/miriam/.
SBO WEB SERVICES
The SBO is a set of controlled vocabularies especially designed for computational modelling. Its content includes terms describing types of entities (whether functional or material, like ‘macromolecule’ or ‘channel’), types of events (‘catalysis’, ‘addition of a chemical group’, etc.), roles of reaction participants in events (‘substrate’, ‘catalyst’, etc.), mathematical expressions (‘mass action kinetics’, ‘Henri-Michaelis-Menten equation’, etc.), parameters used in mathematical expression (‘Michaelis constant’, ‘forward unimolecular rate constant’, etc.), and modelling frameworks in which to use the mathematical expressions (‘continuous modelling’, ‘discrete modelling’, etc.). SBO is made available online to the community through an ontology browser, a search engine and several exports (OBO, OWL and XML). A tracker is also at hand for users to submit new term requests and report errors.
In addition to these facilities, a set of Web Services give access to the ontology from other software tools. These allow retrieval of any details of a given term, or of part of the ontology being the subtree of a given term. For example, using getTermById( ) will return the requested term in a form of an object containing all the details stored about it, while getTreeOWL( ) will return a piece of OWL describing the whole subtree having the given term as its root. Several searches are also available, with methods like: searchOWL( ), searchString( ), etc. Whatever the request is, the data can be retrieved in any of the supported formats: XML, OWL or just as a simple object.
In order to help users accessing these services, we developed and distribute a Java library. It allows very easy creation of clients as the whole communication processes are handle by the library. Therefore, the user is able to query the services by using simple methods like any other local classes. Some documentation about the library, including several examples of use, can be found at: http://www.ebi.ac.uk/sbo/main/static?page=library.
These services can be queried via the following endpoint: http://www.ebi.ac.uk/sbo/main/services/SBOQuery, and the WSDL can be accessed at: http://www.ebi.ac.uk/sbo/main/services/SBOQuery?wsdl. For bug reports or feature requests, please use the SourceForge tracker at: http://sourceforge.net/projects/biomodels/.
IMPLEMENTATION
For exchanging information between the clients and the server, all these Web Services rely on the SOAP protocol (http://www.w3.org/TR/soap/) [which uses the Extensible Markup Language (XML) (http://www.w3.org/XML/) as its message format]. SOAP messages use HTTP (RFC 2616) for message negotiation and transmission, which makes communication between different networks and through firewalls possible without any specific configuration. The Web Services Description Language (WSDL) (http://www.w3.org/TR/wsdl/) is used to fully describe the available methods and the parameters that they need. Since all these elements are standards or recommendations from the World Wide Web Consortium (http://www.w3.org/) these services are highly interoperable, even between different platforms and programming languages.
Although independent, the different BioModels.net Web Services rely on the same infrastructure and set of software tools. First, the Java programming language (http://www.java.com/) is used for all the aspects of these services. Apache Axis (http://ws.apache.org/axis/) is used for all the SOAP related work. It runs inside an Apache Tomcat server (http://tomcat.apache.org/), which provides a Java Servlet and JavaServer Pages (JSP) running environment. In order to provide redundancy in case of hardware failure or crash of the application, two similar Tomcat servers are actually used. Finally, the backend uses a MySQL database (http://www.mysql.com/), which is the same than the one used for the Web application of each project.
The choice of these technologies was primarily driven by what was available when we started to work on these services (in early 2006), and by the software and programming language (Java) used in the group and the community. At that time, Axis was the obvious choice for such a work: standard, interoperable, reliable, easy to develop solution, and supported by a large and strong community.
The development of these services followed a bottom-up approach. This means that the work started by creating the Java classes, including the requested methods, and then, Axis has been used to generated the missing layers of the services (conversion and communication), as well as the WSDL. This puts the focus on the features of the service and its implementation.
INTEGRATION
The Web Services from MIRIAM, SBO and BioModels Database can be used as a unified toolkit when analysing computational models. As an example, let us create and analyse a simple putative workflow, involving requests to all these services, in order to gain a better understanding of a model distributed by a public repository.
For instance, Hong et al. [9] studied the molecular mechanism underlying how DNA damage causes predominantly phase shifts (mostly advances) in the circadian clock. In their publication, the authors describe the study and the model they developed in order to simulate the processes involved. As PLoS Computational Biology advises their authors to deposit their data into a public repository, Hong CI et al. submitted their model to BioModels Database, where it is now freely available and referenced under the identifier BIOMD0000000216.
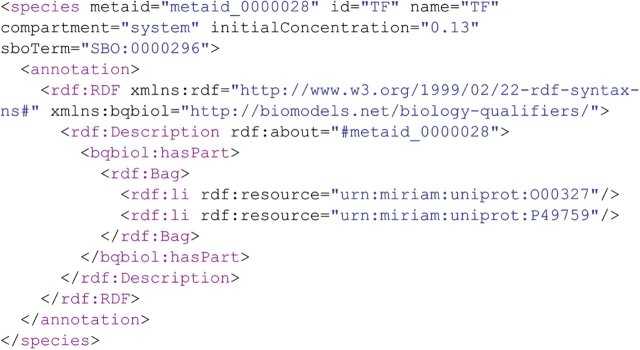
Therefore, by using the method getModelSBMLById(‘BIOMD0000000216’) of BioModels Web Services, one can retrieve this model encoded in SBML. Figure 1 presents a snippet taken from this SBML file. This extract shows the definition of a SBML species, identified as ‘TF’.
Figure 1:
Piece of the SBML encoded model from Hong CI. Please note the usage of SBO terms and MIRIAM URIs in the annotation of the model components.
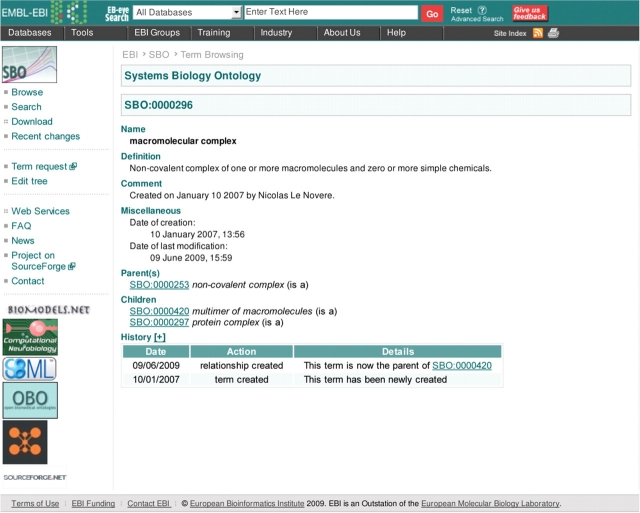
In order to know what the nature of this entity is, one can look at the SBO term associated to it. Here the SBML species is associated with the sboTerm ‘SBO:0000296’. The getTermById(‘SBO:0000296’) method from SBO Web Services, will allow the user to retrieve more information about it. The query returns the full record of this term, including its name, which is, in this case: ‘macromolecular complex’ (cf. Figure 2).
Figure 2:
Details of the SBO term: macromolecular complex (SBO:0000296).
However, this is not enough, as this does not explain what this complex is actually made of. Fortunately, this entity is associated with some annotations (between the ‘annotation’ tags) encoded in RDF (http://www.w3.org/RDF/), following the SBML specifications. These show that ‘TF’ is actually a complex [because the BioModels.net qualifier (http://biomodels.net/qualifiers/) ‘hasPart’ is used] made of two entities identified by the following URIs: ‘urn:miriam:uniprot:O00327’ and ‘urn:miriam:uniprot:P49759’.
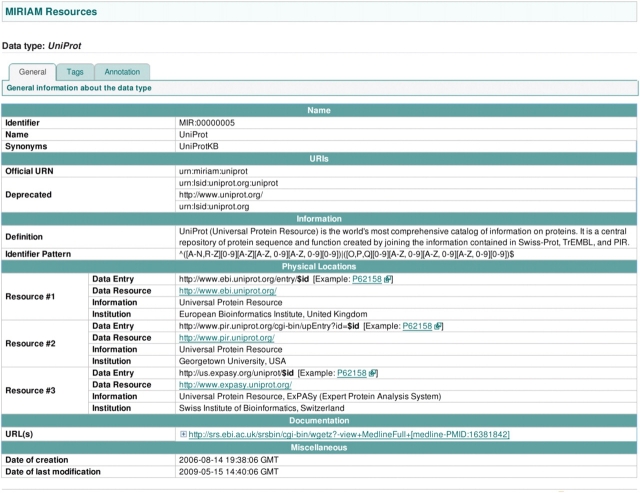
In order to understand what these two entities actually are, one needs to resolve these URIs. This can be done by querying MIRIAM Web Services. For example, the method getLocations(‘urn:miriam:uniprot:O00327’) can be invoked, which returns the physical locations where further information about this specific entity (in this case a protein sequence) can be found. Currently this query returns three different uniform resource locators (URLs), all of them pointing to the same entity in different databases (cf. Figure 3). Therefore, the user learns that ‘TF’ is a macromolecular complex made of ‘Aryl hydrocarbon receptor nuclear translocator-like protein 1’ (referenced by UniProtKB/Swiss-Prot, under the identifier O00327) and ‘Dual specificity protein kinase CLK1’ (referenced by UniProtKB/Swiss-Prot, under the identifier P49759). This brings a much better biological understanding of the model.
Figure 3:
Uniprot record from MIRIAM Resources: there are three different resources or physical locations where the data about this dataset is stored and can be retrieved.
Ultimately, by sequentially querying the proper BioModels.net services, one can fully understand the biology behind this model. Moreover, by setting up a workflow in Taverna or a small script, one could make this whole chain of queries automatic and efficiently extract knowledge out of any annotated model. There lies the power of such services.
AVAILABILITY
Every service is freely accessible to anyone (no restrictions regarding commercial and academic organisations). Moreover, the source code of all these projects is available under the GNU General Public License (GPL) and can be downloaded from their associated project on SourceForge (http://sourceforge.net/).
We recommend users with specific needs, like particular queries not already provided, or heavy usage (high frequency and/or number of queries), to contact us. In this case, we may design specific queries which will be efficient for their needs while still keeping a high quality of service for the other users. Moreover, we always welcome comments or requests from ours users, in order to provide the best service for the community. In case one needs any help for using these services or has any questions related to them, one can use the following email address: biomodels-net-support@lists.sourceforge.net.
To always provide the best access to our data, we are working several updates of the current services. For example, a new module has been added to MIRIAM Resources, which checks every day the state of the referenced resources. This will allow us to provide new methods to, for example, get the most reliable resource for a given dataset. In BioModels Database, a full set of converters has been developed to cope with the various formats handled. New methods could give external access to these useful tools.
Moreover, in order to follow the best practices for Web Services interoperability, established by the industry consortium Web Services Interoperability Organization (WS-I) (http://www.ws-i.org/), we are working on updating our services. This will rely on the tools developed by the Web Services Interoperability Technology (WSIT) project: the Java API for XML Web Services (JAX-WS) and the Java Architecture for XML Binding (JAXB). We already have some prototypes working and available for testing purposes (for example at: http://www.ebi.ac.uk/sbo/demo/). Moreover, we started working on services using new ways of communication, like REST (Representational State Transfer) [10]. The way to query such services (a simple GET to a classic URL) makes them much easier and flexible to use. We are also working on Semantic Web Services, which ultimately would ease the integration and composition of diverse Web Services.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/.
Key Points.
Description of the Web Services provided by the BioModels.net resources.
How to access the whole content of BioModels Database.
How to use the URI generation and resolving services of MIRIAM Resources.
How to query the Systems Biology Ontology and retrieve terms of interest.
Complete example showing how these services can be combined into a workflow in order to understand and analyse computational models.
Supplementary Material
Acknowledgements
The services have been developed with the support of the European Molecular Biology Laboratory (EMBL); the National Institute of General Medical Sciences (NIGMS, grant R01 GM070923-02); and the UK Biotechnology and Biological Sciences Research Council (BBSRC, grant numbers BB/E005748/1 and BB/F010516/1). Authors are grateful to the Open Source and free Software community, which provided many software tools and libraries used in the course of these projects.
Biographies
Camille Laibe and Chen Li are the main developers of the BioModels.net project. They work in the Computational Neurobiology Group, at the European Bioinformatics Institute (an outstation of the European Molecular Biology Laboratory), Hinxton, UK.
Mélanie Courtot developed SBO and the associated Web Services while working in the Computational Neurobiology Group at the European Bioinformatics Institute. She is now pursuing a PhD at the Terry Fox Laboratory, BCCRC, Vancouver, Canada.
Nicolas Le Novère is leading the Computational Neurobiology Group at the European Bioinformatics Institute, Hinxton, UK.
References
- Le Novère N, Finney A, Hucka M, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotechnol. 2005;23:1509–15. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Laibe C, Le Novère N. MIRIAM resources: tools to generate and resolve robust cross-references in systems biology. BMC Syst Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N. Model storage, exchange and integration. BMC Neurosci. 2006;7(Suppl. 1):S11. doi: 10.1186/1471-2202-7-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N, Bornstein B, Broicher A, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–D91. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinn T, Addis M, Ferris J, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–54. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Lloyd CM, Halstead MDB, Nielsen PF. CellML: its future, present and past. Prog Biophys Mol Biol. 2004;85:433–50. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Calzone L, Thieffry D, Tyson JJ, et al. Dynamical modeling of syncytial mitotic cycles in Drosophila embryos. Mol Syst Biol. 2007;3:131. doi: 10.1038/msb4100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CI, Zámborszky J, Csikász-Nagy A. Minimum criteria for DNA damage-induced phase advances in circadian rhythms. PLoS Comput Biol. 2009;5:e1000384. doi: 10.1371/journal.pcbi.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding RT. Architectural styles and the design of network-based software architectures. PhD thesis, University of California, Irvine, 2000. http://www.ics.uci.edu/∼fielding/pubs/dissertation/top.htm. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.