Abstract
In recent years, progress in the study of the lateral organization of the plasma membrane has led to the proposal that mammalian cells use two different organelles to store lipids: intracellular lipid droplets (LDs) and plasma membrane caveolae. Experimental evidence suggests that caveolin (CAV) may act as a sensitive lipid-organizing molecule that physically connects these two lipid-storing organelles. Here, we determine the sequences necessary for efficient sorting of CAV to LDs. We show that targeting is a process cooperatively mediated by two motifs. CAV's central hydrophobic domain (Hyd) anchors CAV to the endoplasmic reticulum (ER). Next, positively charged sequences (Pos-Seqs) mediate sorting of CAVs into LDs. Our findings were confirmed by identifying an equivalent, non-conserved but functionally interchangeable Pos-Seq in ALDI, a bona fide LD-resident protein. Using this information, we were able to retarget a cytosolic protein and convert it to an LD-resident protein. Further studies suggest three requirements for targeting via this mechanism: the positive charge of the Pos-Seq, physical proximity between Pos-Seq and Hyd and a precise spatial orientation between both motifs. The study uncovers remarkable similarities with the signals that target proteins to the membrane of mitochondria and peroxisomes
Keywords: ALDI, caveolin, endoplasmic reticulum, lipid droplets, sorting signal
Lipid droplets (LDs) are dynamic lipid stores present in all eukaryotic cells (1,2). Efficient assembly and metabolization of LDs into energy probably represents a dominant evolutionary advantage for cells. Excessive and long-term accumulation of LDs is a cellular hallmark of widespread human diseases including obesity, diabetes and arteriosclerosis.
Initially considered to be inert cytoplasmic lipid inclusions, LDs are now understood as complex organelles with a central role in the regulation of cellular lipids, key players in the complex dance of diverse lipids moving through cells. When redistribution of subcellular lipids fails, overall lipid homeostasis is impaired, with severe cellular consequences. Exactly how such lipid distributions are controlled is unclear, but proteomic and other studies suggest significant exchanges of proteins between LDs and a variety of other organelles such as mitochondria and endoplasmic reticulum (ER), potentially facilitating lipid redistribution.
To understand how droplets might exchange proteins and lipids with other cellular compartments, it is important to understand droplets' formation and also how proteins are targeted to them. The prevalent model (1,2) suggests that LD biogenesis occurs by esterification and gradual gathering of neutral lipids within the bilayer of the ER. However, some evidence is inconsistent with a model of uncontrolled random gathering: nascent LDs likely originate in specific subdomains within the ER with a unique phospholipid composition (3–5); genes encoding enzymes that synthesize phospholipids are determinants of lipid-droplet size and number. Phospholipids details of the monolayer likely represent a first level of regulation of LD function (6).
LD formation and lipid mobilization are also likely orchestrated by proteins on the droplets surface (7–17). Topologically, there are two structural classes of LD-resident proteins. The first contains proteins without a membrane-integral hydrophobic domain (Hyd), which may be targeted to LDs from the cytosol [see alternative model in (18)]. This group includes LD-resident proteins such as perilipins (19), adipose differentiation-related protein (ADRP) (20) or Rab18 (21,22). In contrast, the second group has a Hyd that is most likely inserted in the ER, allowing the protein to laterally diffuse to droplets. This group includes oleosins and caleosins in plants (23,24), viral components such as hepatitis C virus (HCV) and GB virus B (GBV-B) core proteins (25,26), hairpin membrane-anchored proteins such as caveolins (CAVs) and stomatin (12,27–29), and S-adenosylmethionine (SAM) methyltransferases such as erg6 in Saccharomyces cerevisiae or ALDI and AAM-B in mammalian cells (11,30,31). This complex protein composition likely confers to the LD, a second level of functional regulation. Indeed, some LD-resident proteins – including CAV – dynamically associate with the organelle upon specific and temporary requirements of the cell (9,32,33). Unfortunately, it is unclear how such dynamic association is controlled: a consensus-targeting motif necessary and sufficient for sorting proteins into LDs has not been demonstrated, and thus the regulation of the LD by a specific and timely targeting of proteins remains largely unexplored.
CAV's dynamic association and regulation is particularly important. Mammalian cells likely use two different organelles to store lipids – intracellular LDs and plasma membrane caveolae (34) – and thus regulatory circuits to coordinate their functions are critical. Biophysically, caveolae organize as stable flask-shaped ordered domains with a high density of lipids such as glycosphingolipids, sphingomyelin and cholesterol.
CAVs likely provide a critical link between these two storage organelles. They are essential components of caveolae, with a unique capacity to assemble and stabilize caveolae at the plasma membrane. CAVs could also act as sensitive lipid-organizing molecules not only at the cell surface but also on LDs (9,27–29,32,35–37). Importantly, CAVs associate with LDs following particular cell requirements. They thus may regulate LD function under specific conditions, potentially even contributing to transfer of lipids either to, or away from, the droplets.
Consistent with such a role, CAV1 redistributes from the cell surface to LDs upon treatment with cholesterol or fatty acids, and during liver regeneration. Inhibition of caveolae internalization also inhibits cholesterol-stimulated association of CAV1 with LDs (32). Further, adipocytes from CAV1 gene-disrupted mice show decreased levels of free cholesterol in LDs and CAV1−/− hepatocytes cannot form LDs during liver regeneration (38). Consequently, a link between the trafficking of CAV and a lipid sensing and storage function of the protein has been convincingly proposed (34).
Additional support for the hypothesis that appropriate control of CAV's localization could contribute to control of cellular lipids comes from our studies with CAV mutants (29,36,37). Unlike normal CAVs with regulated LDs association (see above), the truncation mutant CAVDGV (N-terminal deletion of 54 residues of CAV3) constitutively accumulates on LDs. Consistent with a role for CAVs in lipid homeostasis, although the mutant protein is irreversibly confined to the droplets, the expression of CAVDGVpromotes a lipid imbalance in other cellular compartments. CAVDGV causes the accumulation of free cholesterol in late endosomes and the irreversible assembly of neutral lipids in LDs. CAVDGV also decreases free cholesterol at the cell surface and in the Golgi complex, and the induced perturbation of cellular lipid regulation alters specific signaling pathways at the cell surface such as the H-Ras-mediated activation of Raf-1. The inhibitory effect of CAVDGV on signaling is completely reversed by replenishing the cell membrane with cholesterol and mimicked by depletion of membrane cholesterol (39). Therefore, by correct regulation of its localization, it is likely that CAV facilitates multiple cellular processes.
How is CAV's localization controlled? It is clearly essential to identify the molecular determinants involved in the transport of CAVs into the droplets. The prevalent model postulates that the transfer of CAVs into LDs is not regulated, but that simple overaccumulation in the ER diverts the protein into nascent LDs. Proper packing of the central hairpin Hyd is proposed as the only molecular determinant during this process (35). Certainly, truncation of the Hyd of CAVs, stomatin, oleosin, the HCV core protein and AAM-B completely abolishes their association with LDs (12,26,27,31,35,40), demonstrating that the Hyd is required. However, is the Hyd sufficient for targeting CAV1 into droplets? Increased cholesterol promotes trafficking of CAV from the plasma membrane into LDs (32), and CAV leaves LDs when fatty acids are removed or at later stages of liver regeneration when LDs are still abundant (36). Thus, additional levels of regulation appear likely. Multiple lines of evidence suggest that the Hyd alone may not be sufficient. First, similar to CAV, the Hyd of the AAM-B protein was postulated to be sufficient (although with a low efficiency), to partially target proteins into droplets (31), but this appears incorrect because the same study found that deletion of a short sequence C-terminally flanking the Hyd is sufficient to completely abrogate transport of AAM-B into LDs, although the mutant protein still included an intact Hyd. Second, a green fluorescent protein (GFP)-tagged Hyd of CAV2 does not localize to LDs (27). Thus, a Hyd alone is likely insufficient to efficiently sort CAVs within the ER into LDs and additional information is required.
Here, we use mutational analysis to investigate the CAVs region(s) necessary and sufficient for sorting to LDs. We show that the Hyd is necessary but not sufficient for targeting CAVs to droplets; instead, targeting of CAVs into droplets requires two cooperative/auxiliary sequences. First, the Hyd anchors CAVs to the ER. Next, CAVs are specifically sorted by positively charged sequences (Pos-Seqs) into LDs. To gain further mechanistic insight into sorting, we identified functionally interchangeable sorting information within ALDI, and use the information to retarget the cytosolic protein GFP to droplets.
Results
The Hyd is necessary but insufficient for targeting CAVs into LDs
Consistent with a possible role in redistributing lipids between different cellular compartments, CAV redistributes from the plasma membrane to LDS upon treatment with cholesterol, fatty acids or during liver regeneration. We investigated the molecular determinants necessary and sufficient for an efficient transport of CAV to LDS. Such identification paves the way for future studies aimed at understanding how these domains can be dynamically modified to control its subcellular localization.
The CAV family includes three members: CAV1 and CAV2 are abundant in most non-muscle cells and CAV3 is specifically expressed in skeletal muscle and some smooth-muscle cells [see for a recent review (34)]. Upon expression, only CAV1 and CAV3 promote the assembly of caveolae at the cell surface. CAV1 and CAV3 share a high sequence identity. In contrast, CAV2 shows a modest identity with the other two members. As in previous studies, to minimize possible interference with endogenous CAV1, we analyzed CAV3 ectopically expressed in non-muscle COS-1 cells. Owing to high homology, we expect CAV3 findings are relevant for CAV1. Because of its divergence from CAV1 and CAV3, we also studied CAV2. First, we carried out a mutational analysis of the murine CAVs. Mutant proteins were N-terminally tagged with GFP (unless otherwise indicated), expressed during 24 h in lipid-loaded cells and their cellular distribution analyzed by fluorescence microscopy and biochemical methods. Past studies showed that addition of an N-terminal GFP tag to wild-type CAV does not affect its function or localization (29). LDs were routinely visualized by Nile Red staining. None of the mutants significantly affected formation or accumulation of LDs (see examples in Figures 2 and 4).
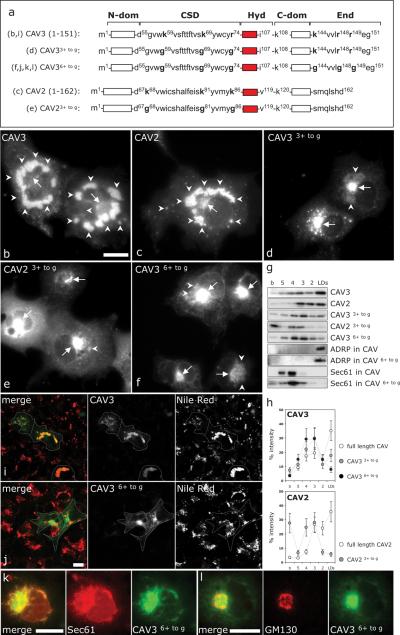
Figure 2. Analysis of the Pos-Seqs in full-length CAVs.
A) Schematic representation of mutants analyzed in the figure. Letters between brackets indicate the corresponding panel for each mutant. Numbers on the residues indicate the relative position of each residue within the full-length protein with respect to the N-terminal end (see in addition Table 1). The Hyd has been colored in red. Mutants were N-terminally tagged with GFP, expressed in lipid-loaded cells for 24 h, additionally treated with cycloheximide for additional 6 h and their final distribution was analyzed by fluorescence microscopy (B–F). Scale bar is 20 μm. G and H) Distribution of the mutants in sucrose density gradients as in Figure 1K. The distribution of the mutants along the gradients was quantified as the relative intensity of each band with respect to the total intensity of the six bands. Endogenously expressed ADRP and Sec61 were used to define the relative position of LD and the ER membranes within the gradients. I and J) Low magnification panels to show LD formation (red) in cells transfected with CAV3 or CAV36+ to g (in green). Discontinuous lines indicate the edge of transfected cells. K and L) Colocalization of CAV36+ to g (green) with the endogenous ER-resident protein, Sec61 (red in K) and the endogenous Golgi marker GM130 (red in L). Scale bar is 20 μm.
Figure 4. The Hyd and the Pos-Seq cooperate for targeting to LDs.
A) Schematic representation of the mutants used in this figure. The three studied sequences have been colored: Hyd (red), the Jux (blue) and the End (orange). Numbers on the residues indicate the relative position of each residue within the full-length protein with respect to the N-terminal end (see in addition Table 1). Letters between brackets indicate the corresponding panel for each mutant. Mutants were N-terminally tagged with GFP, expressed in lipid-loaded cells and their final distribution was analyzed by fluorescence microscopy (B–H). Arrows mark the selected areas for high magnification (G and H). Shows the colocalization between the Hyd (green) and endogenous Sec61 (red) and the Hyd-End3 (green) with Nile Red (red). I) Distribution of the mutants in sucrose density gradients as described in Figure 1. J and K) Low magnification panels to illustrate that cells transfected as above with Hyd and with ER (in green) display a normal formation of LDs (in red), although the mutants accumulate in the ER or the cytosol, respectively. Discontinuous lines indicate the edge of transfected cells. Scale bar is 20 μm.
An N-terminal truncation (first 33 residues deleted) of CAV3 (CAV3NED) distributes to the cell surface similar to wild-type protein (41). A larger 54 residue N-terminal truncation CAV3DGV, converts the protein into a bona fide LD-resident protein [Figure 1A; see also (29)]. Similarly, truncation of its equivalent 66 residues, converted CAV2 (CAV2DKV) into an LD-resident protein (29). As expected, GFP-CAV3DGV and GFP-CAV2DKV also accumulated on the LDs of COS-1 cells (Figure 1B,C). The intracellular distribution of GFP-CAV3DGV and GFP-CAV2DKV was further corroborated by subcellular fractionation in sucrose density gradients (Figure 1K). Although both proteins accumulated in the LD fraction (cf. LD marker GFPADRP in Figure 1K), CAV3DGV associated with LDs more efficiently than CAV2DKV. Further, while CAV3DGV localized almost entirely to droplets, CAV2DKV was present on droplets and membrane gradient fractions (cf. endogenous ER marker Sec61 in Figure 1K).
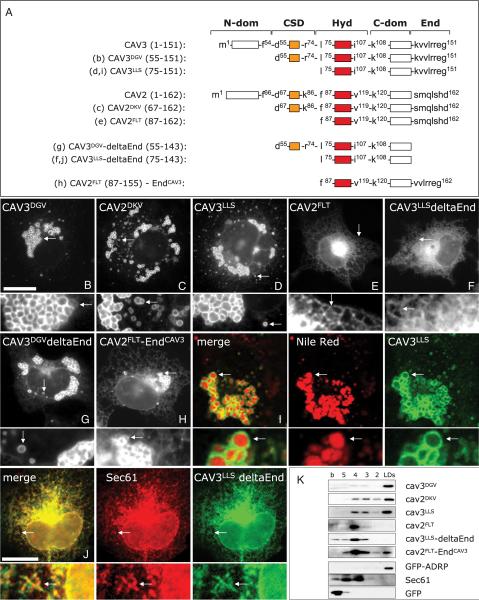
Figure 1. Identification of the LD-targeting motifs contained in CAVs.
A) Schematic representation of the mutants of CAV3 and CAV2 used in this figure. The Hyd has been colored in red and the CSD in orange. Numbers on the residues indicate the relative position of each residue within the full-length protein with respect to the N-terminal end (see in addition Table 1). Letters between brackets indicate the corresponding panel for each mutant. Mutants were N-terminally tagged with GFP, expressed in lipid-loaded cells and their final distribution was analyzed by fluorescence microscopy (B–J). I) Shows the colocalization between CAV3LLS (green) and Nile Red-positive LDs (red). J) Shows the colocalization of CAV3LLS-deltaEnd (green) and the endogenous ER-resident protein, Sec61 (red). Arrows mark the selected areas for the high magnification panels. Scale bar is 20 μm. K) Distribution of the mutants in sucrose density gradients. Cells were treated as indicated above and additionally incubated for 12 h with cycloheximide. Fractions were collected from the top of the gradient and the distribution of the mutants (detected by means of western blotting and anti-GFP antibodies) was compared with GFP, Sec61 and GFP-ADRP. `LDs' is the top fraction of the gradient in which LDs fractionate and `b' corresponds to the bottom fraction of the gradient. See text and tables for more details.
This continued localization of the CAV mutants to LDs enabled us to dissect the LD targeting of CAVs, which is otherwise hard to study because of its transient nature. Because CAV3DGV and CAV2DKV efficiently localize to droplets, they contain the necessary LD-targeting information. Because the CAV scaffolding domain (CSD) (Figure 1A) is critical for CAVs role in the regulation of cell signaling (34), CAV3DGV and CAV2DKV included intact CSDs. The CSD is a highly conserved segment of 20 amino acids that N-terminally flanks the Hyd (Tables 1 and 2), characterized by positively charged residues and thus a markedly basic isoelectric point (pI). The positive charge of the CSD is especially evident in the proximity of the Hyd (pI 9.63 for CAV3 and pI 9.05 for CAV2). We hypothesized that the CSD might be required for targeting CAVs to LDs. Indeed, deletion of the CSD of CAV2DKV (CAV2FLT; Figure 1A) completely inhibited its targeting into LDs and the protein accumulated in the membranes of the ER (Figure 1E), consistent with it playing a role in LD targeting, and also consistent with a previous report (27). However, deletion of the CSD of CAV3DGV (CAV3LLS; Figure 1A) did not apparently modify association with LDs [Figure 1D; (29)]. These results were confirmed and quantified by fractionation in sucrose density gradients (Figure 1K). Intriguingly, although clearly on LDs, CAV3LLS now demonstrated reduced association with the organelle when compared with CAV3DGV. The distribution of CAV3LLS in the density gradient was similar to that of CAV2DKV, and was found now in ER fractions. This was in strong contrast to CAV2FLT which is completely excluded from LDs, although the mutant protein (with an intact Hyd) accumulates to high levels in the ER membranes. Overall, the CSD appeared to playing a LD-targeting role in both CAVs, but was more important for CAV2. To explain this, we hypothesized that, unlike CAV2, perhaps CAV3 has a second LD-targeting sequence, in addition to the CSD.
Table 1.
Complete sequence of the proteins analyzed in this work
| Name | Sequence |
|---|---|
| CAV2 | mgletekadv qlfmaddays hhsgvdyadp ekyvdsshdr dphqlnshlk lgfedliaep etthsfdkvw |
| NP_058596 | icshalfeis kyvmykfltv flaiplafia gilfatlscl hiwilmpfvk tclmvlpsvq tiwksvtdvv igplctsvgr sfssvsmqls hd |
| CAV3 | mmteehtdle ariikdihck eidlvnrdpk ninedivkvd fedviaepeg tysfdgvwkv sfttftvsky |
| NP_031643 | wcyrllstll gvplallwgf lfacisfchi wavvpciksy lieiqcishi yslcirtfcn plfaalgqvc snikvvlrre g |
| ALDI | mdvlvpllql lvllltlplh llallgcwqp icktyfpylm atltarsykk meskkrelfs qikdlkgtsn evtllelgcg |
| Q562C4 | tganfqfypp gckvtcvdpn pnfekfltks maenrhlqye rfivaygenm kqladssmdv vvctlvlcsv qsprkvlqev qrvlkpggll ffwehvsepq gsqallwqrv leptwkhigd gchltretwk diekaqfsev qlewqpppfk wlpvgphimg kavk |
The table shows the complete amino acid sequence of CAVs and ALDI. The Hyd has been underlined and the sequences analyzed in this work have been highlighted with bold letters (CSD and C-terminal sequence of CAVs and the C-terminal region of ALDI).
Table 2.
Sequences analyzed in this work
| Name | Sequences | pI |
|---|---|---|
| CAV2 CSD | -d kvwicshalfeiskyvmyk- | 9.05 |
| CAV3 CSD | -d gvwkvsfttftvskywcyr- | 9.63 |
| CAV2 C-terminal end | -p lctsvgrsfssvsmqlshd | 7.13 |
| CAV3 C-terminal end | -p lfaalgqvcsnikvvlrreg | 9.56 |
| CAV3LLS-deltaEnd | -l cirtfcnplfaalgqvcsni | 7.97 |
| CAV2FLT-EndCAV3 | -p lctsvgrsfssvvvlrreg | 10.30 |
| ALDI C-terminal end | -pppfkwlpvgphimgkavk | 10.31 |
| ALDI-deltaEnd | -qlewqpppfkwlpvgphim | 6.75 |
| CAV2FLT-EndALDI | -plctsvgrsfssvsmgkavk | 10.07 |
| Hyd of ALDI | mdvlvpllqllvllltlplhllallgcwqpick- | 6.49 |
| End3 | -pick-gkavkgkavkgkavk | 10.70 |
| End3 (k to d) | -picd-gdavdgdavdgdavd | 3.25 |
| End3 (k to g) | -picg-ggavgggavgggavg | 5.91 |
| Jux ALDI | -tyfpyfmamltarsykkmeskkrelfsqi | 9.87 |
| Jux delta14 | -ykkmeskkrelfsqi | 9.82 |
The table shows the sequences and pI of the mutant proteins analyzed in the text. Positively charged residues are highlighted as bold letters.
The CAV3 protein encloses a second LD-targeting motif
We searched for the hypothesized second LD-targeting motif within the CAV3 molecule, not present in CAV2. By similarity with the CSD, we looked for a positively charged sequence (Pos-Seq). The comparison between CAV2 and CAV3 revealed a candidate at the C-terminal end of CAV3 (Table 2). Thus, as the CSD, this region was arbitrarily defined by the presence of a highly conserved Pro residue situated at 20–21 amino acids upstream from the end of CAV2 (Pro−20) and CAV3 (Pro−21) [also found in CAV1 (Pro−21)]. The sequence beyond this Pro residue is positively charged in the case of CAV3 (pI 9.56), but not in the case of CAV2 (pI 7.13) (Table 2). The position of the positive residues was similar between CAVs of different species and the sequence identity of this segment was relatively conserved in the case of CAV1 and CAV3. We then tested whether this sequence is required for targeting CAV3LLS into droplets. Thus, we deleted the last eight amino acids of CAV3LLS (CAV3LLS-deltaEnd). This truncation highly reduces the pI of the N–terminal segment of CAV3 (pI 7.97; Table 2). The resulting mutant CAV3LLS-deltaEnd was observed in the ER membranes and now was excluded from LDs (Figure 1F), in agreement with the proposed model. The distribution of the mutants was confirmed in density gradients (Figure 1K), by colocalization of CAV3LLS with Nile Red (a fluorescent LD marker) and CAV3LLS-deltaEnd with Sec61 (Figure 1I,J). This truncation had little effect on CAV3DGV (CAV3DGV-deltaEnd), most probably because in addition it contains an intact LD-targeting motif within the CSD (Figure 1G). Therefore, as we concluded for CAV2FLT, the distribution of CAV3LLS-deltaEnd shows that the transport of CAVs to LDs is not only determined by the accumulation of the protein in the ER. On the contrary, because CAV2FLT and CAV3LLS-deltaEnd accumulate in the ER membranes but are completely excluded from LDs, these results extend the prevalent model (see Introduction) and show that the Pos-Seq mediates sorting of CAVs within the ER before entering the droplets, and also that targeting contributions from multiple Pos-Seq domains can improve a protein such as CAV3's targeting to LDs.
The Pos-Seqs identified here show a highly variable amino acid composition and thus complicate the identification of equivalent sequences in other LD-targeted proteins. However, because targeting motifs attached to all proteins having the same destination must be interchangeable, these sequences can be functionally defined by their ability for conducting the sorting of a non-resident protein to LDs. Thus, to further validate the sorting role of the Pos-Seq, we aimed to convert CAV2FLT into an LD protein. To do so, the last seven residues of CAV2FLT were replaced with the equivalent residues of CAV3 (CAV2FLT-EndCAV3; Figure 1A; Table 2) that had been identified as part of one of the functional Pos-Seqs. This exchange converts the C-terminal end of CAV2FLT into a Pos-Seq (pI 10.30). As expected, the last seven residues of CAV3 conferred to CAV2FLT the capacity to accumulate on LDs (Figure 1H,K). These results validate the Pos-Seq as functionally interchangeable information for targeting CAVs to LDs. As deletion of the Hyd completely abolished the association with LDs (27,35), we conclude that targeting of CAVs into droplets requires two cooperative/auxiliary sequences, the Hyd and at least one Pos-Seq.
Role of the Pos-Seq within full-length CAVs
We next analyzed the significance of the Pos-Seqs described here in the context of the full-length CAVs. Our mutational analysis determined that the Pos-Seq contained within the CSD is essential for targeting CAV2 to droplets and cooperative in the case of CAV3. Therefore, the three positive residues within the CSD of CAV3 and CAV2 were substituted by neutral Gly residues (CAV33+ to g and CAV23+ to g) (Figure 2A). The proteins were expressed in fatty acid-loaded cells for 24 h and additionally treated with cycloheximide for 6 h to reduce the pool of newly synthesized proteins. As previously described (29), full-length CAV3 and CAV2 were seen in a punctate staining corresponding to the plasma membrane and largely accumulated on LDs (Figure 2B,C). The density gradients showed that more than 35% of CAV3 and CAV2 were on LDs (Figure 2G,H for quantification). In striking contrast, the mutants CAV3+ to g were only seen occasionally on the plasma membrane and accumulated in the ER (Figure 2D,E). Contrary to full-length CAVs, both mutants accumulated in a perinuclear region (see arrows in Figure 2 and colocalization with an ER marker in Figure 2K), which was similar to the distribution of CAV3LLS-deltaEnd and CAV2FLT (cf. Figure 1). In addition, the mutants partially overlapped with GM130, a Golgi marker (Figure 2L). Surprisingly, a minor pool of the mutants, especially clear in the case of CAV3, was observed on LDs (arrowheads in Figure 2). The density gradients confirmed such redistribution and only 17.8% of CAV33+ to g and 5.7% of CAV23+ to g floated on droplets (Figure 2G,H). Thus, we next constructed a mutant in which the six positively charged residues contained within the CSD and the End of CAV3 were substituted by Gly residues (CAV36+ to g) (Figure 2A). By immunofluorescence, the overall distribution of CAV36+ to g was similar to CAV33+ to g (Figure 2F,K,I). However, the sucrose density gradients showed that only 7.8% of CAV36+ to g was on droplets (Figure 2G,H). These results clearly show a crucial role of the Pos-Seqs identified here in the efficient targeting of full-length CAVs into droplets. The fact that the mutants dramatically decrease LD localization of CAV, but a residual small amount of localization remains, may suggest the existence of an additional Pos-Seq in the N-terminal region of CAV.
Expression of the full-length CAVs increased the levels of Nile Red fluorescence associated with LDs (Figure 2I), confirming a role of CAV in droplet formation or stabilization. On the contrary, the expression of CAV+ to g mutants did not significantly affect the formation of droplets (see example of CAV36+ to g in Figure 2J). However, we noticed that the CAV+ to g mutants, especially CAV36+ to g, modified the overall flotation of the ER membranes in sucrose density gradients (Figure 2G, cf. Sec61 in CAV and CAV36+ to G, note increase in Sec61 in upper portion of gradient), suggesting that accumulation of CAVs promotes a concomitant accumulation of lipids in the ER. In fact, as commented in the Introduction section, CAV is a fatty acid binding protein. Such observation paves the way for future studies aimed at understanding the role of CAVs in the regulation of the intracellular trafficking of lipids.
The LD-targeting Pos-Seq of ALDI is functionally interchangeable with CAVs
To determine whether the findings might be somewhat general, we translated our analysis to ALDI, a recently identified SAM methyltransferase (Q562C4) which is expressed in liver and kidney (11). ALDI is a convenient second protein to study because: (i) it is a naturally occurring bona fide LD-resident protein; (ii) similar to CAVs, ALDI moves within the ER into LDs; (iii) ALDI is formed by a relatively simple sequence of 244 amino acids of which almost half comprises the methyltransferase activity of the protein; (iv) the topology of ALDI is highly similar to CAV3LLS (the minimal CAV3 sequence targeted to droplets) and (v) most importantly, our previous characterization suggests that ALDI may be targeted to the initial deposits of lipids that eventually form LDs in the ER and is thus a promising marker for studying the biogenesis of LDs.
To investigate the LD-targeting motifs of ALDI, the amino acid sequence of the protein (Table 1) was divided into five discrete regions (Figure 3A): an N-terminal Hyd (Hyd: 1 to 33), a juxtamembrane region (Jux: 34 to 62), the highly conserved SAM-methyltransferase domain (63 to 177) and a C-terminal segment (178 to 244) at the end of which, similarly to CAV3, we detected positively charged residues (End: 240 to 244). As expected, full-length ALDI largely accumulated on LDs (Figure 3B,H). In contrast, when the Hyd was deleted (ALDI-deltaHyd), the protein was diffusely distributed in the cytosol and highly concentrated inside the nucleus (Figure 3C,H), which was similar to GFP (Figure 3D). Next, the Pos-Seq located at the end of the protein was analyzed. Once more by similarity with the length of the CSD, this region was arbitrarily defined by the presence of a Pro residue (Pro−19) close to the C-terminal end of the protein (pI 10.31; Table 2). When the last five amino acids of ALDI were truncated to decrease the overall positive charge of the C-terminal segment (ALDI-deltaEnd, pI 6.75; Table 2), the protein was totally excluded from LDs (Figure 3E,H) and accumulated in the ER membranes (Figure 3G,H). Therefore, identically to CAV3LLS, a C-terminal Pos-Seq is required for sorting ALDI to droplets.
Figure 3. Identification of the LD-targeting motifs contained in ALDI.
A) Schematic representation of the mutants used in the figure. The Hyd has been colored in red and the juxtamembrane region (Jux) in blue. Numbers on the residues indicate the relative position of each residue within the full-length protein with respect to the N-terminal end (see in addition Table 1). Letters between brackets indicate the corresponding panel for each mutant. Mutants were N-terminally tagged with GFP, expressed in lipid-loaded cells and their final distribution was analyzed by fluorescence microscopy (B–G). G) Shows the colocalization between ALDI-deltaEnd (green) and the ER-resident protein, Sec61 (red). Arrows mark the selected areas for high magnification. Scale bar is 20 μm. H) Distribution of the mutants in sucrose density gradients as in Figure 1K.
Because the proteins sorted within the ER to LDs do not seem to share any conserved sequence, the targeting information must be encoded in the structural features of the proteins. If this is true, the information contained in the Pos-Seq of ALDI is expected to be functionally interchangeable with CAVs. We tested this hypothesis when the End of ALDI was inserted within the equivalent sequence of CAV2FLT (CAV2FLT-EndALDI, pI 10.07; Table 2), the new protein accumulated on LDs as efficiently as previously did with EndCAV3 (Figure 3F,H).
Cooperative/auxiliary role of Hyd and Pos-Seq in sorting proteins to LDs
Targeting peptide sequences and targeting patches are defined as the continuous (peptide) or distant regions of the protein (patches) that are necessary and sufficient for a correct targeting to the organelle. In CAVs and ALDIs sequences, experiments described earlier have identified two necessary regions for targeting to LDs: the Hyd and the Pos-Seq, with the general model that the Hyd domain localizes the protein to the ER, and then the Pos-Seq domain is used to load it into droplets. To further test this idea, we investigated whether we could use this knowledge to retarget a cytosolic protein to the droplets. As it was conveniently fluorescent, we used GFP which is known to distribute throughout the cytosol when expressed by itself. Neither Hyd nor the End of ALDI was able to target GFP to LDs when expressed individually. Consistent with our model, the Hyd alone accumulated in the ER (Figure 4B,I) and the End of ALDI showed a similar distribution to GFP alone (Figure 4C,I). However, a protein that contained the Hyd and the End of ALDI (Figure 4A) was partially targeted to the droplets, although the protein was still abundant in the ER (Figure 4D). Guided by the fact that in the CSD the positive charge are distributed in a sequence of 20 residues, and that the End sequence is only 5 residues long, we investigated the effect of using more than one End. The addition of a double repeat (Hyd-End2) increased the localization of GFP to LDs (Figure 4E). Finally, a triple repeat of End efficiently targeted the GFP-fusion protein (Hyd-End3) to LDs (Figure 4F,I). The cellular distribution of the Hyd within the ER membranes and Hyd-End3 on LDs was confirmed by colocalization with Sec61 (Figure 4G) and Nile Red staining (Figure 4H). Similar to the rest of the mutants, GFP-Hyd and GFP-End did not significantly alter the ability of cells to accumulate LDs (Figure 4J,K). The Hyd N-terminally tagged with myc and the Hyd-End3 N-terminally tagged with hemagglutinin (HA) display an identical distribution to the equivalent GFP-tagged mutants, ruling out any non-specific effect of GFP on the location of the mutants (data not shown). In conclusion, sorting of proteins within the ER to droplets depends on two cooperative/auxiliary regions: (i) the Hyd for association with the ER membranes and (ii) the Pos-Seq for sorting within the ER into LDs. Accordingly, from this point the sequence End3 will be also referred as a Pos-Seq because it is functionally equivalent to the CSD.
Proximity and proper spatial orientation between Hyd and Pos-Seq are likely required for targeting to LDs
Having determined that both the Hyd and the Pos-Seq were critical for LD localization, we wondered whether their position in relation to each other was important. Homology modeling, against other methyltransferases, suggests that for ALDI the two domains are likely close to each other (not shown). With this suggestion that physical proximity and possibly orientation matter, we investigated the effect of keeping the two domains separate in a model protein formed by the combination of Hyd and Jux-Seq from ALDI. Similarly to End3, the Jux-Seq is a positively charged segment (pI 9.87; Table 2), but in contrast to End3, it contains bulky aromatic Tyr and Phe residues placed between the Hyd and the positive residues (Figure 5A; Table 2), preventing close apposition of the two domains. The resulting protein, Hyd-Jux (Figure 5B,E) was excluded from LDs, suggesting that physical proximity may be important. Consistent with such a hypothesis, when we removed the bulky residues placed between the positive charges and the Hyd, the resulting protein (Hyd-Jux delta14) was then observed to efficiently accumulate on droplets (Figure 5C,E), although it showed a very similar charge to Hyd-Jux (pI 9.8; Table 2).
Figure 5. Spatial determinants for sorting the model protein to LDs.
A) Schematic representation of mutants analyzed in the figure. Negative numbers on the residues indicate the relative position with respect to the C-terminal end. Letters between brackets indicate the corresponding panel for each mutant. The Hyd has been colored in gray. The residues that separate the Hyd and the positives charges of the Jux-Seq are underlined. Mutants were N-terminally tagged with GFP, expressed in lipid-loaded cells and their final distribution was analyzed by fluorescence microscopy (B–D). E) Distribution of the mutants in sucrose density gradients. F) Schematic representation of the proteins analyzed in this figure. Negative numbers on the residues indicate the relative position with respect to the C-terminal end. The Hyd has been colored in gray. Letters between brackets indicate the corresponding panel for each mutant. All the constructs were N-terminally tagged with GFP, expressed in lipid-loaded cells and their final distribution was analyzed by fluorescence microscopy (G–K). Arrows mark the selected areas for the high magnification panels. l) Distribution of the mutants treated as in Figure 1 in sucrose density gradients. M) Mutants were transfected for 12 h and then fractionated in sucrose density gradients. Scale bar is 20 μm.
Finally, we noticed that there was a Pro residue situated at 20 or 21 amino acids upstream of the C-terminal end of CAV3, CAV2, CAV1 and ALDI, between the Hyd and the Pos-Seq. Pro residues contain an unusual ring in the N-end amine group that forces amino acid sequences connected by the Pro into a tight turn, and thus most likely introduces a change in the spatial orientation of the Pos-Seq with respect to the Hyd. To investigate whether this might be important, we studied the importance of the central Pro residue that marks the change in polarity of the Hyd-End3 (Pro−19), in our GFP-Hyd-End3 protein. The Pro residue was substituted with a Leu residue (p to l in Figure 5A). Interestingly, the mutant Hyd-End3(p to l) accumulated in the membranes of the ER and was completely excluded from LDs (Figure 5D,E). Overall, these results suggest that both physical proximity and orientation between the Hyd and the Pos-Seq are likely important for targeting proteins into droplets, but the exact details of this placement remain to be explored in future work.
Mutational analysis of the model protein
The Hyd-End3 is a synthetic 48 amino acid polypeptide efficiently targeted to LDs. As the Pos-Seq of ALDI and the CAVs are functionally interchangeable, the sorting information must be encoded in the structural features of the sequence. Therefore, as a model LD-resident protein we further analyzed the Hyd-End3. First, we analyzed the importance of the charge within the model protein, by mutation of the positive residues (Figure 5F). The seven Lys residues were substituted by negative Asp residues (k to d) or by neutral Gly residues (k to g) (Table 2). Both mutants accumulated in the ER membranes and were excluded from droplets (Figure 5G,H,L). This result further supports the hypothesis that the positive charge of the End3, rather than the polarity, or the absence of negatively charged residues is essential for sorting to LDs. Next, we evaluated the importance of the Val residues within the End3. The three hydrophobic Val were substituted with low hydrophobic Gly residues (v to g) or with hydrophobic Leu or Ile residues (v to l and v to i). The switch of Val residues did not inhibit the association of the proteins with LDs (Figure 5I,J,K). However, when compared with the Hyd-End3, the mutated proteins showed a relatively higher accumulation in the membranes of the ER (see v to g in Figure 5L). To further investigate the possibility that Val residues cooperate during the targeting of Hyd-End3 to droplets, the mutant proteins were transfected for a short time period and their appearance on LDs was analyzed in sucrose density gradients. After 12 h, only the Hyd-End3 that contained the Val residues efficiently accumulated on LDs (Figure 5M), suggesting that although it is not essential for LD targeting, Val residues likely support efficient transport to the droplets.
LD-resident proteins accumulate in the ER before entering the LD
As commented earlier, the prevalent model postulates that transport of proteins within the ER into LDs is a passive diffusion movement driven by overaccumulation of the proteins in the ER. However, the results presented here are inconsistent with this model, because a number of proteins that accumulate in the ER (that lack Pos-Seq domains) fail to be transferred to LDs. To test in a controlled way whether there is active transfer into restricted regions of the ER before the proteins enter the droplets, we wanted to compare similar proteins (with and without the Pos-Seq that we hypothesize mediates the transfer) under identical conditions. We therefore used our model proteins Hyd and Hyd-End3 to visualize by means of real-time video microscopy the differential simultaneous ER sorting between a LD-resident and a non-resident protein. To do this, GFP-Hyd and orange-Hyd-End3 were cotransfected in serum-starved cells. Under these experimental conditions, GFP-Hyd and orange-Hyd-End3 were observed in the membranes of the ER (Figure 6A and fixed cells in Figure 6D). The addition of fatty acids caused a gradual separation of the proteins. Whereas orange-Hyd-End3 partially accumulated on the newly formed LDs (after 4 h, arrows in Figure 6B), GFP-Hyd remained in the ER membranes excluded from the new droplets. The expression levels of the mutants were comparable (see also Figure 6C), demonstrating that transport of proteins into LDs is not caused by a different accumulation within the ER. On the contrary, because in the same cell the Hyd does not enter the droplets, although it accumulates in the ER, and the Hyd-End3 efficiently moves into the newly formed LDs, these results further show that the Pos-Seq specifically mediates transport of proteins into restricted regions of the ER before entering the droplets.
Figure 6. Differential sorting of Hyd and Hyd-End3 within the ER.
GFP-Hyd and orange-Hyd-End3 were cotransfected in starved cells and cells were selected under the microscope for coexpression of both proteins (A). Next, fatty acids were added to the cells and images were captured every 2 min. B) The figure shows the selected image 4 h after the treatment with fatty acids and arrows mark the selected areas for high magnification. C) The similar expression levels of the mutants were confirmed by western blotting. D) Cotransfected GFP-Hyd (green) and orange-Hyd-End3 (red) in PFA-fixed starved cells. Scale bar is 20 μm.
Discussion
Here, we identify a combined targeting motif for localizing proteins to lipid droplets, which includes a Hyd for initial localization to the ER, followed by a Pos-Seq-mediated sorting of proteins from the bulk of the ER to LDs. The LD is probably the last cellular organelle for which a consensus sorting signal has not yet been defined. Thus, identification of this motif is an indispensable first step to fully recognize the cell biology of the multifunctional LD and to ascertain how this organelle is regulated under specific cellular conditions or perturbed during disease. While we only directly investigate the targeting motif in the context of CAV and ALDI, it is likely much more general (see below) and can be used to retarget cytosolic proteins such as GFP to LDs.
Proteins are sorted in the ER before entering the LD
The ER is a dynamic pleiomorphic organelle organized in continuous but distinct subdomains, which form in response to the changes in the levels of proteins and lipids (42). This could be the case for LDs, which form in response to lipid loading of the ER. Differences in phospholipid composition of LDs when compared with bulk ER (3–5), and video microscopy experiments performed in living cells (11,36), suggest that LD biogenesis occurs in specialized subdomains within ER membranes (43). However, molecular determinants and mechanisms involved in the transport of proteins into these subdomains were previously unknown.
Others have suggested that the Hyd was the only molecular determinant required for targeting proteins within the ER to LDs (31,35) [see other proteins in Refs. (44–46)]. In this model, lateral diffusion of CAVs into LDs required only a proper packing of the central Hyd, and was driven by accumulation of the protein in the ER membranes. This model extended to proteins with a similar topology, because truncation of the Hyd of CAV2, stomatin, oleosins, the HCV core protein, AAM-B and ALDI completely abolished its transport into LDs (12,26,27,31,35,40).
Our findings are inconsistent with this model: we show that the Hyd is necessary but not sufficient for efficient targeting of proteins to LDs. This was true for ALDI, for the CAV truncation mutants and for otherwise cytosolic GFP, consistent with the finding that a GFP-tagged Hyd of CAV2 did not localize to LDs (27). Consistent with the insufficiency of the Hyd domain alone, we show that multiple proteins with intact Hyd, including CAV2FLT, CAV3LLS-deltaEnd, ALDI-deltaEnd, Hyd, and also constructs reported by other authors (26,27,31,35), accumulate to high levels in ER membranes but are completely excluded from LDs. Finally, real-time video microscopy experiments showed that in the same cell, the differential sorting of two proteins from the ER to the LD is not determined by their expression levels, but rather is dependent on the presence of the Pos-Seq. These observations strongly suggest that (i) integral membrane proteins with an intact Hyd that reside in the ER can be totally excluded from LDs, (ii) LDs originate within specific subdomains of the ER that are not accessible to those proteins and (iii) active sorting mechanisms must exist for transferring proteins from the bulk of the ER to LDs or alternatively to exclude proteins from the LD-forming sites. Thus, in addition to signals contained in the Hyd, additional information – that potentially allows additional regulation of localization – must be in place to mediate in this transfer.
The Pos-Seq mediates the transfer of proteins from the bulk of the ER to LDs
We identify the Pos-Seq as this additional information. We show that the two motifs cooperate: first, the Hyd is necessary for insertion in the ER membranes and second, at least one Pos-Seq (by itself or forming a sorting signal with Hyd), conducts the efficient sorting of the protein within the bulk of the ER to droplets. Loss of either feature results in miss-sorting and accumulation of the proteins in the cytosol or the ER membranes, but in either case their exclusion from LDs. Hyd molecular determinants were extensively characterized in previous studies (31,35,47); we focused on properties of the Pos-Seq. The average Pos-Seq identified here shows a length of 20 residues, 3 positively charged residues (Lys and/or Arg), a pI of 9.8 and it is functionally defined by its ability to mediate an efficient transport of the Hyd into LDs. Analysis of the model protein Hyd-End3 suggests that, in addition to the positive charge of the residues, the proximity to the Hyd and a specific spatial orientation are critical molecular determinants for the sorting process conducted by the Pos-Seq. More generally, additional studies on the model protein support a model where, rather than simply occurring in a linear organization, sorting sequences are organized as patches to target proteins to LDs; this remains to be more fully explored.
While we favor the proximity/orientation hypothesis, it is also possible that deletion of the Pos-Seqs results in improper folding or aggregation that may block the ER exit of newly synthesized mutants. Although we cannot rule out this possibility for all the mutants, it seems unlike in the particular case of the small model proteins (Figures 4, 5 and 6). Similarly, the Pos-Seq could also participate in packing the Hyd that must be accommodated in the monolayer of phospholipids that encircles the LD and most probably is in contact with the hydrophobic core of the droplets. This possibility was proposed in the case of proteins with an uncommonly long Hyd, such as the 75 residues long Hyd of oleosin (26). In principle, this could be the role of the positive charges contained within the Jux-Seq of ALDI. However, because the Jux-Seq of ALDI did not efficiently target the Hyd to LDs (Figure 5), something more than simply assuring proper folding of the Hyd is required for sorting within the ER into droplets.
While we have only carefully evaluated the targeting of CAVs and ALDI, a preliminary analysis suggests that this targeting is likely used by other LD-resident proteins as well (Table 3); additional studies will be important to investigate some of these candidates, in particular to those that are sometimes cytosolic. The Pos-Seq-mediated targeting of cytosolic proteins to LDs (as tested with GFP here) requires the proteins to be first localized to the ER (18). We show that this could occur via a Hyd domain, but in principle it could also be because of recruitment by a second ER-bound protein.
Table 3.
Analysis of the presence of Pos-Seq within identified LD-targeting regions
| Definition | Sequence | pI |
|---|---|---|
| ADRP 1 (48) (mouse: 55–81) | - k55tvtsaamtsalpiiqklepqiavan - | 8.6 |
| ADRP2 (48) (mouse: 155–181) | - r150tksvvngsintvlgmvqfmnsgvd - | 8.8 |
| Perilipin H1 (45) (mouse: 242–260) | - k242qghslamwipgvaplssl - | 8.8 |
| Perilipin H2 (45) (mouse: 320–342) | - v320lrevtalpnprgllggvvh - | 9.6 |
| Perilipin H3 (45) (mouse: 349–364) | - i349savtwapaavlgtvgrilh - | 9.8 |
| TIP-47 (46) (human: 154–174) | - t154rgavqsgvdktksvvtggvq - | 10.0 |
| PRP19P (49) (mouse: 167–250) | - g167mtpeiiqklqdkatvltterkkrg - | 10.0 |
| Stomatin (12) (human: 21–88) | - i54kiikeyeraiifrlgrilqggakg - | 10.4 |
| MLDP (50) (human: 137–183) | - v137aksvtgmvdlaqrgrrwsgelrrs - | 11.8 |
| ApoA-V (51) (human: 146–191) | - rvvhhtgrfkelf191hpyaeslvsgigr– | 10.0 |
| AAM-B protein (31) (human: 39–61) | - f39lvrftviyneqmaskkrelfsn - | 9.7 |
Analysis of the regions recognized as necessary for targeting proteins to LDs (see additional references). When the regions (numbers between brackets) were longer than 30 residues, only the first 25 amino acids were analyzed. In the case of stomatin, the HyD (residues from 21 to 53) was not considered. All the mapped fragments contain without exception one or more Pos-Seq.
The Pos-Seq is a common sorting signal for mitochondria, peroxisomes and lipid droplets
We note an interesting similarity between the proteins sorted from the ER to LDs and the proteins anchored to other specific membranes by a single Hyd close to the C-terminus (tail anchored) or the N-terminus (signal anchored) (52,53). Such proteins do not share any conserved targeting sequence; instead, sorting information is encoded in structural features of the proteins. For these proteins, the specific transport within the ER, or to peroxisomes, chloroplasts and mitochondria, is finely determined by a combination of the length and hydrophobicity of the Hyd domain and the presence of one or two flanking Pos-Seqs (see examples in Table 4). For example, sorting due to the Pos-Seq is seen in Bcl-2 family proteins, the key regulators of cellular apoptosis. Bcl-xL and Bcl-2 have a similar Hyd in terms of hydrophobicity and length (Table 4), but Bcl-xL (with two positively charged residues at either side of the Hyd) localizes exclusively to mitochondria whereas Bcl-2 (with only one positively charged residue at either side) can also be found in the ER (54). Because of these similarities, specific mutations within the charges of the Pos-Seq are sufficient for targeting mitochondrial proteins into the membranes of the ER or the cytosol. Similarly, specific deletions of LD-resident proteins target the mutants into the mitochondria (48,55).
Table 4.
Sequences of proteins targeted to mitochondria and peroxisomes
| Name | Sequences | ||||
|---|---|---|---|---|---|
| (Orientation) | Mitochondria | Hyd | Cytosol | pI | |
| Signal-anchored mitochondria | Tom70 | gagtlpr whvalaigaplllgagamylws rrrrrreag | 9.8/Hyd/12.3 | ||
| Tom20 | mvgr nsaiaagvcgalfigyciyf drkrrsdpnfk | 9.5/Hyd/10.9 | |||
|
| |||||
| (Orientation) | Cytosol | Hyd | Mitochondria | pI | |
| Tail-anchored mitochondria | Tom5 | tektlkq aayvaaflwvspmiwhlv kkqwk | 8.3/Hyd/10.3 | ||
| Bcl-2 | fswlslk tllslalvgacitlgaylg hk | 8.8/Hyd/8.8 | |||
| Bcl-xL | gqerfnr wfltgmtvagvvllgslfs rk | 9.6/Hyd/11.0 | |||
|
| |||||
| (Orientation) | Peroxisome | Hyd | Cytosol | pI | |
| Signal-anchored peroxisome | Pex3p | rhkkk llfgtgviavsyavssfv snkl | 12.0/Hyd/8.5 | ||
|
| |||||
| (Orientation) | Cytosol | Hyd | Peroxisome | pI | |
| Tail-anchored peroxisome | pAPX | rskvmvkdst vlaqgavgvavaaavvilsyf yevrkrmk | 10.0/Hyd/10.3 | ||
The table shows the sequences that target signal- and tail-anchored proteins into the mitochondrial outer membrane and peroxisomal membrane. The Hyd of these proteins has been underlined and the protein orientation indicated. The pI corresponding to the flanking regions is respectively indicated. Positively charged residues are highlighted as bold letters. See text in Discussion for details and references.
A more detailed discussion of the similarities and differences between LD-sorting sequences and those for other destinations remains for future work. Unfortunately, even after years of extensive studies, a common role of the Hyd/Pos in supporting efficient and specific targeting of proteins to mitochondria or peroxisomes remains unclear (52). However, we note that the average permissive Hyd of LD-resident proteins is longer (approximately 30 residues) and more hydrophobic than the Hyd found in mitochondrial proteins (16–20 residues). In conclusion, targeting proteins into specific organelles is likely similar, and organized by complex and multiple mechanisms. More detailed studies in each case, combined with careful comparison between cases, should help understand how specificity is achieved.
Not all LD localization occurs at the same rate
While there is interest in how proteins localize to LDs, such localization has previously been viewed as an all-or-none process. However, frequently the cell achieves a distribution of a protein, where the protein localizes simultaneously to both LDs and other locations; the localization of many LD-bound proteins changes quite dramatically in time. To control the relative amounts of protein on the droplet versus other locations, or control the rate of accumulation on (or release from) the droplet, the affinity of the protein for LDs is clearly important. Until these studies, however, there has been no indication of how it might be possible to tune such affinities. We show that the number, spatial disposition and/or length of Pos-Seq can contribute to such affinity. Further, an intriguing conclusion of the analysis of the model protein is that the presence of Val residues, but not other hydrophobic amino acids, accelerates transport to LDs. In fact, a single C-terminal Val residue also accelerates the transport of inefficiently exported proteins from the ER, and hence operates as an export signal (56). The Sec24C/D components of the coat protein II (COPII) complex selectively mediate the export of these Val-tagged proteins (57). In the proteomic study where ALDI was identified, we also reported Sar-1 (a component of the COPII complex) as a major protein of hepatic LDs (11). Hence, it is possible that the LD-targeting sequences identified here assist during the interaction with coat proteins to finely regulate the formation/budding and functioning of LDs.
Concluding remarks
We demonstrate that the efficient sorting of proteins within the bulk of the ER to LDs is conducted by two cooperative/auxiliary motifs: the Hyd and at least one flanking Pos-Seq. Importantly, these findings relate to the proteins reaching the LD from the ER into a large group of diverse and functionally important proteins targeted to specific organelles by a similar sorting signal. Accordingly, we propose that the LD could be part of a complex membrane system organized by analogous but exclusive sorting mechanisms that finely preserve the identity and function of organelles such as the ER, mitochondria and peroxisomes. Further work is necessary to test the attractive possibility that this sorting signal is recognized by membrane-bound, cytosolic or lipidic receptors. Nevertheless, the significant progress achieved during the characterization of signal- and tail-anchored proteins provides a completely new perspective for studying the poorly understood cell biology of the multifunctional LD.
Methods
Plasmids
Mutant proteins were N-terminally tagged with the GFP (unless otherwise indicated). Standard YUX cloning, using primers containing EcoR1 (forward) and XmaI (backward) restriction sites, was used to synthesize GFP-CAV3DGV deltaPos, GFP-CAV3LLS and GFP-CAV3LLS deltaPos. Murine GFP-CAV2FLT was kindly provided by Dr Toyoshi Fujimoto (Nagoya, Japan) (27). The cDNA encoding mouse full-length CAV2 and its mutants were acquired from GenScript Corporation. These were subcloned after cutting with EcoR1 and Xma1 restriction enzymes into the pEGFPC2. Expression vector encoding the full-length mouse ALDI cDNA fused to the enhanced GFP has been previously described (11). Primers containing BspE1 (forward) or SmaI (backward) restriction sites were designed to clone different fragments and mutants of CAVs and ALDI into pEGFP-C1 by standard YUX recombinant techniques. The orange-tagged Hyd-Pos3 mutant was obtained following subcloning of the GFP-Hyd-Pos3 into m-Orange expression vector kindly provided by Dr Roger Y. Tsien (Howard Hughes Medical Institute). GFP-CAV3 and GFP-CAV3DGV have been previously described (29). The GFP-ADRP was kindly provided by Dr John McLauchlan (Institute of Virology, Glasgow). Sequences of the primers are available upon request. The theoretical isoelectric point (pI) was calculated following the method described by Gasteiger et al. [http://www.expasy.ch/tools/pi_tool.html; (58)].
Cell culture, transfections and fluorescence microscopy
Nile Red and BSA-conjugated arachidonic acid-linoleic acid-oleic acid (LOA; L-0163) was obtained from Sigma Chemical Co and Mowiol was from Calbiochem. COS-1 cells were maintained in DMEM supplemented with 10% v/v fetal calf serum, L-glutamine (2 mM), penicillin (50 U/mL) and streptomycin sulphate (50 μg/mL) (Biological Industries). Sixteen hours before transfection, cells were grown on glass coverslips and fusion proteins were transfected following the manufacturer's instructions for an additional 24 h using Qiagen Effectene Transfection Reagent (IZASA SA). The precise concentration of each construct was specifically adjusted to get a similar expression and transfection efficiency (see example in Figure 6). To promote the formation of LDs, cells were transfected and after 6 h incubated with a medium that contained 25 μL/mL of LOA for an additional 24 h. Under these experimental conditions, LDs were routinely observed by Nile Red staining and by bright field microscopy (36). Then cells were fixed in 4% paraformaldehyde (PFA) diluted in PBS for 1 h at room temperature. PFA-fixed cells were then washed twice in PBS and mounted in Mowiol. In some experiments, Nile Red was added to Mowiol 1/1000 (from saturated stock solution in acetone). The cells were observed using an oil immersion Plan-Apo63x/1.4 objective in an Axio-plan or Axiovert 200M Zeiss microscope (Zeiss). Images were captured with an AxioCam HRc camera and then digitally treated with AxioVision 3.1 software. Image analysis was performed with Adobe-Photoshop 5.5 software (Adobe Systems Inc). Rabbit anti-Sec61 alpha (07–204) was from Upstate Biotechnology, mouse anti-GM130 (610823) from BD Transduction Laboratories and anti-ADRP (610102) from Progen Biotechnik (Germany) and inmunofluorescence was performed exactly as described in (11).
Time-lapse video microscopy
Sixteen hours before transfection, cells were grown on glass coverslips and GFP-Hyd and orange-Hyd-Pos3 were transfected using the Qiagen Effectene Transfection Reagent. To accumulate proteins within the ER membranes, cells were transfected for 6 h and then serum starved for an additional 24 h. Under these experimental conditions, cells lacked LDs, as shown by Nile Red staining and by bright field microscopy (11). Under these experimental conditions, GFP-Hyd, orange-Hyd-Pos3 and GFP-ALDI were found mostly in the ER. Finally, 25 μL/mL of LOA was added to the cells to promote the formation of LDs. Images were then captured using a Leica TCS SL laser scanning confocal spectral microscope (Leica Microsystems Heidelberg GmbH), with argon and HeNe lasers attached to a Leica DMIRE2 inverted microscope equipped with an incubation system with temperature and CO2 control. Images were taken every 20 seconds for a total period of 4 h with a 63× oil immersion objective lens [numerical aperture (NA) 1.32], a 488-nm laser line, an appropriate excitation beam splitter and emission range detection and the confocal pinhole set at 2–3 Airy units to minimize changes in fluorescence efficiency due to proteins/structures moving away from the plane of focus.
Isolation of lipid droplets
Two 10-cm dishes of COS-1 cells were transfected as described above for 24 h (12 h in the indicated experiments) in a medium that contained 50 μL/mL of LOA. Next, unless the contrary is indicated, the cells were additionally treated for 12 h with 10 μg/mL of cycloheximide (Sigma Chemical Co). The cells were washed twice with cold PBS and resuspended in 500 μL of homogenization buffer [50 mmol/L Tris–HCl, pH 7.5; 150 mmol/L sodium chloride and 5 mmol/L ethylenediaminetetraacetic acid (EDTA)], supplemented with 10 μg/mL leupeptin and 10 μg/mL aprotinin. Cells were disrupted at 4°C by nitrogen cavitation in a cell disruption bomb (ref. 46,39, Parr Instrument Co) at 800 psi for 15 min and collected dropwise. Cells were in addition disrupted by passing though a 22-gauge needle 10× at 4°C. Nuclei and unbroken cells were removed by centrifugation at 1600 × g in a TLS-55 rotor (Beckman Coulter Inc) for 5 min at 4°C. The resulting supernatant (500 μL) was mixed with an equal volume of 2.5 M sucrose and loaded at the bottom of a discontinuous sucrose gradient formed by layers of 200 μL of 30, 25, 20, 15, 10 and 5% sucrose (w/v) freshly prepared in homogenization buffer. Gradients were centrifuged in a TLS-55 rotor at 166 000 × g for 3 h at 4°C and finally stopped without the brake. A total of five aliquots of 280 μL and one last aliquot (700 μL) containing the rest of the sample were collected from the top by using a Centritube Slicer (Beckman Coulter Inc). Finally, gel electrophoresis and western blotting were performed as described previously (11). The polyclonal anti-GFP was from Abcam, rabbit anti-Sec61 alpha (07–204) was from Upstate Biotechnology and the anti-ADRP (#610102) from Progen Biotechnik. The gradients shown in the figures are representative of at least two independent experiments.
Acknowledgments
A. P. is ICREA Professor at IDIBAPS and is supported by the grant (BFU2008-00345) from Ministerio de Ciencia e Innovación (Spain). R. G. P. is a Principal Research Fellow of the National Health and Medical Research Council (NHMRC) of Australia and this work was supported by an NHMRC Project grant. S. P. G. acknowledges support from the NIH, grant GM64624. C. E. is supported by grants (BFU2006-01151) and FT (BFU2006-15474) from MCI. T. G. is supported by the National Heart Foundation of Australia (G06S2559) and NHMRC project grants (510293, 510294). C.E. is supported by grants (BFU2006-01151 and V-2006-RET2008-O). We thank Dr Maria Calvo for help with the confocal microscopy (Unitat Microscopia Confocal, Serveis Cientificotècnics, Universitat de Barcelona-IDIBAPS) and Maria Molinos for technical assistance.
References
- 1.Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Ohsaki Y, Cheng J, Suzuki M, Shinohara Y, Fujita A, Fujimoto T. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim Biophys Acta. 2009;1791:399–407. doi: 10.1016/j.bbalip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 4.Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- 5.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 9.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 10.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 11.Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 12.Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- 13.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem (Tokyo) 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 16.Wu CC, Howell KE, Neville MC, Yates JR, III, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 17.Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. Faseb J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 20.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 21.Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 23.Chen JC, Tsai CC, Tzen JT. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 1999;40:1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490. [DOI] [PubMed] [Google Scholar]
- 24.Qu RD, Huang AH. Oleosin KD 18 on the surface of oil bodies in maize. Genomic and cDNA sequences and the deduced protein structure. J Biol Chem. 1990;265:2238–2243. [PubMed] [Google Scholar]
- 25.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci U S A. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem. 2002;277:4261–4270. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 27.Fujimoto T, Kogo H, Ishiguro K, Tauchi K, Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J Cell Biol. 2001;152:1079–1085. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostermeyer AG, Paci JM, Zeng Y, Lublin DM, Munro S, Brown DA. Accumulation of caveolin in the endoplasmic reticulum redirects the protein to lipid storage droplets. J Cell Biol. 2001;152:1071–1078. doi: 10.1083/jcb.152.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pol A, Luetterforst R, Lindsay M, Heino S, Ikonen E, Parton RG. A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J Cell Biol. 2001;152:1057–1070. doi: 10.1083/jcb.152.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci. 2008;121:1852–1860. doi: 10.1242/jcs.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Lay S, Hajduch E, Lindsay MR, Le Liepvre X, Thiele C, Ferre P, Parton RG, Kurzchalia T, Simons K, Dugail I. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 2006;7:549–561. doi: 10.1111/j.1600-0854.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 33.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 34.Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 35.Ostermeyer AG, Ramcharan LT, Zeng Y, Lublin DM, Brown DA. Role of the hydrophobic domain in targeting caveolin-1 to lipid droplets. J Cell Biol. 2004;164:69–78. doi: 10.1083/jcb.200303037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, Luetterforst R, Enrich C, Parton RG. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol Biol Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pol A, Martin S, Fernandez MA, Ingelmo-Torres M, Ferguson C, Enrich C, Parton RG. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. 2005;16:2091–2105. doi: 10.1091/mbc.E04-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez MA, Albor C, Ingelmo-Torres M, Nixon SJ, Ferguson C, Kurzchalia T, Tebar F, Enrich C, Parton RG, Pol A. Caveolin-1 is essential for liver regeneration. Science. 2006;313:1628–1632. doi: 10.1126/science.1130773. [DOI] [PubMed] [Google Scholar]
- 39.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 40.Hope RG, McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J Gen Virol. 2000;81:1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 41.Luetterforst R, Stang E, Zorzi N, Carozzi A, Way M, Parton RG. Molecular characterization of caveolin association with the Golgi complex: identification of a cis-Golgi targeting domain in the caveolin molecule. J Cell Biol. 1999;145:1443–1459. doi: 10.1083/jcb.145.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgese N, Francolini M, Snapp E. Endoplasmic reticulum architecture: structures in flux. Curr Opin Cell Biol. 2006;18:358–364. doi: 10.1016/j.ceb.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Listenberger LL, Brown DA. Lipid droplets. Curr Biol. 2008;18:R237–238. doi: 10.1016/j.cub.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian V, Garcia A, Sekowski A, Brasaemle DL. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J Lipid Res. 2004;45:1983–1991. doi: 10.1194/jlr.M400291-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Ohsaki Y, Maeda T, Maeda M, Tauchi-Sato K, Fujimoto T. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem Biophys Res Commun. 2006;347:279–287. doi: 10.1016/j.bbrc.2006.06.074. [DOI] [PubMed] [Google Scholar]
- 47.Abell BM, High S, Moloney MM. Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J Biol Chem. 2002;277:8602–8610. doi: 10.1074/jbc.M103712200. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura N, Fujimoto T. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem Biophys Res Commun. 2003;306:333–338. doi: 10.1016/s0006-291x(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 49.Cho SY, Park PJ, Lee JH, Kim JJ, Lee TR. Identification of the domains required for the localization of Prp19p to lipid droplets or the nucleus. Biochem Biophys Res Commun. 2007;364:844–849. doi: 10.1016/j.bbrc.2007.10.091. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem. 2006;281:14232–14240. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- 51.Shu X, Ryan RO, Forte TM. Intracellular lipid droplet targeting by apolipoprotein A-V requires the C-terminal and signal peptide segments. 2008;49:1670–1676. doi: 10.1194/jlr.M800111-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 53.Rapaport D. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaufmann T, Schlipf S, Sanz J, Neubert K, Stein R, Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki R, Sakamoto S, Tsutsumi T, Rikimaru A, Tanaka K, Shimoike T, Moriishi K, Iwasaki T, Mizumoto K, Matsuura Y, Miyamura T, Suzuki T. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79:1271–1281. doi: 10.1128/JVI.79.2.1271-1281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nufer O, Guldbrandsen S, Degen M, Kappeler F, Paccaud JP, Tani K, Hauri HP. Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J Cell Sci. 2002;115:619–628. doi: 10.1242/jcs.115.3.619. [DOI] [PubMed] [Google Scholar]
- 57.Wendeler MW, Paccaud JP, Hauri HP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Humana Press; New Jersey, USA: 2005. [Google Scholar]