Abstract
Protein prenylation occurs in the protozoan that causes African sleeping sickness (Trypanosoma brucei), and the protein farnesyltransferase appears to be a good target for developing drugs. We have cloned the α- and β-subunits of T. brucei protein farnesyltransferase (TB-PFT) using nucleic acid probes designed from partial amino acid sequences obtained from the enzyme purified from insect stage parasites. TB-PFT is expressed in both bloodstream and insect stage parasites. Enzymatically active TB-PFT was produced by heterologous expression in Escherichia coli. Compared with mammalian protein farnesyltransferases, TB-PFT contains a number of inserts of >25 residues in both subunits that reside on the surface of the enzyme in turns linking adjacent α-helices. Substrate specificity studies with a series of 20 peptides SSCALX (where X indicates a naturally occurring amino acid) show that the recombinant enzyme behaves identically to the native enzyme and displays distinct specificity compared with mammalian protein farnesyltransferase. TB-PFT prefers Gln and Met at the X position but not Ser, Thr, or Cys, which are good substrates for mammalian protein farnesyltransferase. A structural homology model of the active site of TB-PFT provides a basis for understanding structure-activity relations among substrates and CAAX mimetic inhibitors.
African trypanosomiasis occurs in 36 countries in sub-Saharan Africa and has resurged in recent years with as many as 300,000 annual cases. Human African trypanosomiasis is caused by Trypanosoma brucei gambiense or Trypanosoma brucei rhodesiense. The cattle form of trypanosomiasis, known as nagana, is caused by T. brucei brucei. Despite the long standing medical and veterinary importance of African trypanosomiasis, there has never been an ideal drug for treating these diseases. The arsenical melarsoprol was first introduced for human trypanosomiasis in the 1940s and remains standard therapy for infections that have invaded the central nervous system. Not surprisingly, this arsenical is extremely toxic. Drugs such as suramin and pentamidine do not eradicate central nervous system infection, and eflornithine lacks activity against T. brucei rhodesiense. All clinically useful drugs for human trypanosomiasis require parenteral administration and thus are impractical in areas where medical facilities are limited.
The need for better drugs to treat African trypanosomiasis has motivated the search for novel drug targets in T. brucei. We (2) and Field et al. (1) have described the occurrence of protein prenylation in T. brucei. Protein prenylation in mammals and yeast involves the attachment of 15-carbon farnesyl or 20-carbon geranylgeranyl groups to the C-terminal cysteine residues of a subset of cellular proteins. Many of these prenylated proteins are small GTPases, including Ras, Rac, Rab, and Rho, that have roles in cellular signal transduction and intracellular vesicle trafficking (3, 4). Other examples of prenylated proteins include the γ-subunits of heterotrimeric G proteins, nuclear lamins, multiple proteins in the phototransduction cascade, and viral antigens (5). The known functions of prenyl groups attached to cellular proteins is to anchor proteins to membranes and to serve as molecular handles for mediating protein-protein interactions (4).
Three enzymes have been identified in eukaryotic cells including those from mammals, plants, and yeast that attach prenyl groups to proteins: protein farnesyl transferase (PFT),1 protein geranylgeranyl transferase I (PGGT-I), and protein geranylgeranyl transferase II (PGGT-II) (6, 7). PFT transfers the farnesyl group from farnesyl pyrophosphate to the cysteine SH of the C-terminal protein sequence CAAX (is usually, but not necessarily, an aliphatic residue, and X is usually Ser, Met, Gln, or Cys). There is particular interest in PFT because selective inhibition of this enzyme suppresses transformation induced by oncogenic forms of Ras that are farnesylated and arrests the growth of human tumors in rodents (8–10). Over the past several years, hundreds of potent PFT inhibitors have been synthesized with the primary goal of developing anticancer drugs (11). This effort has afforded an excellent opportunity to couple drug development for human trypanosomiasis (see below) to work underway in cancer chemotherapy.
Protein prenylation in T. brucei was detected by showing that cells cultured with radiolabeled mevalonic acid, the precursor of prenyl groups, leads to radiolabeling of a specific set of proteins when analyzed by gel electrophoresis (1, 2). The cytosolic fraction of T. brucei was observed to contain PFT activity when assayed with the yeast RAS1 mutant protein with C-terminal sequence CVIM (2). Because sequences of CAAX-containing farnesyl acceptors for T. brucei were not known, we prepared mixtures of CAAX-containing peptides to find one that had high affinity for T. brucei PFT (12). The hexapeptide SSCALM bound T. brucei PFT with submicromolar affinity, and this peptide formed the basis for affinity chromatography of the parasite enzyme and overall ~60,000-fold purification. The enzyme (TB-PFT)1 is a heterodimer of subunits with apparent molecular masses of 61 and 65 kDa when analyzed by denaturing gel electrophoresis. The subunits of mammalian protein farnesyltransferases are considerably smaller with molecular masses of 48 and 46 kDa for the α- and β-subunits, respectively. A radiolabeled photoaffinity analog of farnesyl pyrophosphate bound the 61-kDa protein, strongly suggesting that this protein is the β-subunit of TB-PFT.
We observed that TB-PFT follows different substrate specificity rules than the mammalian homolog (12). For example, Ras proteins with C-terminal sequences CVLS and CVIM are good substrates for mammalian PFT, whereas only Ras-CVIM could be detectably farnesylated by the purified TB-PFT. This finding encouraged our efforts to identify compounds that would selectively inhibit the parasite enzyme and not the mammalian enzyme. When CAAX mimetics were tested against purified TB-PFT and mammalian PFT, they were found to have variable activity against the two homologs (12). Interestingly, these CAAX mimetics as well as farnesyl pyrophosphate analogs were consistently more toxic to live parasites than to mammalian cells, suggesting the possibility that inhibition of PFT is less tolerated by parasite cells (12, 13). It was also demonstrated that CAAX mimetics directly inhibited prenylation of specific protein in vivo as analyzed by SDS-PAGE (12).
In the present study, we subjected purified TB-PFT to tryptic digestion and peptide sequencing. The partial sequence data were used to obtain the full-length clones for the α- and β-subunits of parasite PFT. Recombinant TB-PFT, produced by heterologous expression of both subunits in Escherichia coli, was examined for its CAAX substrate specificity, and it was found to be very similar to the specificity of the enzyme isolated from parasites but substantially different than that of rat PFT. A three-dimensional homology model of TB-PFT was constructed based on the x-ray structure of rat PFT (14), and this provides structural insight for the unique CAAX and CAAX mimetic selectivity of TB-PFT.
EXPERIMENTAL PROCEDURES
Parasite Culture
The procyclic form of T. brucei brucei strain IsTar 1.7 was cultured in upright 75-cm2 tissue culture flasks (Corning Inc.) in SDM-79 medium containing penicillin (100 units/ml), streptomycin (100 µg/ml), and 10% heat inactivated fetal bovine serum (Atlanta Biologicals) at 27 °C with shaking (2). Bloodstream form T. brucei brucei, strain 427, was cultured in HMI-9 medium containing penicillin, streptomycin, and 10% fetal bovine serum in stationary flasks at 37 °C with 5% CO2 (13).
Purification of TB-PFT
Large scale purification of TB-PFT was carried out with T. brucei brucei EATRO 140 (procyclic form) cells from 50 liters of culture (about 1010 cells/liter) as described previously (12). Briefly, cells were lysed in 1 mm Tris-HCl, 1 mm EDTA, pH 8.0, and the cytosolic fraction was subjected to protein precipitation with 60% saturated ammonium sulfate at 0 °C. After dialysis against 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm DTT, the sample was loaded onto a column (4.8 × 24 cm) of Q-Sepharose (Amersham Pharmacia Biotech), and the protein was eluted with a 50–600 mm NaCl gradient in the same buffer. The fractions containing TB-PFT activity were pooled and subjected to hydrophobic interaction chromatography on a column (2.6 × 14 cm) of phenyl-Sepharose 6 (low substitution; Amersham Pharmacia Biotech). The enzyme solution was adjusted to 1.2 m KCl by adding 4 m KCl and then loaded on to the column. After extensive washing with 1.5 m KCl in the same buffer, elution was performed with a gradient of 1.5–0 m KCl in 20 mm Tris-HCl, 1 mm DTT, pH 7.4. SSCALM-Sepharose (prepared as described in Ref. 12) affinity chromatography was carried out at room temperature using a small Pasteur pipette column (0.9 ml of gel). TB-PFT-active fractions from phenyl-Sepharose were concentrated, and the buffer was replaced with 1 ml of 30 mm potassium phosphate, 1 mm DTT, 0.1% β-octylglucoside, pH 7.7, prior to application to the column. After washing with 60 bed volumes of the same buffer as above, elution of TB-PFT was carried out by washing successively with the same buffer containing 20, 40, 60, 80, 100, 150, and 300 mm NaCl. Most of the TB-PFT activity was eluted with 20–80 mm NaCl. The sample was concentrated to about 40 µl and subjected to gel filtration chromatography on a Superdex 200 HR10/30 (1 × 30 cm, Amersham Pharmacia Biotech) using ice-cold buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm DTT, 0.2% β-octylglucoside, pH 8.0). TB-PFT-active fractions (320 µunits) were combined and concentrated to 30 µl using a Microcon 10 (Amicon). A small portion of the sample (1 µl) was analyzed on SDS-PAGE, the purity of TB-PFT was estimated to be >90%, and the yield was about 2 µg of protein for each of the α- and β-subunits (estimated by comparison of the silver staining intensity with that of known amounts of recombinant rat PFT subunits) (15).
TB-PFT Assay
The standard assay for TB-PFT activity was performed with 5 µm RAS1-CVIM (12) and 0.75 µm (0.3 µCi) [3H]FPP (20 Ci/mmol; American Radiolabeled Chemicals), 30 mm potassium phosphate, 5 mm DTT, 0.5 mm MgCl2, and 20 µm ZnCl2, pH 7.7, in a total volume of 20 µl. After incubation at 30 °C for 15 min, the reaction was stopped by adding 200 µl of 10% HCl in ethanol. The amount of radioactivity transferred to the protein substrate was quantified by the glass fiber filter method (12) to give microunits (pmol/min product).
Microsequencing of TB-PFT Subunits
Purified TB-PFT was submitted to SDS-PAGE on a 10% gel, and the proteins were electrotransferred at 100 V for 1 h to a nitrocellulose membrane using ice-cold buffer (25 mm Tris, 192 mm glycine in 20% methanol, pH 8.3). The membrane was stained with 0.1% Amido Black in 45% methanol/10% acetic acid, destained with 10% acetic acid, rinsed with water, and then air-dried. The stained protein bands of TB-PFT α- and β-subunits (apparent masses of 65 and 61 kDa) on the membrane were cut out and submitted to microsequencing.
Membrane-bound protein was digested with trypsin in situ (16, 17), and peptides were fractionated by reverse phase HPLC (18) with the use of a 1-mm Reliasil C18 column (Column Engineering, Canada). Selected peak fractions were analyzed by a combination of automated Edman chemical sequencing (model 477A/120A from PE Biosystems, Foster City, CA) and matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (Reflex III from Bruker-Franzen, Bremen, Germany) (16, 19). Four and five peptide sequences were obtained from the 65- and 61-kDa gel bands, respectively (see Fig. 1).
FIG. 1. Amino acid sequence of TB-PFT α-subunit (A) and β-subunit (B).
The alignment of TB-PFT and rat PFT sequences was carried out as described in the text. Identities are in bold type, and similarities are underlined. Amino acid segments that were found by Edman sequencing of tryptic peptides are shown with lowercase letters. Large inserts (>25 residues) present in TB-PFT are shown (αI–αV and βI–βIII).
Molecular Cloning of TB-PFT α- and β-Subunits
The Institute for Genomic Research T. brucei data base was subjected to a BLAST search (tBlastN) with TB-PFT peptide sequences. Clones with high homology scores (see “Results”) were obtained from the Institute for Genomic Research. Sequencing of BAC clones was performed as recommended on the Institute for Genomic Research website. Rapid amplification of cDNA ends was used to clone the 5′-ends of the α- and β-subunits of PFT (20). First, mRNA was isolated from T. brucei procyclics using the QuickPrep Micro mRNA Purification kit (Amersham Pharmacia Biotech). The mRNA was reverse transcribed to first strand cDNA using Superscript II (Life Technologies, Inc.) and random primers. The cDNA was used as template for semi-nested PCR using one sense oligonucleotide to the conserved splice leader sequence of the mature mRNA (5′ -aacgctattattagaacagtttc-3′) and two antisense oligonucleotides to internal portions of the genes for which there was known sequence. For the PFT α-subunit, the following antisense oligonucleotides were used sequentially: 5′-gtatcaacttcgcctcttccgtg-3′ and 5′-gcgcttcgtgaccccgttactgc-3′. For the PFT β-subunit, the following antisense oligonucleotides were used sequentially: 5′-agttcccactcaagtcacggcc-3′ and 5′-ccatcaacccaccttcttccctg-3′. PCR products were gel purified, cloned into pGEM-T (Promega), and sequenced using Big Dye Terminator chemistry and an ABI 377 DNA sequencer (PE Biosystems).
RNA Blots
Total RNA was extracted from mid-logarithmic phase parasites using the UltraSpec RNA Isolation system (Biotecx Laboratories, Inc., Houston, TX). Approximately 4 µg of RNA was loaded per lane, electrophoresed on a 1% formaldehyde agarose gel, and then blotted to Hybond-N nylon membrane (Amersham Pharmacia Biotech). 32P-Labeled probes were prepared as follows. Full-length PFT α- and β-genes were amplified by PCR (as described above). The products were isolated from agarose gels and purified with the Genecleanp® III kit (Bio101, Vista, CA) and 32P-labeled by random priming (21). The T. brucei β-tubulin probe was prepared from the EcoRI-HindIII fragment of pTbαβT-1 (22). Again, the gel purified fragment was labeled by random priming. The blots were washed under high stringency conditions and analyzed by phosphorimaging.
Production of Antiserum to TB-PFT α-Subunit
The α-subunit was amplified by PCR from procyclic genomic DNA using the sense oligonucleotide, 5′-aggagatctagtactatgaataagagcgcggttcgtag-3′, and the antisense oligonucleotide, 5′-ggataagctttcaaacatagtcccggttc-3′. The resulting PCR fragment were designed to include the BglII and HindIII restriction sites at its 5′- and 3′-ends, respectively, which allowed for directional cloning into the same restriction sites of the expression vector pRSETB (Invitrogen, Carlsbad, CA). This expression vector produces a fusion protein containing a six-histidine tag attached to the N terminus of the expressed protein. Several clones of the new plasmid, pRSETB-TbPFTα, were sequenced with the T7 promoter primer (Amersham Pharmacia Biotech) to verify the proper assembly of the construct. DNA from one clone was transformed into E. coli strain BL21 (DE3) pLysS, and transformants were obtained with ampicillin and chloramphenicol selection. A 0.5-liter culture was grown to a cell density of A600 = 0.5, and protein was induced with 1 mm IPTG at 37 °C. After 4 h the cells were centrifuged, sonicated, and analyzed by SDS-PAGE. The recombinant fusion protein was purified by nickel chromatography (23).
The purified protein was used to produce polyclonal antiserum in a single rabbit (Cocalico Biologicals). The material was injected with Freund’s adjuvant using the following schedule: 100 µg (day 1), 50 µg (day 14), 50 µg (day 21), and 50 µg (day 49). Test bleeds were obtained on days 35 and 56, and exsanguination was on day 91.
Expression Vector for TB-PFT
The α-subunit of TB-PFT was amplified by PCR from procyclic genomic DNA using the following sense and antisense oligonucleotides (5′-cgggatcctttggaggtaaaataaatgaataaaagcgcggttcgtagtgaagaaagccgc) and (5′-gaagatcttcatatgttaaaactcctcaacgtagtcgcggttcataacg). The high fidelity polymerase, Pwo (Roche Molecular Biochemicals), was used, which produces blunt end PCR products. The ~1800-base pair PCR product was ligated into the vector, pCR-Blunt II TOPO (Invitrogen). The PFT β-subunit was similarly amplified using the oligos (5′-atacttccatatgtcttttgtaccacctccagc) and (5′-gaagatcttcaacacataaaagtctttgcgc). The ~1800-base pair product was also cloned into the pCR-Blunt II TOPO vector. Next the α-subunit gene was excised from its vector with BamHI and NdeI and gel purified. This fragment was cloned into the BamHI/NdeI sites immediately upstream of the β-subunit in the vector containing the β-subunit. The resulting junction between the α- and β-subunit genes was composed of the following DNA sequence: PFTα-GTTGAGGAGTTTTAACATATGTCTTTT-PFTβ). This junction provided a Glu-Glu-Phe epitope tag on the extreme C terminus of the translated α-fragment. Within the DNA sequence encoding this tag, GAGGAGTTT, is a ribosomal binding site (GGAG) nine bases upstream of the start ATG of the PFT-β gene. The resulting plasmid was digested with BamHI and Bgl2 to release a ~ 3600-base pair fragment containing the α-and β-genes in tandem. The expression plasmid, pRD578 (containing the human PGGT-1 subunit genes in tandem) was kindly provided by C. Omer (Merck) (24). This expression plasmid is driven by the T7 ribosomal promoter. The human genes were removed from the vector by digesting with BamHI, and the 4.6-kb plasmid fragment was gel purified. The 3600-base pair TB-PFT α-β fragment was ligated into the BamHI site of the prepared expression plasmid, thus replacing the human PGGT-1 genes. Clones were identified with proper orientation of the insert and were designated pRD578-TbPFT. Two clones that contained α-subunits generated in independent PCR reactions and β-subunits generated in independent PCR reactions were sequenced.
Heterologous Expression of TB-PFT and Immunoblotting
pRD578-TbPFT was transformed into the E. coli strain BL21 pLysS, and clones were selected with ampicillin and chloramphenicol. Bacteria were grown, and protein was induced as described in the legend to Fig. 6.
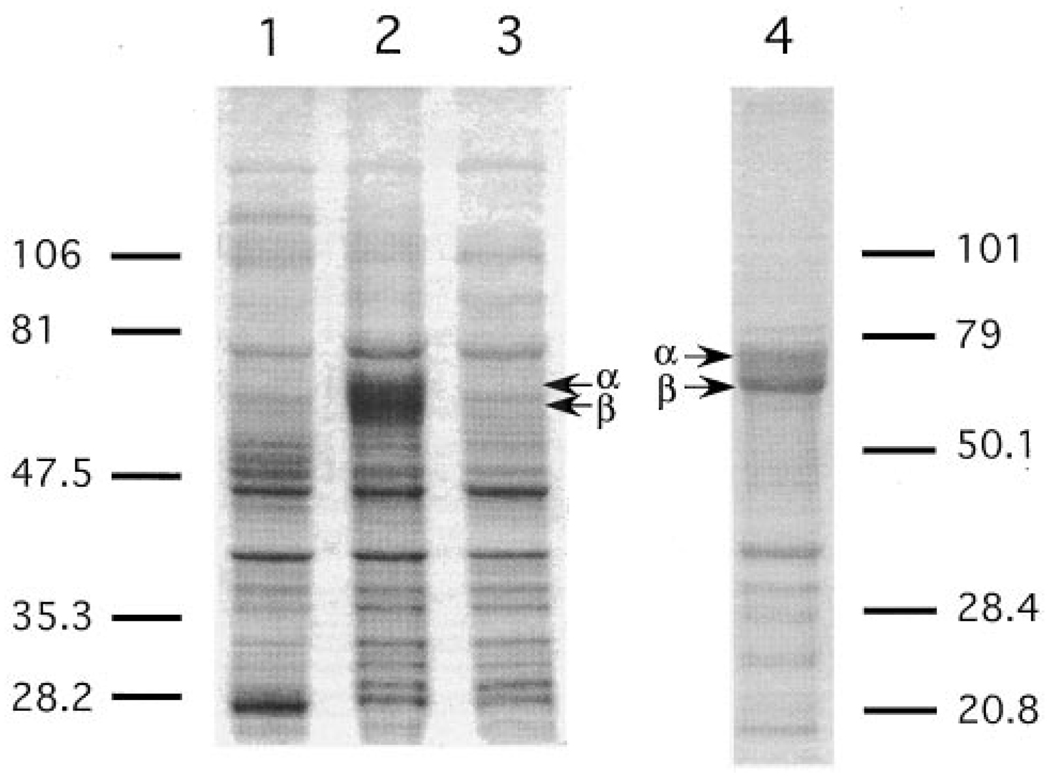
FIG. 6. Expression of TB-PFT in E. coli.
E. coli BL21 pLysS was cultured in 5 ml of medium at 37 °C. Cells were collected before or after induction with 0.4 mm IPTG at room temperature for 2 h. Cell pellets were treated with 100 µl of Laemmli sample buffer, and sample aliquots (1.5 µl for lanes 1 and 2, and 4 µl for lane 3) were subjected to SDS-PAGE analysis using a 10% gel. Protein bands were visualized with Coomassie Blue staining. Lane 1, untransformed E. coli; lane 2, E. coli transformed with pRD578-TbPFT expressing T. brucei α- and β-subunits that was harvested after IPTG induction for 2 h; lane 3, same as lane 2 but without IPTG induction. Lane 4 was from a separate gel with different molecular mass markers and shows protein from the 120,000 × g pellet obtained from a lysate of E. coli expressing TB-PFT.
To measure enzymatic activity of recombinant TB-PFT, expression was induced at room temperature for 30 min with 0.4 mm IPTG. E. coli from a 5-ml culture was harvested and sonicated at 0 °C using a microtip (10 s ×10 times) in 0.6 ml of lysis buffer (5 mm sodium phosphate, pH 8.0, 75 mm NaCl, and 5 mm DTT, containing protease inhibitors, 0.5 mm phenylmethylsulfonyl fluoride, 2 µg/ml each of anti-pain, leupeptin, and pepstatin A, and 10 µg/ml of aprotinin). The homogenate was centrifuged at 120,000 × g for 1 h at 4 °C. The resulting supernatant (cytosolic fraction) contains typically 400–500 µg of total protein and was used as a source of recombinant TB-PFT.
For immunoblot analysis, E. coli total protein were resolved on a 10% Laemmli gel and electrotransferred to a nitrocellulose membrane. The standard immunoblot analysis was carried out using nonfat milk as a blocking agent, a 1/2000 dilution of the anti-TB-PFT-α antiserum, and the ECL detection method (Amersham Pharmacia Biotech) using a 1/1,000 dilution of the secondary antibody.
Substrate Specificity Studies with SSCALX Peptides
A mixture of peptides of the type SSCALX where X is all 20 naturally occurring amino acids was prepared by SynPep (Dublin, CA). A mixture of 20 solid phase synthesis resins, each loaded with a different amino acid, was placed in the reaction cup, and standard solid phase peptide extension was carried out to add the SSCAL segment (Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry). The peptide mixture was fractionated by C18 reverse phase HPLC (Vydac 218TP1010 column) using solvent gradients of water/0.06% trifluoroacetic acid to acetonitrile/0.06% trifluoroacetic acid. Partially purified peptides were repurified using appropriate shallower gradients. By this method, 11 of the 20 peptides (X = Gly, Ala, Ser, Thr, Cys, Asn, Trp, Pro, Lys, Arg, or Asp) were obtained in pure form (confirmed by electrospray mass spectrometry). The remaining 9 peptides, which could not be obtained in pure form, were individually synthesized (United Biochemicals Research, Seattle, WA) and purified by HPLC. Peptides were dissolved in water, and concentrations were determined by assay for SH groups with Ellman’s reagent. Peptides containing N-terminal Biotin-CONH(CH2)5CO were prepared as described (13).
Reaction mixtures contained 10 µm each of the 20 different SSCALX peptides and 0.75 µm (0.3 µCi) [3H]FPP under standard assay conditions (see above) and were incubated at 30 °C for 30 min (or 15 min for rat PFT reactions) and then terminated by the addition of 200 µl of methanol. The mixture was applied to an Spe + anion exchange column (J. T. Baker) to remove [3H]FPP. The flow-through and methanol washes (200 µl × 6 times) were combined and counted for radioactivity to determine the amount of [3H]farnesylated peptides as described previously (25). Other conditions are given in the legend to Fig. 7. Formation of radioactive farnesylated peptides was confirmed by analyzing the samples on silica gel thin layer chromatography in a solvent of 1-propanol/NH4OH/H2O (6:3:1, v/v/v) followed by fluorography as described previously (25) (not shown). For acidic peptides (X = Asp and Glu), the methanol extract from the reaction mixture was directly applied to the thin layer chromatography plate to detect the radioactive peptide spots.
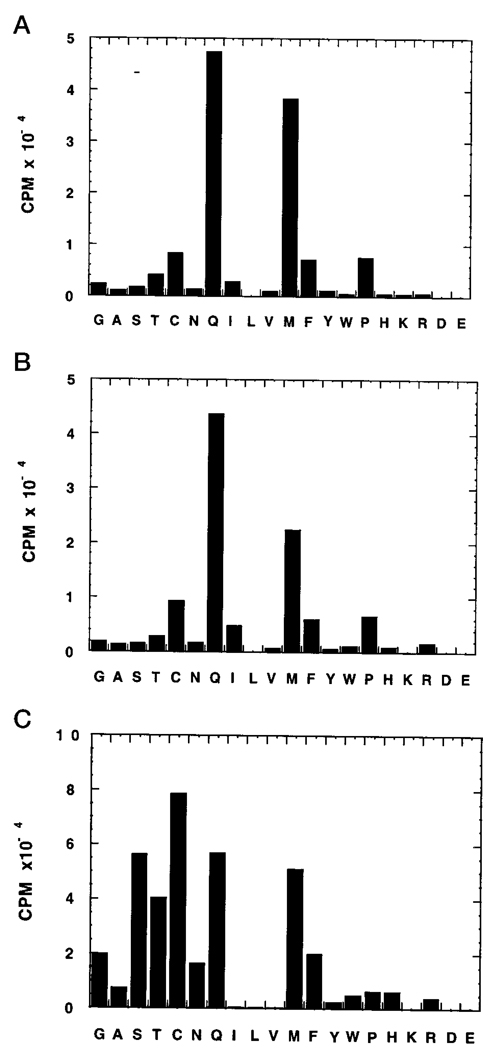
FIG. 7. CAAX specificity of TB-PFT.
Purified TB-PFT (0.025 microunits) from procyclic parasites (A) or recombinant (about 3 µg of cytosolic protein from E. coli transformed with pRD578-TbPFT) (B) was incubated with 10 µm each of the 20 different SSCALX peptides and 0.75 µm (0.3 µCi) [3H]FPP for 30 min at 30 °C under standard assay conditions. Results are expressed as the radioactivity above the level measured with the no peptide control (6,000–10,000 cpm). For comparison, 0.05 µg of recombinant rat PFT (prepared by expression in the baculovirus/Sf9 cell system) (C) was also incubated with the peptides for 15 min at 30 °C. Based on replicate analyses, estimated errors are <15% for all cpm values.
RESULTS AND DISCUSSION
Cloning of TB-PFTα- and β-Subunits
Four peptide sequences, shown in Fig. 1A, were obtained from the purified parasite protein 65-kDa gel band and were used to search parasite DNA data bases for homologous DNA sequence. The 15-amino acid peptide, SQFLLDNTHQVYTAR, matched perfectly to a translated portion of end sequence from BAC clone 26E18 in the Institute for Genomic Research data base. The deposited sequence contained 513 bases and, when translated, was found to be homologous to a portion of the middle segment of known PFT α-subunits. This BAC clone was obtained from the Institute for Genomic Research and directly sequenced to determine the remaining 3′-end of the gene. Because the BAC clone did not contain the 5′-end of the putative PFT α-subunit, the remaining portion of the gene was amplified with rapid amplification of cDNA ends. The procedure produced a single fragment of approximately 0.8 kb that was cloned and sequenced. This fragment contained an ATG 63 bases downstream of the miniexon splice site, and this ATG was assumed to represent the start codon of the gene (26). A contig of the above fragments was compiled to reveal an open reading frame of 1839 bases.
Five peptide sequences (Fig. 1B) obtained from the purified parasite protein 61-kDa gel band were used to search the T. brucei genomic data bases. The 16-amino acid peptide, NCLRPTNPIFNINQSK, matched perfectly to a translated portion of clone 28H5 in the Institute for Genomic Research data base. The identified sequence of 699 bases was from a library of sheared T. brucei DNA. This data base sequence contained the whole 3′-end of the gene, but it did not contain the 5′ portion. The 5′ fragment of the gene was amplified with the rapid amplification of cDNA ends method, as above. The putative start ATG was located 78 bases downstream from the miniexon splice site. The contig of overlapping sequences gave an open reading frame of 1788 bases. The GenBank™ accession numbers are AF230368 for the α-subunit and AF230369 for the β-subunit.
Expression of TB-PFT in Parasites
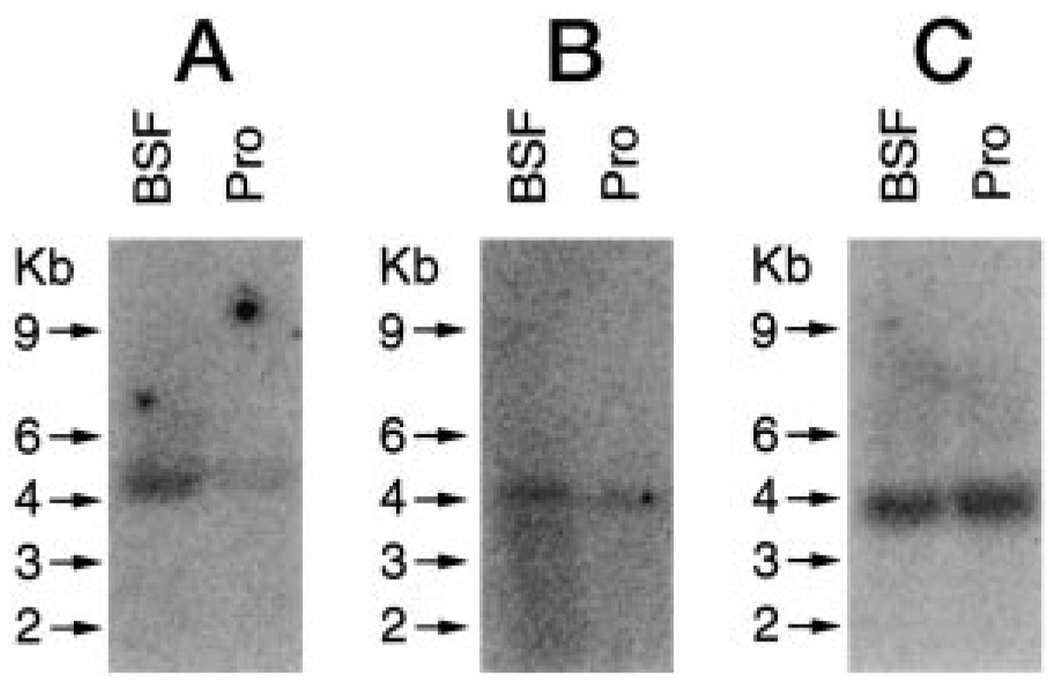
DNA probes for the α- and β-subunits of TB-PFT were specifically hybridized to RNA isolated from both procyclic and bloodstream forms of T. brucei (Fig. 2). The mRNA for the α-subunit migrated slightly above the 4-kb standard, and the mRNA for the β-subunit migrated slightly below the 4-kb standard. The level of mRNA expression for both TB-PFT subunits was moderately greater in bloodstream form T. brucei than in procyclic form. Because protein expression in trypanosomes is primarily controlled by steady state RNA levels, these data indicate that TB-PFT is expressed in both life-cycle stages of T. brucei. This evidence that TB-PFT is expressed in the bloodstream form of T. brucei underscores the potential to target this enzyme for therapeutic purposes in human and animals.
FIG. 2. Northern blot analysis of bloodstream form (BSF) and procyclic (Pro) T. brucei.
Total RNA was electrophoresed, blotted onto a nylon membrane, and hybridized with a 32P-labeled probe to TB-PFT α-subunit (A). The blot was stripped and probed for TB-PFT β-subunit (B). The membrane was stripped again and probed for β-tubulin to demonstrate equivalent loading (C). Transcript sizes are marked in kilobases (Kb).
Comparison of TB-PFT and Rat PFT
The amino acid sequences of the α- and β-subunits of rat PFT and TB-PFT were aligned using ALIGN2D of the MODELER package (Molecular Simulations Inc.) (27) followed by minor manual adjustments (Fig. 1). Because TB-PFT subunits contain a number of large insertions, use was made of the x-ray structure of rat PFT (Protein Data Bank identification number 1qbq.pdb) (14, 28) to evaluate the alignment of protein core residues. In the automated alignment procedure, core residues including the repetitive motif QX13NX3WX2R in the α-subunit, which provides residues that pack adjacent α-helices together, as well as the repetitive motif PXNY in the same subunit that resides in the turns joining adjacent α-helices and that pack against the β-subunit (14, 28), were recognized and matched. In addition, Phe and Tyr residues of the β-subunit that pack against each other in the central core of the protein (14, 28) were also aligned automatically. Because TB-PFT subunits contain a number of large insertions, use was made of the x-ray structure of rat PFT to make minor manual adjustments at the insertion termini.
After removing the large insertions in TB-PFT (>25 residues), the sequence identity between the core regions of TB-PFT and rat PFT is 21% for the α-subunit and 36% for the β-subunit. A homology model of the TB-PFT α- and β-subunits was constructed using the appropriate regions of the x-ray structure of rat PFT α- and β-subunits as templates and the alignments shown in Fig. 1 as templates. The amino acid conformations for the core of the individual subunits were built using MODELER.
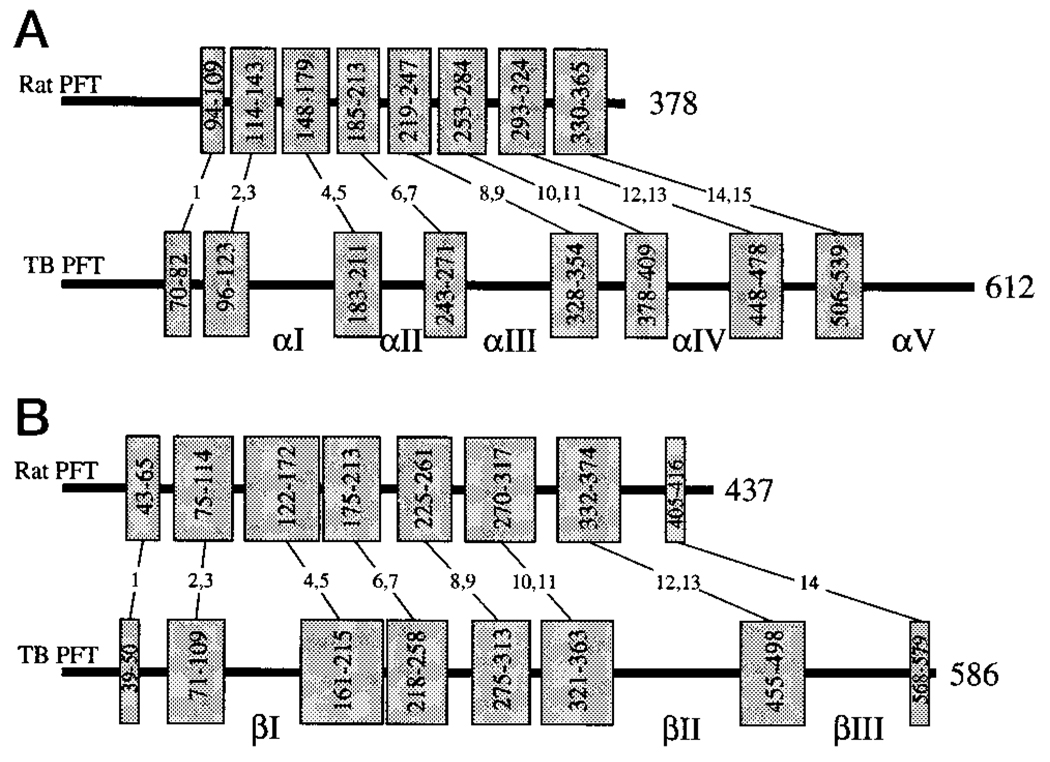
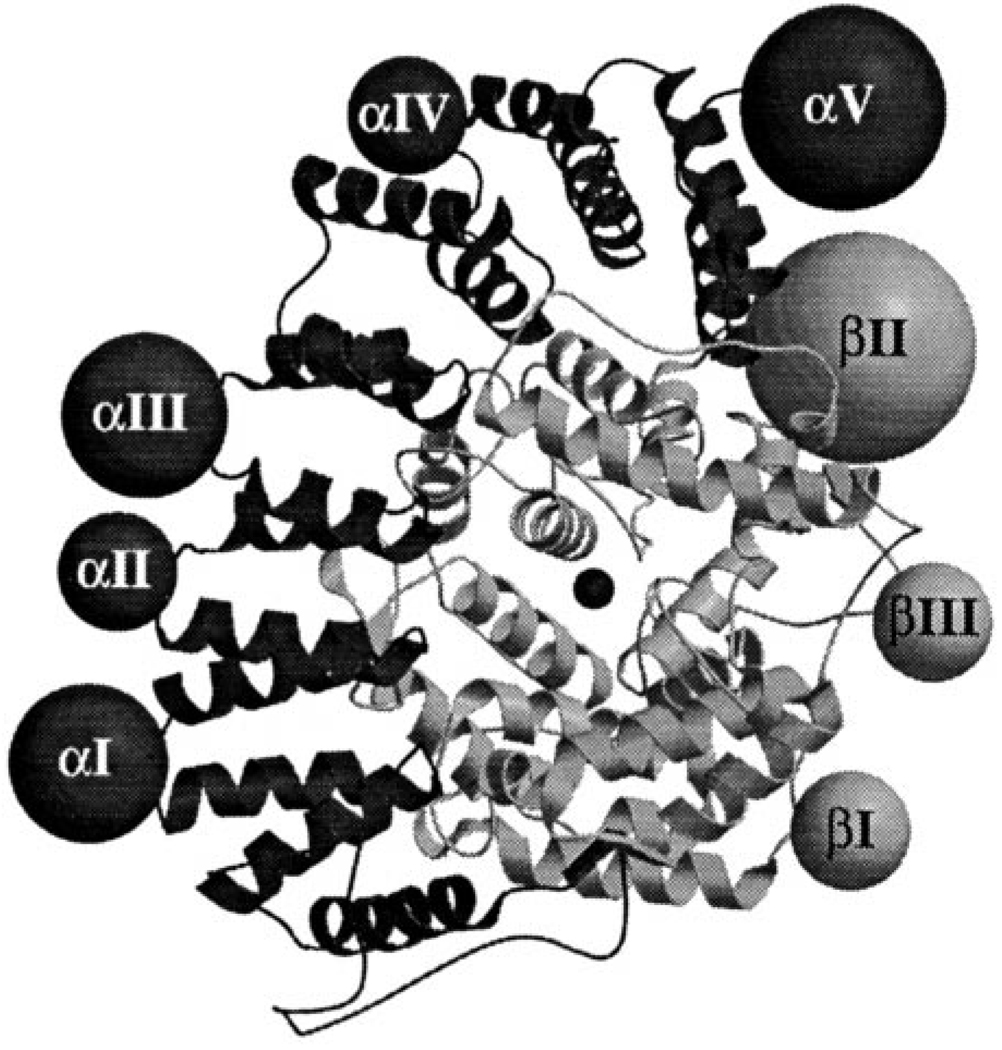
The homology model and predicted locations of the insertions in the structure are shown in Fig. 3. The sequence alignments show that there are eight major insertions in TB-PFT and no major deletions (Fig. 1). The α-subunit contains five insertions (αI- αV) ranging from 26 to 71 amino acids in length, whereas the β-subunit has only three insertions (βI–βIII) ranging from 39 to 85 residues. Fig. 3 shows the relationship between the secondary structure elements in rat PFT versus TB-PFT and the location of the TB-PFT insertions. These insertions result in α- and β-subunits that are 234 and 149 residues larger than those in rat PFT, respectively. The tertiary structure of both PFT subunits is built around a core of six or seven pairs of anti-parallel α-helices. Each of the eight insertions fall in the loops connecting the pairs of helices and not within the helical pairs themselves (Fig. 4). All of these insertions occur in loops that are on the surface of the enzyme rather than at the interface between subunits or near the active site.
FIG. 3. Sequence comparison of rat PFT and TB-PFT α- (A) and β-subunits (B).
The thick bars indicate the full amino acid sequence. The α-helices are represented by the shaded boxes, with the paired helices underlying the tertiary structure combined for clarity. The starting and ending residue of each helix or pair of helices is shown in the box. The helices are numbered (1–15 α-subunit; 1–14 β-subunit) as described previously (14). Insertions greater than 25 residues in TB-PFT relative to rat PFT are numbered (αI–αV and βI–βIII).
FIG. 4. Structural comparison of rat PFT and TB-PFT.
A ribbon diagram of rat PFT α-subunit (dark shading) and β-subunit (light shading) is shown. The active site is marked by the catalytic zinc in the center of the figure. The eight large TB-PFT insertions positioned relative to rat PFT are shown as spheres (larger diameter for larger inserts) and labeled according to Fig. 3.
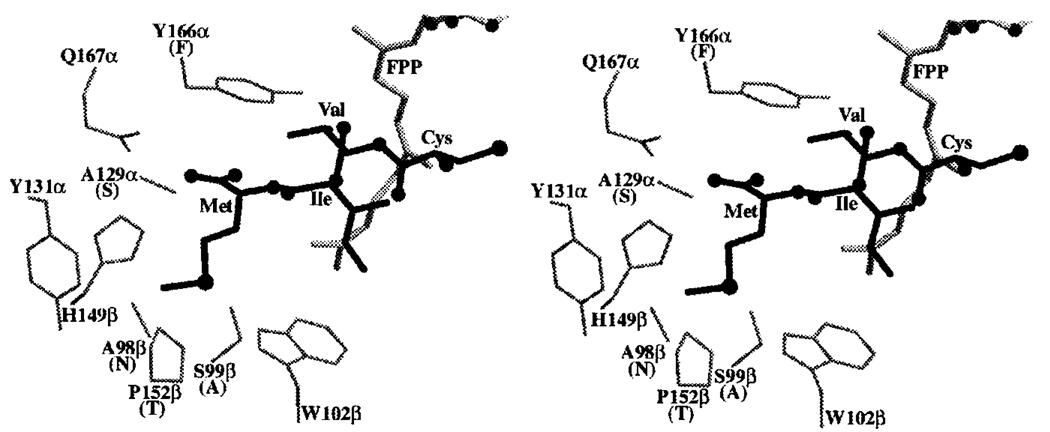
Rat PFT contains a hydrophobic cavity that accommodates FPP substrate (14, 28, 29). All of the residues that line this cavity are identical in rat PFT and TB-PFT except for the modest change of Gly-250 in the β-subunit of rat PFT being replaced by Ala in TB-PFT. The x-ray structure of rat PFT containing bound CVIM tetrapeptide (28) identifies the residues that are in direct contact with the CAAX substrate. All residues surrounding the CVI portion of CVIM in rat PFT and TB-PFT are identical except for the modest change of Tyr-166 in the α-subunit of rat PFT being replaced by Phe in TB-PFT. The hydroxyl group of Tyr-166 does not engage in hydrogen bonding to CVI (28), and so replacement by Phe should have no effect on CAAX binding. Interestingly, significant amino acid changes between rat PFT and TB-PFT occur in the portion of the active site that contacts the Met residue of CVIM. Fig. 5 shows the portion of rat PFT surrounding this Met residue. In TB-PFT, Ala-129 of the α-subunit of rat PFT is replaced by Ser, and Ser-99 and Pro-152 of the β-subunit of rat PFT are replaced by Ala and Thr, respectively. The Ala-129 to Ser change probably has little effect on CAAX binding because both side chains are probably too far from Met to contact it. Replacement of Pro-152 with Thr still allows the Met to bind, because both side chains would be in van der Waals’ contact with Met. The most significant change appears to be the replacement of Ser-99 with Ala. This change is expected to deepen the pocket for the X residue of CAAX in TB-PFT versus rat PFT (Fig. 5). In particular, this substitution may allow amino acid side chains with branching at the γ-carbon to bind to the X pocket.
FIG. 5. X-ray structure of rat PFT residues in the vicinity of the Met residue of bound CVIM (28).
Stereo diagram showing enzyme-bound CVIM (bold sticks) and rat PFT α- and β-subunit residues close to the Met of CVIM as well as Tyr-166α (see text for a discussion of these residues). Also shown is bound FPP (gray sticks, based on the x-ray structure of the ternary complex of rat PFT with CVIM and a hydroxyphosphonate FPP analog).
Heterologous Expression of Enzymatically Active TB-PFT in E. coli and Immunoblot Analysis
The TB-PFT α- and β-subunits were cloned in tandem into the expression vector pRD578 (24) to give pRD578-TbPFT. As shown in Fig. 6, induction of protein expression with IPTG leads to the appearance of two new protein bands, when analyzed by SDS-polyacrylamide electrophoresis. These bands were not seen in extracts from nontransformed E. coli or from transformed E. coli in the absence of IPTG (Fig. 6). These bands co-migrate with those of the native PFT subunits isolated from T. brucei (data not shown and Ref. 12). This result argues that the correct translation start sites were identified in the cloned genes for TB-PFT α- and β-subunits. The slower migrating gel band is selectively detected by immunoblot analysis with antiserum raised against recombinant α-subunit (not shown). This result is consistent with our earlier demonstration that the faster migrating TB-PFT subunit band is the β-subunit because it is selectively labeled by a photoaffinity analog of FPP (the FPP binding cavity resides in the β-subunit) (12, 14).
A crude extract (cytosolic fraction) from E. coli transformed with pRD578-TbPFT showed significant PFT catalytic activity (specific activity of 16 microunits/mg protein using RAS-CVIM as substrate, whereas no detectable activity was measured using extracts from nontransformed E. coli or from transformed bacteria in the absence of IPTG). Enzymatic activity reached the highest level 30 min after induction with 0.4 mm IPTG at room temperature; thereafter the activity declined accompanied by increased amounts of an insoluble form of TB-PFT. Additional studies show that most of the expressed TB-PFT is found in the insoluble fraction of the E. coli lysate (Fig. 6) but that >90% of the enzymatic activity is in the soluble fraction.
CAAX Specificity Studies
We have previously reported that TB-PFT displays distinct CAAX substrate specificity from that of the mammalian enzyme. Whereas Ha-Ras with C terminus CVLS and RAS-CVIM are good substrates for rat PFT, only RAS-CVIM is farnesylated by TB-PFT (2). In the present study, we systematically investigated the recognition of the X residue of CAAX by TB-PFT using a series of 20 peptides SSCALX (X = all 20 naturally occurring amino acids). As shown in Fig. 7A, the peptides where X = Met and Gln are greatly preferred by TB-PFT. Formation of radio-farnesylated peptide separated from [3H]FPP by anion exchange chromatography was confirmed by thin layer chromatography followed by fluorography (not shown). The peptides where X = Cys, Phe, and Pro gave small but significant activity above the level of the no peptide control. Peptides with other X residues were farnesylated poorly by TB-PFT (Fig. 7A). As shown in Fig. 7B, the CAAX specificity of TB-PFT produced in E. coli is virtually identical to that of native TB-PFT isolated from parasites. This argues that the genes for TB-PFT cloned in this study code for the same protein isolated from T. brucei procylic cells. As shown in Fig. 7C, rat PFT behaves very differently than TB-PFT with respect to X residue recognition. The hydrophilic X residues including Ser, Thr, Cys, and Asn are well tolerated by rat PFT but poorly recognized by TB-PFT. This may be due to the predicted increased hydrophobicity of the X binding pocket in TB-PFT versus rat PFT (Ser-99 goes to Ala). The greater ability of TB-PFT compared with rat PFT to accept X = Q may be due to the Ser-99 to Ala change that is predicted to allow γ-carbon branching on the X residue side chain (as described above).
Interestingly, the N-terminally biotinylated peptide NPFREKKFFCAIL (γ-subunit of human trimeric G protein) where X = Leu is farnesylated by TB-PFT as efficiently as Ha-Ras-CVIM (2), whereas mammalian PFT farnesylates this peptide poorly (25). Thus, it was surprising to find that SS-CALL is a poor substrate for TB-PFT (Fig. 7). This suggests that enzyme recognition of the X residue can be modulated by changes in the internal residues (a of CAAX). The only T. brucei CAAX-containing protein that we have been able to find in the genome data bases is a small GTP-binding protein with homology to Ras and Rap proteins (GenBank™ accession number AJ002963). A dodecapeptide Biotin-CONH(CH2)5CONH-ANRKKKSGCTML patterned after the GTP-binding protein was found to be a good substrate for TB-PFT, comparable with RAS-CVIM when tested at 5 µm. This again shows that CAAX peptides with a C-terminal leucine can be good substrates for TB-PFT, which is surprising given that X = Leu specifies gerangeranylation by PGGT-I in mammals (6).
Implications of the TB-PFT Amino Acid Sequence and Homology Model for Design of CAAX Mimetic Inhibitors
In our previous study, we found that the CAAX mimetic FTI-276 where X = Met inhibits rat PFT and TB-PFT with comparable affinities (12). This is expected based on the suggestion that Met should be well tolerated in the X binding site of TB-PFT, even with the residue changes (Fig. 7), and residues in contact with portions of FTI-276 other than Met are identical in the two enzymes. As noted above, replacing Ser-99 of rat PFT with Ala in TB-PFT may allow for γ-carbon branching of the side chain X residue of CAAX mimetics. This may explain why the CAAX mimetic where X = Leu, GGTI-297, displays virtually identical potency as does FTI-276 (X = Met) against TB-PFT, but GGTI-297 is 10-fold less active than FTI-276 against rat PFT (12). The homology structural model of TB-PFT described here provides an initial framework for structure-based design of parasite-selective PFT inhibitors. Expression of active recombinant TB-PFT may provide quantities of enzyme suitable for x-ray structural analysis.
Acknowledgments
We are grateful to Ken Stuart for providing large volume parasite cultures. We thank Lynn Barrett and Martin Sadilek for excellent technical assistance and Caroline Cameron for many helpful discussions.
Footnotes
This work was supported by National Institutes of Health Grant CA52874 (to M. H. G.) and Core Grant P30 CA08748 (to P. T.). This work was supported in part by NIAID, National Institutes of Health Grant AI43062 and National Science Foundation Chemistry Instrumentation Grant CHE 9807748. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: PFT, protein farnesyl transferase; PGGT, protein geranylgeranyl transferase; FPP, farnesyl pyrophosphate; DTT, dithiothreitol; IPTG, isopropyl β-d-thiogalactopyranoside; TB-PFT, PFT from T. brucei; PAGE, polyacrylamide gel electrophoresis; HPLC, high pressure liquid chromatography; PCR, polymerase chain reaction; kb, kilobase(s); contig, group of overlapping clones.
REFERENCES
- 1.Field H, Blench I, Croft S, Field MC. Mol. Biochem. Parasitol. 1996;82:67–80. doi: 10.1016/0166-6851(96)02723-5. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama K, Lin Y, Stuart KD, Gelb MH. Mol. Biochem. Parasitol. 1997;87:61–69. doi: 10.1016/s0166-6851(97)00043-1. [DOI] [PubMed] [Google Scholar]
- 3.Glomset JA, Gelb MH, Farnsworth CC. Trends Biochem. Sci. 1990;15:139–142. doi: 10.1016/0968-0004(90)90213-u. [DOI] [PubMed] [Google Scholar]
- 4.Glomset JA, Farnsworth CC. Annu. Rev. Cell Biol. 1994;10:181–205. doi: 10.1146/annurev.cb.10.110194.001145. [DOI] [PubMed] [Google Scholar]
- 5.Clarke S. Annu. Rev. Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset J, Gelb MH. Biochem. Soc. Trans. 1992;20:479–484. doi: 10.1042/bst0200489. [DOI] [PubMed] [Google Scholar]
- 7.Casey PJ, Seabra MC. J. Biol. Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 8.Njoroge FG, Doll RJ, Vibulbhan B, Alvarez CS, Bishop WR, Petrin J, Kirschmeier P, Carruthers NI, Wong JK, Albanese MM, Piwinski JJ, Catino J, Girijavallabhan V, Ganguly AK. Bioorg. Med. Chem. 1997;5:101–113. doi: 10.1016/s0968-0896(96)00206-4. [DOI] [PubMed] [Google Scholar]
- 9.Koblan KS, Kohl NE, Omer CA, Anthony NJ, Conner MW, deSolms SJ, Williams TM, Graham SL, Hartman GD, Oliff A, Gibbs JB. Biochem. Soc. Trans. 1996;24:688–692. doi: 10.1042/bst0240688. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Qian Y, Hamilton AD, Sebti SM. Oncogene. 1998;16:1467–1473. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- 11.Leonard DM. J. Med. Chem. 1997;40:2971–2990. doi: 10.1021/jm970226l. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama K, Trobridge P, Buckner FS, Van Voorhis WC, Stuart KD, Gelb MH. J. Biol. Chem. 1998;273:26497–26505. doi: 10.1074/jbc.273.41.26497. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama K, Trobridge P, Buckner FS, Scholten J, Stuart KD, Van Voorhis WC, Gelb MH. Mol. Biochem. Parasitol. 1998;94:87–97. doi: 10.1016/s0166-6851(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 14.Park H-W, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama K, Zimmerman KZ, Scholten JD, Gelb MH. J. Biol. Chem. 1997;272:3944–3952. doi: 10.1074/jbc.272.7.3944. [DOI] [PubMed] [Google Scholar]
- 16.Erdjument-Bromage H, Lui M, Sabatini DM, Snyder SH, Tempst P. Protein Sci. 1994;3:2435–2446. doi: 10.1002/pro.5560031227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lui M, Tempst P, Erdjument-Bromage H. Anal. Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 18.Elicone C, Lui M, Geromanos S, Erdjument-Bromage H, Tempst P. J. Chromatogr. 1994;676:121–137. doi: 10.1016/0021-9673(94)00089-1. [DOI] [PubMed] [Google Scholar]
- 19.Tempst P, Geromanos S, Elicone C, Erdjument-Bromage H. Methods Companion Methods Enzymol. 1994;6:248–261. [Google Scholar]
- 20.Weston D, Patel B, Van Voorhis WC. Mol. Biochem. Parasitol. 1999;98:105–116. doi: 10.1016/s0166-6851(98)00152-2. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Thomashow LS, Milhausen M, Rutter WJ, N. Agabian N. Cell. 1983;32:35–43. doi: 10.1016/0092-8674(83)90494-4. [DOI] [PubMed] [Google Scholar]
- 23.Kroll DJ, Abdel-Malek I, Abdel-Hafiz H, Marcell T. DNA Cell Biol. 1993;12:441–453. doi: 10.1089/dna.1993.12.441. [DOI] [PubMed] [Google Scholar]
- 24.Omer CA, Diehl RE, Kral AM. Methods. Enzymol. 1995;250:3–12. doi: 10.1016/0076-6879(95)50057-x. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset JA, Gelb MH. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez R, Sali A. Curr. Opin. Struct. Biol. 1997;7:206–214. doi: 10.1016/s0959-440x(97)80027-9. [DOI] [PubMed] [Google Scholar]
- 28.Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Biochemistry. 1998;37:16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- 29.Long SB, Casey PJ, Beese LS. Biochemistry. 1998;37:9612–9618. doi: 10.1021/bi980708e. [DOI] [PubMed] [Google Scholar]