Abstract
Effective cell seeding throughout the tissue scaffold often determines the success of tissue-engineering products, although most current methods focus on determining the total number, not the distribution, of the cells associated with tissue-engineering constructs. The purpose of this investigation was to establish a quick, convenient, and efficient method to quantify cell survival, distribution, and infiltration into degradable scaffolds using a combination of fluorescence cell staining and cryosectioning techniques. After cell seeding and culture for different periods of time, seeded scaffolds were stained with a live cell dye and then cryosectioned. Cryosectioned scaffolds were then recompiled into a three-dimensional (3D) image to visualize cell behavior after seeding. To test the effectiveness of this imaging method, four common seeding methods, including static surface seeding, cell injection, orbital shaker seeding, and centrifuge seeding, were investigated for their seeding efficacy. Using this new method, we were able to visualize the benefits and drawbacks of each seeding method with regard to the cell behavior in 3D within the scaffolds. This method is likely to provide useful information to assist the development of novel materials or cell-seeding methods for producing full-thickness tissue grafts.
Introduction
Tissue engineering has emerged as an important discipline in regenerative medicine aimed at developing transplantable tissues and organs.1–3 The tissue-engineering approach involves the combination of cells, a support biomaterial construct, and micro-environmental factors to induce differentiation signals into surgically transplantable formats and promote tissue repair, functional restoration, or both.4 Using approved materials by the Food and Drug Administration, poly glycolic acid, poly-L-lactic acid, and their co-polymers, such as poly(lactide-co-glycolide) (PLGA), many researchers have sought to design porous constructs that can be seeded with cells in vitro, expanded in culture, and ultimately integrated into a segment of functional tissue through cell migration and proliferation and extracellular matrix (ECM) production to replace the degradable biomaterial. Although many successful developments have been made in the area of tissue engineering skin and bladder,3,5–7 many limitations still hamper the ability to produce thick and complex structured tissues. One main hurdle is the inability to observe the distribution and migration of seeded cells throughout the scaffold.8 This study was thus focused on developing a robust quantitative method that can be used to monitor the migration, distribution, and growth of cells throughout the construct.
Much prior research has relied on different seeding methods to improve seeding efficiency and distribution in scaffold.9–11 These methods can be simply divided into two approaches: static and dynamic seeding techniques.12,13 The most common static seeding techniques include surface seeding14,15 and direct injection16,17 into the scaffold. Dynamic seeding methods, involving movement of the cell solution through and around the scaffold, have been shown to provide better cell coverage and subsequent growth than static seeding.12,13 Also, even cell distribution, leading to cell growth on the surface and the interior of the scaffold, may limit the formation of a necrotic core inside the scaffold.8,18
The most common approach to assess cell seeding in tissue-engineering constructs is by quantification of total cell numbers.19–21 Cell quantification can be performed using many different methods, including trypsinizing adhered cells and counting,22–24 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) or 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay,23–25 and DNA assay of cell lysates.24,26,27 Unfortunately, none of these methods can provide critical information about the degree of cell distribution and infiltration throughout tissue scaffolds.28,29
The morphology of the adherent cells on the surfaces of tissue-engineering constructs can be observed using scanning electron microscopy23,24 and confocal fluorescent microscopy after labeling adherent live cells.27 Transmission electron microscopy has been used to observe detailed cellular–scaffold information.30 However, it would be difficult to translate such high-resolution images into reliable cell-distribution results. Because of the light reflection, opacity, or both of the tissue-engineering constructs, microscopic visualization of viable and apoptotic or necrotic cells without disruption of the scaffold structure is limited to the surface layers of the scaffold. In addition, the visual depth of the confocal microscope is often limited to less than 500 μm depending on the material.31,32 As a result, new methods are needed to analyze the behavior of cells at much deeper depths in the scaffold construct. Some recent approaches to quantify cell distribution in tissue-engineered scaffolds include micro-computed tomography and magnetic resonance imaging of magnetic iron oxide nanoparticles entrapped in seeded cells.33,34 Although these and other studies have investigated the distribution of cells within constructs, the need still exists for a quick and economical method that allows visualization of the three-dimensional (3D) distribution of cells while also allowing quantification of distribution and infiltration in the scaffold.
Using a combination of fluorescence cell staining, cryosectioning, and 3D image compilation, a novel method has been established to quantitatively evaluate cell-seeding efficiency, survival, distribution, and long-term infiltration into PLGA salt-leached scaffolds. Using this technique, we investigated dynamic and static cell seeding methods with PLGA salt-leached scaffolds to monitor the short-term and long-term distribution and infiltration of cells into the polymeric construct. Briefly, after culture, the scaffolds were stained for live cells, fixed, and sectioned, after which all of the individual scaffold sections were imaged for live cell fluorescence and and the images compiled to form a 3D rendering of the living cells in the scaffold. The densities and distributions of cells within the individual sections were then analyzed. Our results have shown that a limited amount of cells infiltrate and that low cell densities exist with increasing depth into the center of the scaffold. This is in agreement with previous observations/assumptions.
Methods
PLGA salt-leached scaffold fabrication
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. PLGA salt-leached scaffolds were fabricated following established procedure.35 Briefly, PLGA (75:25, 113kDa, Medisorb Inc., Birmingham, AL) was dissolved in chloroform at 10% (w/v). The PLGA polymer solution was then added evenly over a Petri dish. Sodium chloride (porogen weight fraction of 90%, sieved at 150–250 μm) was then spread evenly on PLGA solution with continuous stirring in a fume hood until the solvent–polymer solution became pasty. After 72 h, the scaffold was placed under vacuum to complete solvent evaporation overnight. For the salt-leaching process, all scaffolds were submersed in distilled water and placed on an orbital shaker at 100 rpm. The water was changed every 30 min at room temperature until chlorides could not be detected by addition of 0.1 M silver nitrate.36 After salt leaching, the scaffolds (3-4 mm thick) were dried and cut into circular pieces 5 mm in diameter. The scaffolds were then sterilized by submersion in 70% ethanol. After 24 h, the ethanol was exchanged by submerging the scaffolds in phosphate buffered saline (PBS) and orbital shaking three times for 5 to 10 min. The scaffold was then incubated with protein free Dulbecco's modified Eagle medium (DMEM) for 24 h.
Scaffold characterization
Before cell seeding, scanning electron microscopy (SEM) was used to observe the cross-sectional morphology of the 3D PLGA scaffolds. The scaffolds were freeze fractured to expose the cross-section. Briefly, the scaffolds were immersed in liquid nitrogen and then cut using a sharp blade along their cross-sections. The scaffold specimens were sputter coated with silver before observation using a Hitachi S3000N scanning electron microscope (Hitachi High Tech Inc., Tokyo, Japan). The pore sizes were calculated from SEM images using the Image J program (Public Domain Image Processing Program, National Institutes of Health, Bethesda, MD).
Cell culture and scaffold seeding
Mouse 3T3-Swiss albino fibroblasts were obtained from ATCC (CCL-92, Manassas, VA). The cells were cultured in DMEM containing 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% antibiotic solution (100 U/mL penicillin, 100μg/mL streptomycin, Atlanta Biologicals), henceforth referred to as complete DMEM. After expansion for four passages, the cells were detached for the cell-seeding experiments. To determine cell-seeding efficiency on unmodified PLGA salt-leached scaffolds, four common seeding methods (static surface seeding, cell suspension injection, cell seeding with an orbital shaker, and cell seeding using a centrifuge) were investigated. A cell density of 1 × 106 cells/scaffold was used for all methods. Static surface seeding was performed as described in many previous works.37–39 Briefly, a concentrated cell solution (100 μL) containing complete DMEM was evenly added on top of the scaffold. The scaffold was allowed to incubate for 3 h, which is the time required for cell attachment, as observed in control wells. At the end of incubation, the scaffold was submersed in complete DMEM. Cell suspension injection was performed by injecting a concentrated cell solution (1 × 106 cells/10 μL) into the center of the scaffold using a 25-gauge needle, as described in other reports.16,17 Cell seeding with the use of an orbital shaker was performed similar to previous reports.40,41 Scaffolds were incubated with the cell solution (in complete medium containing 1 × 106 cells/3 mL per scaffold) in a 50-mL tube that was placed on an orbital shaker for 1 h at room temperature. After orbital shaking, the scaffold was returned to a well plate and submersed in complete medium. Centrifuge seeding was accomplished as described previously.42 The scaffold was mixed with a cell solution containing complete medium (1 × 106 cells/5 mL) in a 50-mL tube. The tube was then centrifuged at 2000 rpm at room temperature for 1 min. The process was performed three times, with re-suspension of the test tube–bound cells after each centrifuge cycle. The scaffold was then returned to a well plate with complete culture medium.
MTS assay
To determine cell-seeding efficacy, cell numbers on some seeded scaffolds were determined after 6 h of incubation using an MTS assay (Aqueous One Cell Proliferation Assay, Promega, Madison, WI). Cells in 2D culture seeded with equivalent numbers were used as total cell number controls. The cell-seeded scaffolds and controls (cells seeded on culture plates) (n = 3 for all groups) were thoroughly rinsed in PBS and then submersed in 20 μL of MTS solution with 200 μL of fresh medium. For the subtraction of medium background, three wells were filled with 200 μL of medium and 20 μL of MTS only. The samples were then incubated for 2.5 h to begin the formation of the soluble formazan product. The optical density (OD) of the formed product was then analyzed using a microplate reader (SpectraMAX 340, Molecular Devices, Sunnyvale, CA) at 490 nm with background subtraction at 630 nm. The average medium background was then subtracted from the OD. The OD of the formazan product produced by the scaffolds was compared with that of control cultures seeded with numbers equivalent to the scaffold seeding density to determine percentage of cell-seeding efficiency following manufacture recommended procedures.
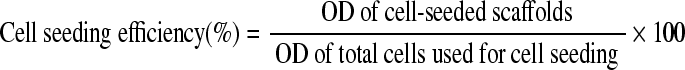 |
To determine the long-term effects of the cell-seeding method, similar experiments were conducted with a 7-day culture time. To avoid cell migration from tissue culture plate to scaffold, the scaffolds were moved into a new well plate after 3 days of culture. The medium was refreshed every third day until the 7-day time point was reached. At this point an MTS assay was performed to determine total cell numbers for each scaffold, as described above.
Fixable live cell staining and scaffold sectioning
To observe the effects of seeding method on cell viability, scaffolds were stained using a Live/Dead Cytotoxicity/Viability kit (Invitrogen, Carlsbad, CA) 3 h after seeding and imaged on the top and bottom surfaces. The numbers of live and dead cells, expressing green and red fluorescence, respectively, were counted. Live and dead cell percentages were determined based on the cell counts as described in the manufacturer's instructions. Three scaffolds for each seeding method were cryosectioned and stained for live cells using a modification of the manufacturer's protocol. Stock solutions were prepared for carboxylfluorescein diacetate, succinimidyl ester (CFDA-SE) (5 mM in dimethylsulfoxide, Invitrogen). At the end of the experiments, scaffolds were rinsed and then submersed in 3 mL of PBS. CFDA-SE (final concentration 2.5 μM) was then added to the PBS and incubated for 15 min to label live cells green (emission at 517 nm) by labeling intracellular esterase as recommended by the manufacturer. After verification of live cells using a fluorescent microscope, the scaffolds were rinsed in PBS and then fixed in methanol for 10 min. After multiple rinses in PBS, the scaffolds were frozen and embedded in OCT and then sectioned using a cryostat (CM 1850 Leica Microsystems, Bannockburn, IL) at 20 μm. The scaffold sections were then used in image analyses.
Image compilation and data analysis
The stained scaffold sections were imaged using a Leica DMLP microscope equipped with a Nikon E500 camera (8.4V, 0.9A, Nikon Corp., Japan). The optimal camera exposure settings and microscope settings were fixed and remained the same for all scaffold groups and time points. Two-dimensional images of each section were then taken using a 4× objective. The scaffolds were cut to a specific diameter (5 mm) and thickness (3-4 mm) so as to fit in the visible area of the 4× objective as determined using hemacytometer measurements of the visible area. For imaging scaffold sections, the oval scaffold sections were centered in the 4× visible area, and a centering mark on the Nikon E500 camera image output was verified before imaging.
The 2D cross-sections of the scaffold were built into 3D images using the following protocol. To eliminate any edge effects and to focus on cell behavior in the bulk of the scaffold, the edge exterior sections were removed from the image sets. The sections of the scaffold were loaded and then stacked using ImageJ.43 To visualize cell distribution in the center of the scaffold, the low-surface-area cross-sections on the ends of the scaffold were removed from the stack. Approximately 100 sections at 20 μm per section were compiled for each scaffold, representing the bulk of the scaffold. The pre-aligned image sections were then stacked and recompiled using the Volume Builder plug-in. The optimal Volume Builder plug-in settings were determined in pilot experiments and remained fixed throughout the experiments. The image was then converted to 16-bit and thresholded to remove any background fluorescence. The threshold values were then set for the duration of the scaffold sections. The particle counter settings were adjusted to account for only a specified range consistent with 3T3 cell sizes determined by measuring the maximum pixilated area of individual cells on the scaffold sections. The circularity was left at default value to account for spherical and oval-shaped areas. Each scaffold section was then scanned using the particle counter. Data for total particle number were acquired for each section and compared with physical cell counts to verify correct program settings. Using these settings, cell density counts were taken for each scaffold section for all four methods after 7 days.
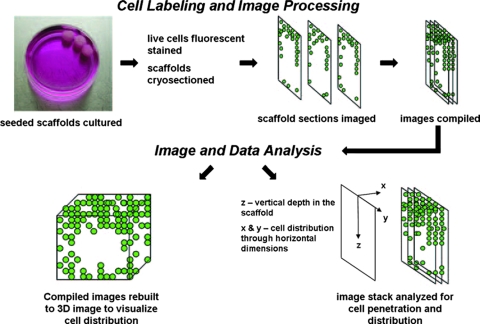
Two methods were used to assess the location of cells within the sections. Emphasis was placed on analyzing the vertical penetration of cells into the scaffold. First, the stacked scaffold sections were organized into five vertical zones of equivalent dimensions measuring 650 μm horizontally across the cross-sections and 1000 μm vertically on the sections. The process was repeated for each replicate scaffold of the same seeding method (n = 3). Each zone was then individually scanned using the particle counter to determine the number of cells occupying the particular zone. Total cell values were expressed as cell density based on the zone area. The process was repeated for each zone and the zones compared to assess vertical cell density through the vertical depth of the scaffold. The sections were then scanned for live cells with recording of the vertical distance of each cell from the scaffold top surfaces. The values for each scaffold and seeding method were then collected and expressed as a percentage of the total penetration distance to the center of the scaffold. A flowchart depicting image compilation and data analysis is shown in Figure 1.
FIG. 1.
Flowchart of three-dimensional (3D) cell distribution and infiltration analyses. After culture, the live cells were fluorescent stained and the scaffold cryosectioned. Aligned sections were then compiled and stacked. These stacked images were then rendered into a 3D object depicting the distribution of cells throughout the scaffold. Finally, the stacked images were analyzed for cell horizontal distribution in the x-y plan and cell vertical penetration through the scaffold in the z-plane. Color images available online at www.liebertonline.com/ten.
Statistical analyses
One-way analysis of variance (ANOVA) was used to compare groups (p < 0.05). Afterward, Student t-tests were used to compare each method, with surface seeding assigned as control with Bonferroni adjustment (p < 0.05).
Results
Scaffold morphology
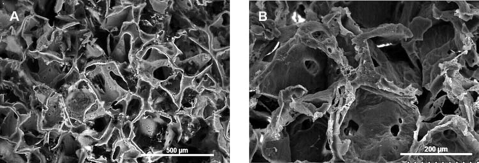
SEM images of the PLGA scaffold surface and cross-sections were collected at different magnifications (Fig. 2A, B). The scaffolds exhibited well-defined, interconnected pore structures. Pore sizes were measured using ImageJ from 10 different cross-sectional SEM micrographs representing different regions of the scaffold. The dimensions of more than 30 pores were measured and averaged to obtain a mean pore size of 212.87 ± 35.9 μm. This was in good agreement with the 150- to 250-μm range of salt sizes used to fabricate these scaffolds.
FIG. 2.
Scanning electron micrographs of the cross section of poly(lactide-co-glycolide) (scaffolds prepared by salt leaching at different magnifications. (A) 80×; (B) 180×. Pore sizes ranged from 150 to 250 μm. Scale bars are shown in the micrographs.
Conventional evaluation of scaffold seeding
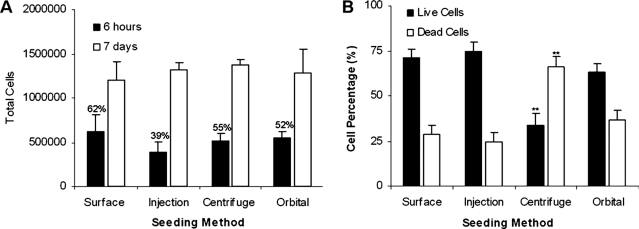
Scaffolds were seeded using four different methods with the same seeding density (1 × 106 cells/scaffold), and initial cell seeding efficiency was determined using MTS assay 6 h after seeding. Surface seeding, orbital seeding, and centrifuge seeding all had respectable efficiencies (62%, 55%, and 52%, respectively, Fig. 3A). Although the seeding efficiency of injection seeding was 39%, the differences between various cell-seeding methods were not statistically significant. Cell numbers on the scaffolds were also determined at 1 week (Fig. 3A). Overall, despite minor differences in early cell-seeding efficiencies, cell-seeding methods had insignificant influences on cell growth in all scaffolds based on the assay.
FIG. 3.
Effect of seeding methods on cell accumulation in scaffolds. (A) Total cell results determined 6 h after seeding (seeding efficiency reported as percentage above 6-h columns) and after 1 week of culture using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay. Assays of equivalent cell numbers on well plates were used as control values for total available cells. (B) Percentage of live and dead cells 3 h after different seeding methods using live/dead cell fluorescence staining. Vertical lines denote ± 1 standard deviation (n = 3 in all cases), **p < 0.01 versus surface-seeded scaffolds.
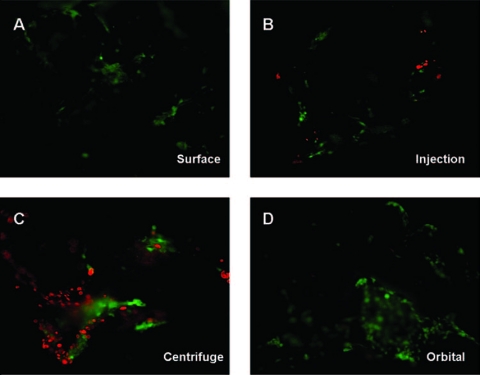
To determine the effects of seeding method on cell viability, the scaffolds were also stained for live and dead cells (Fig. 3B). Cells seeded on well plates were used as controls and represent 100% viability. The seeding methods affected the survival of seeded cells. Specifically, scaffolds seeded using surface, injection, and orbital seeding methods resulted in approximately 25% cell death 3 h after seeding. Rather unexpectedly, the centrifuge seeding method led to the greatest reduction in viability, with an average cell death of more than 50% (Fig. 3B). Representative images used to determine live and dead cell percentages are presented in (Fig. 4).
FIG. 4.
Image of live and dead cells on scaffolds using different cell seeding methods. Areas depicted are those of the densest regions for each seeding method. (A) Surface seeding, (B) injection seeding, (C) centrifuge seeding, and (D) orbital seeding. Color images available online at www.liebertonline.com/ten.
3D cell organization and central distribution in scaffolds
Surface seeding method
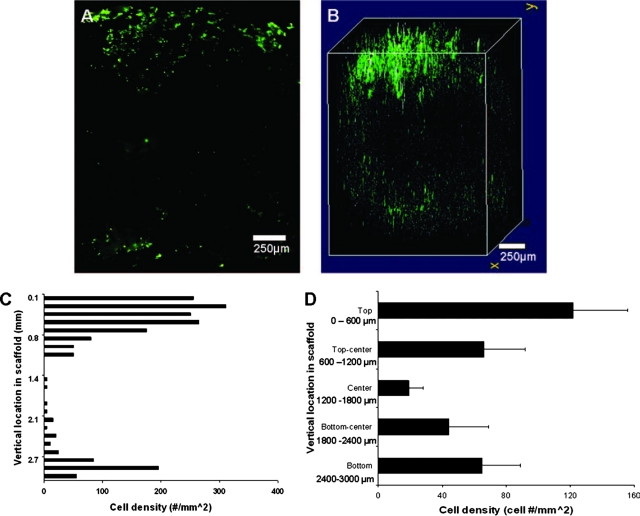
To visualize cell distribution in the center of the scaffold, a section representing the exact middle of the scaffold (top, through the center, to the bottom) was imaged (Fig. 5A). We find a dense layer of cells on the top of scaffold and, to a lesser extent, on the bottom of the scaffold. As expected, few cells were found in the center of the scaffold. Next, we compared this central distribution with a 3D compiled live cell image (Fig. 5B). From this merged image, we observed a thick and spread-out distribution of cells along the upper (seeded side) surface of the scaffold. In the center, a large void space was observed, whereas the bottom (non-seeded side) had a sporadic distribution of cells, with a density lower than observed on the seeded side. Finally, the vertical distribution through the center was determined (Fig. 5C). Consistent with the 3D images, we detected a high cell density (120 cells/mm2) in the surface (top) layer where the cells were seeded (Fig. 5D). This density diminished toward the center of the scaffold (18 cells/mm2) and rose slightly at the bottom edge (65 cells/mm2).
FIG. 5.
Cell images and distribution of surface seeded scaffolds. (A) Two-dimensional (2D) center slice of a surface-seeded scaffold with live cells concentrated along the seeded surface, absent from the center of the slice, and along the non-seeded side of the scaffold. (B) 3D scaffold image showing overall cell distribution, with a large quantity of cells on the seeded surface and fewer cells along the lower surface and nearly no cells in the center of the scaffold. (C) Vertical distribution of the cell density on a cross-sectioned surface-seeded scaffold (n = 1). (D) Scaffold samples (n = 3) were divided into five equal zones representing the exterior surfaces, the exact center of the section, and the transition zones between (zone cross-section area = 1000 μm × 650 μm). From the seeded surface, the cell density diminishes with increasing depth to the center, increasing past the center and below to the lower exterior surface. Vertical lines denote ± 1 standard deviation. Color images available online at www.liebertonline.com/ten.
Injection seeding method
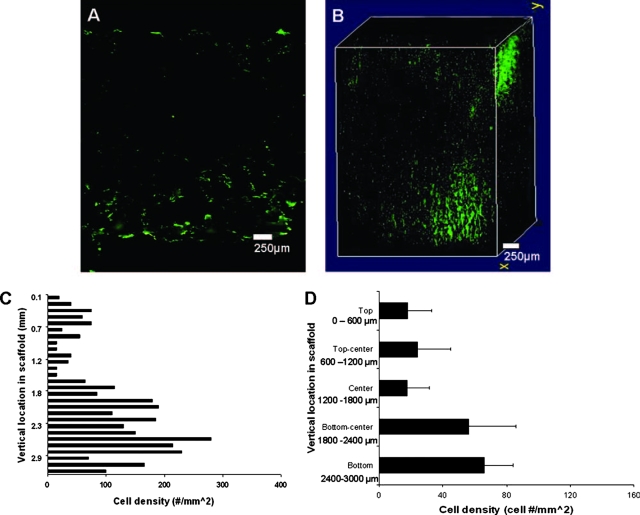
For injection seeding, we observed a dense cell presence along the bottom of the scaffold, with a few cells located near the central region (Fig. 6A). The 3D image showed a dense collection of cells in an isolated area on the top right corner of the scaffold which coincided with the needle entry point into the scaffold. The center portion of the 3D image showed another collection of cells slightly shifted from the scaffold center, with a dense pocket of cells close to the bottom portion of the scaffold that corresponded to the area below the injection site (Fig. 6B). Vertical distribution (Fig. 6C) showed a minimal cell presence in the upper layers through the center, with cell density never exceeding 20 cell/mm2 (Fig. 6D). However, the cell density increased in lower half of the scaffold, rising to 50 cells/mm2 and finally to 70 cells/mm2 in the bottom layer.
FIG. 6.
Cell images and distribution of injection seeded scaffold. (A) Two dimensional (2D) center slice showing minimal cell presence on the upper surface down to the center, with most cells appearing in the area below the center of the scaffold toward the bottom portion of the scaffold. (B) 3D distribution shows a high density of cells along the injection route on the upper corner of the scaffold descending to a high-density area on the bottom portion of the scaffold representing the injection point. (C) Vertical distribution of the cell density on a cross-sectioned injection-seeded scaffold (n = 1). (D) Cell density remains low from the upper exterior surface to the center of scaffold, increasing past the center at the injection point and reaching a maximum at the bottom surface of the scaffold. Vertical lines denote ± 1 standard deviation; n = 3. Color images available online at www.liebertonline.com/ten.
Centrifuge seeding method
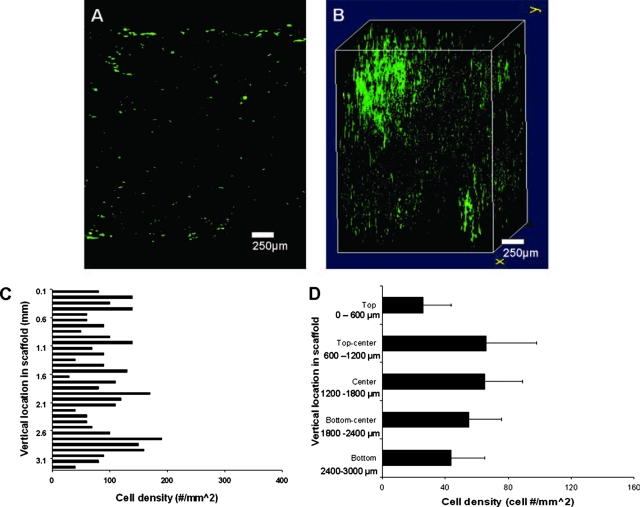
In contrast to the static seeding methods investigated, the centrifuge seeding method distributed cells evenly throughout the scaffold, although at a low density, as evident in the 2D center image (Fig. 7A). The 3D image shows that most of the cells were present in the left side of the scaffold, which corresponds to the bottom of the scaffold as oriented in the centrifuge tubes after seeding (Fig. 7B). A different vertical cell distribution trend was observed with dynamic seeding (Fig. 7C). For centrifuge seeding, the upper exterior surface showed a low cell density of 25 cells/mm2 (Fig. 7D). However, the cell density inside the scaffold was more than twice that, ranging from 55 to 70 cells/mm2 through the center zones. The lower surface density remained moderate, with slightly more than 40 cells/mm2.
FIG. 7.
Cell images and distribution of centrifuge-seeded scaffold. (A) The center slice shows even but light cell distribution throughout the center section. (B) The three-dimensional (3D) distribution appears uniform throughout the section, with a high-density area located on the edge of the scaffold, probably indicative of the portion of the scaffold situated at the bottom of the tube during centrifugation. (C) Vertical cell distribution on a cross-sectioned centrifuge-seeded scaffold (n = 1). (D) For all samples, cell density appears lowest on the exterior surfaces, increasing beyond the exterior surface to similar cell density values in the central regions, consistent with the even distribution seen in 3D images (vertical lines denote ± 1 standard deviation; n = 3. Color images available online at www.liebertonline.com/ten.
Orbital seeding method
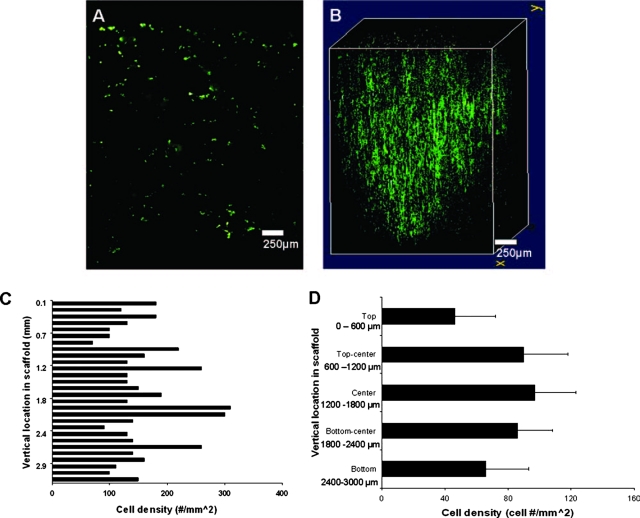
The central 2D section of orbital-seeded scaffolds showed a dense cell presence throughout the entire section (Fig. 8A). Upon merging the sections into a 3D image, we observed that orbital seeding evenly distributed the cells throughout the bulk of the section in large numbers (Fig. 8B). For orbital seeding, all zones had an appreciable cell density (Fig. 8C), with no layer less than 40 cells/mm2 (Fig. 8D). The interior of the scaffold showed enhanced cell penetration, with cell density values greater than 80 cells/mm2 for these zones. The cell density decreased slightly on the bottom surface of the scaffold, with a density of just over 70 cells/mm2.
FIG. 8.
Cell images and distribution of orbital seeded scaffolds. (A) The center slice of the orbital scaffold shows even cell distribution similar to that of the centrifuge seeding but with an apparently higher cell density. (B) The three dimensional image of the orbital scaffold shows a uniform sphere of live cells with good cell distribution and patches of higher cell density randomly located within the scaffold. (C) Vertical cell distribution on a cross-sectioned orbital-seeded scaffold (n = 1). (D) The overall density distribution was similar to that with dynamic centrifuge seeding, with cell density increasing to maximum at the center of the scaffold, although greater cell density was observed in the central regions of the scaffold. Vertical lines denote ± 1 standard deviation; n = 3. Color images available online at www.liebertonline.com/ten.
Comparison of cell distribution between the seeding methods
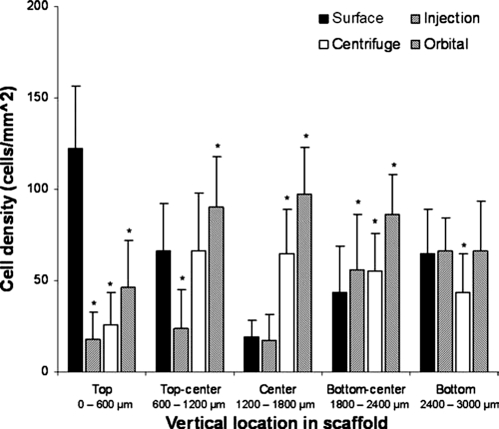
Finally, we compared the central cell density and cell infiltration of all of the seeding methods tested to compare the distribution of cells within the construct after 7 days of incubation. Differing levels of cell in-growth were seen for each method (Fig. 9). ANOVA testing of all groups revealed that there was a significant difference (p < 0.01) for the groups. Surface seeding led to the highest cell density on the top layer. However, as we descended through the scaffold center to the bottom, orbital seeding appeared to achieve the highest cell density through the construct. For all layers analyzed below the surface layer, orbital seeding attained the highest cell density average for each layer. All other methods tested showed variability in cell density in these layers, with only the bottom layer showing little difference between the seeding methods.
FIG. 9.
Comparison of cell vertical distribution with all seeding methods. The upper surface showed the highest cell density for surface seeding, as expected. As depth decreased, the density of surface seeding diminished, as centrifuge and orbital seeding rose to values significantly higher than static seeding methods in the center of the scaffold. The cell density values for all methods were similar with increasing depth to the bottom. Vertical lines denote ± 1 standard deviation (n = 3 in all cases), *p < 0.05 versus surface-seeded scaffolds.
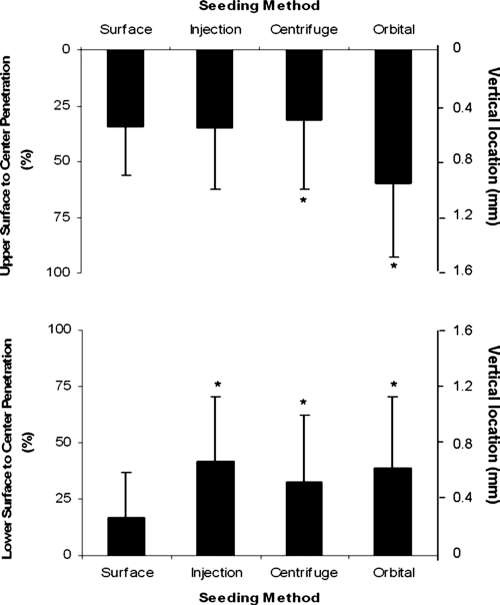
The infiltration distance in both vertical directions toward the center was quantified as a measure of the ability of each seeding method to influence cell penetration. The average vertical cell location was determined for each scaffold for each seeding method (n = 3) as a representation of the average vertical cell location in the scaffold (Fig. 10). The upper surface, representing the seeded side of the scaffold for surface seeding, showed minimal penetration of the cells toward the center, with penetration slightly greater than 25% at 1 week. Minimal penetration was observed from the scaffold bottom toward the center, with infiltration less than 25% toward the center of the scaffold. For injection seeding, where cells were dense, cell solution was introduced into the center of the scaffold, and infiltration was slightly higher from both directions, with both values slightly greater than 40% depth from the exterior surfaces. Centrifuge seeding led to a location similar to surface seeding for the upper portion of the scaffold, at slightly greater than 25%, similar to the location for the bottom portion, also approximately 25%. However, the average vertical location of orbital seeded cells was higher than with other methods for the upper portion of the scaffold: greater than 50% of the distance from the exterior surface to the center of the scaffold. Location in the lower portion of the scaffold was approximately 40% of the distance.
FIG. 10.
Average vertical cell location in scaffolds using different seeding methods. The top graph shows the average cell location for all seeding methods in terms of percentage distance toward the center of the scaffold. Similar values are observed for surface, injection, and centrifuge seeding, whereas the orbital seeding value was nearly twice as far into the scaffold. The bottom graph shows the average cell location as percentage distance from the bottom exterior surface toward the center of the scaffold. Most of the cells in surface-seeded scaffolds resided close to the bottom, whereas the injection, centrifuge, and orbital seeding showed modest cell penetration, between 25% and 50% of the distance toward the center of the scaffold. Vertical lines denote ± 1 standard deviation (n = 3 in all cases), *p < 0.05 versus surface-seeded scaffolds.
Discussion
For analysis of seeded tissue-engineered scaffolds, many studies have not adequately investigated seeded cell distribution and infiltration in in vitro and in vivo settings.44–46 The importance of cell behavior on scaffolds provides the key to ensuring the ultimate functionality in tissue-engineered constructs.47,48 It is specifically important to understand how seeding method selection affects cell distribution.9 It is likely that an even distribution of cells throughout the scaffold and across the exterior surfaces will lead to the most efficient distribution, growth, and infiltration.9,49 Many reports have highlighted the importance of cell organization, orientation, and differentiation state in dictating tissue-engineering implant functionality.9,47,48 As a result, there is a renewed interest in visualizing and quantifying the behavior of cells on 3D scaffolds.8,50 Thus, the purpose of this study was to investigate a novel method of monitoring cell survival, distribution, and infiltration in polymeric tissue-engineered scaffolds.
Recently, several methods have been investigated as tools to qualitatively analyze the distribution of seeded cells in scaffolds. For example, synchrotron radiation micro-computed tomography was used to quantify the 3D distribution of cells on yarn-fiber scaffolds.33 Magnetic resonance microscopy has also been used to visualize interior slices of scaffolds to analyze cell distribution in different seeding methods.51 Magnetic nanoparticles and conventional magnetic resonance imaging have also been used to visualize cell distribution on tissue-engineered scaffolds.34 However, these methods require the use of sophisticated equipment that may not be accessible to some tissue engineers. Thus, there is still a need for a simple, cost effective method to visualize 3D cell distribution and infiltration on a constant basis.
Without a convenient approach to determine cell distribution and penetration, most studies have limited their scope to MTT and MTS assays and trypsinizing scaffold-associated cells to assess cell behavior on tissue-engineered scaffolds.22,24,52,53 Unfortunately, these methods fail to provide information on the location, distribution, and infiltration of cells into constructs. In fact, early research conducted for each seeding method showed little variation, with efficiencies of approximately 50%, consistent with unmodified PLGA salt-leached scaffolds using these methods, although in disagreement with analysis of the sections of the scaffold.19,54–57 In addition, such assays, designed for assaying cell toxicity and cell growth in 96 wells, may not adequately scale up to large 3D constructs, leading to inadequate perfusion of the compound within the large scaffold. This may obscure dye conversion results, failing to provide accurate total cell numbers for cells on the exterior of the scaffold and within the scaffold.
The recent development of fluorescent labeling dyes such as CFDA-SE has provided a simple method of labeling and tracking cell migration and location, as well as determining the total number of scaffold-associated cells.21,46 Results have shown that fluorescence labeling is a powerful tool to investigate cell distribution in tissue constructs.21,58 By expanding the use of fluorescent dye to visualize cell–scaffold interactions, this new method not only analyzes cell density by calculating total cell numbers for each section of the scaffold using the dye CFDA-SE, but also determines how the cells are distributed and, most importantly, how far they infiltrate into the scaffolds. This technique may allow the reconstruction of scaffolds to 3D images after single or multiple immunohistochemical staining to view the distribution and infiltration of different cells types within a scaffold construct in vitro or in vivo.
After labeling scaffolds with CFDA-SE to visualize live cells, we saw differences in the total number of scaffold-associated cells based on the choice of cell-seeding method, inconsistent with our MTS observations. Because most of the scaffold-associated cells were on the top, bottom, and side of scaffold, it is likely that the total cell number measurement according to MTS assay would not be able to assess the infiltrated cells, which are composed of only small numbers of total adherent cells.
Surface seeding has been the most commonly used seeding method in tissue engineering because of its ease of application.14,15,59 Our 3D images and cell location data have shown that cells tend to reside on the seeded surface, with slight penetration into the scaffold. Analysis of cell density in the central regions shows a light cell presence after seeding for 7 days. The lack of cell infiltration into the center of the constructs may be due to excess cell solution flow around the scaffold and pooling at the bottom. Such findings are in agreement with recent published work, suggesting that the localized attachment and growth of seeded cells on scaffold surfaces would hinder the development of large constructs for potential clinical applications.42,59,60
Injection seeding was proposed as a method of delivering cells to the center portions of the scaffold, in hopes that better cell distribution throughout the construct could be achieved.16,17,61 Our results show that few cells were present in the exact center of the constructs. However, analysis of cell density in areas slightly offset from the center showed a spike in cell density consistent with what would be expected with a cell injection to that region. It is possible that, during seeding, the solution injection was slightly off from the exact center of the scaffold. In addition, from the 3D images of live cells on the injection-seeded scaffolds, we observed that cells appeared to be concentrated at the point of needle entry site along the upper surface and densely located in a region below the injection point on the bottom surface of the scaffold. This may be due to the cell solution leaking through the porous scaffold to the bottom of the scaffold and pooling there, resulting in high cell density on the bottom below the injection point, as reported in other cell-injection studies.61 Therefore our results support that cell injection into scaffolds may be a viable method of delivering cells to a specific location within the scaffold, as suggested previously.62 However, this method may not provide the even and reproducible cell distribution needed to facilitate cell organization throughout the scaffold, as desired for most tissue-engineering applications, such as tissue-engineered vasculature, heart, tendon, and muscle.9,63–65
Because of the need for a more-efficient cell delivery method, dynamic seeding techniques were developed based on the principle that moving a cell solution throughout the construct would lead to high cell deposition in the scaffold porosity, as well as on the exterior surfaces.12,66,67 Many investigations of dynamic seeding methods support the ability of dynamic seeding methods to more efficiently seed scaffolds.12,60,66,67 One such commonly investigated dynamic seeding method is centrifuge seeding.19,42,61 After centrifuge seeding, we observed even, improved cell distribution throughout the construct and sections for 3D volumes and 2D sections, consistent with previous observations.19 Analysis of cell density throughout the center of these constructs support our image observations showing greater cell density than with static methods. However, the harshness of this method may limit it during the seeding procedure. Our results have revealed that centrifuge seeding leads to significant levels of cell death after the centrifuge procedure. The potential influence of a large number of dead cells on subsequent cell growth in the tissue scaffold has yet to be determined. The centrifugal force–associated cell death may perhaps be avoided by lessening the speeds used during the procedure, as indicated in previous work.42
The most successful seeding method used in our investigation was orbital seeding. This method provides the benefits of dynamic centrifuge seeding but uses much lower speeds and provides a continuous perfusion of the cell suspension through and around the scaffold. Recent investigations using orbital seeding support that this method can efficiently deliver cells throughout small-scale scaffolds.41,68 Indeed, our 2D and 3D images showed greater cell density and more-even distribution than with all other seeding methods tested. In addition, cell survival after orbital seeding appears to be consistent with other static seeding methods investigated.
In summary, all seeding methods have their unique benefits and drawbacks (Table 1). In the development and characterization of new tissue-engineered scaffolds and seeding methods, the ability to detect and analyze how cells are distributed initially and over the long term in these structures is critically important. Thus, a convenient method of analyzing cell distribution and penetration will aid tissue engineers in evaluating how well cells integrate with their scaffolds, what seeding methods will be required, and how well cells constitute the scaffolds. By analyzing some common seeding methods with PLGA salt-leached scaffolds, we have demonstrated the ability of this method to characterize the distribution and penetration of cells within the scaffolds.
Table 1.
Summary of Benefits and Drawbacks of Four Commonly Used Cell Seeding Methods
| Benefits | Drawbacks | |
|---|---|---|
| Static Methods | ||
| • Surface Seeding | - Homogeneous seeding on scaffold surfaces | - Poor cell penetration into scaffold |
| • Injection Seeding | - Cell seeding in specific area within scaffold | - Damage the scaffold and reduced cell distribution/viability |
| Dynamic Methods | ||
| • Orbital Seeding | - Even surface distribution and deposition of cells within the scaffold | - Cell distribution depends on pore interconnectivity of scaffold |
| • Centrifuge Seeding | - Improved cell penetration | - Damage cells and scaffold architectures by centrifugal force |
In conclusion, we have demonstrated the feasibility of a new method of constructing 3D images from live cell–stained scaffold sections. This method provides a quick and efficient method to visualize and quantify cell distribution in polymeric scaffolds using materials accessible to a wide range of researchers. In addition to visualizing live cell distribution and infiltration in tissue-engineered scaffolds, this method can also be used in conjunction with other methods to acquire data on cell infiltration and distribution in tissue-engineered constructs. Such information cannot be obtained using commonly used cell quantification methods, including trypsinization and MTT and MTS methods. A more comprehensive understanding of cell distribution throughout the tissue construct may lead to a better design of better tissue-engineered products and applications.
Acknowledgment
This work was supported by National Institutes of Health Grant RO1 GM074021.
References
- 1.Gurtner GC. Callaghan MJ. Longaker MT. Progress and potential for regenerative medicine. Annu Rev Med. 2007;119:1735. doi: 10.1146/annurev.med.58.082405.095329. [DOI] [PubMed] [Google Scholar]
- 2.Brown RA. Phillips JB. Cell response to biomimetic protein scaffolds used in tissue repair and engineering. Int Rev Cytol. 2007;262:75. doi: 10.1016/S0074-7696(07)62002-6. [DOI] [PubMed] [Google Scholar]
- 3.Atiyeh BS. Costagliola M. Cultured epithelial autograft (CEA) in burn treatment: three decades later. Burns. 2007;33:405. doi: 10.1016/j.burns.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Malafaya PB. Silva GA. Reis RL. Natural-origin polymers as carrier and scaffolds for biomolecules and cell delivery in tissue engineering applications. Adv Drug Deliv Rev. 2007;59:207. doi: 10.1016/j.addr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Johnsen S. Ermuth T. Tanczos E. Bannasch H. Horch RE. Zschocke I. Peschen M. Schopf E. Vanscheidt W. Augustin M. Treatment of therapy-refractive ulcera cruris of various origins with autologous keratinocytes in fibrin sealant. Vasa. 2005;34:25. doi: 10.1024/0301-1526.34.1.25. [DOI] [PubMed] [Google Scholar]
- 6.Nevins ML. Aesthetic and regenerative oral plastic surgery: clinical applications in tissue engineering. Dent Today. 2006;25:144. [PubMed] [Google Scholar]
- 7.Atala A. Bauer SB. Soker S. Yoo JJ. Retik AB. Tissue-engineering autologous bladders for patients needing cystoplasty. Lancet. 2006;367:1241. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 8.Sengers BG. Taylor M. Please CP. Oreffo RO. Computational modeling of cell spreading and tissue regeneration in porous scaffolds. Biomaterials. 2007;28:1926. doi: 10.1016/j.biomaterials.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Martin I. Wendt D. Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004;22:80. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Wendt D. Marsano A. Jakob M. Heberer M. Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol Bioeng. 2003;84:205. doi: 10.1002/bit.10759. [DOI] [PubMed] [Google Scholar]
- 11.Li Y. Ma T. Kniss DA. Lasky LC. Yang ST. Effects of filtration seeding on cell density, spatial distribution, and proliferation in nonwoven fibrous matrices. Biotechnol Prog. 2001;17:935. doi: 10.1021/bp0100878. [DOI] [PubMed] [Google Scholar]
- 12.Zhao F. Ma T. Perfusion bioreactor system for human mesenchymal stem cell tissue engineering: dynamic seeding and construct development. Biotechnol Bioeng. 2005;91:482. doi: 10.1002/bit.20532. [DOI] [PubMed] [Google Scholar]
- 13.van den Dolder J. Spauwen PH. Jansen JA. Evaluation of various seeding techniques for culturing osteogenic cells on titanium fiber mesh. Tissue Eng. 2003;9:315. doi: 10.1089/107632703764664783. [DOI] [PubMed] [Google Scholar]
- 14.Choong CS. Hutmacher DW. Triffitt JT. Co-culture of bone marrow fibroblasts and endothelial cells on modified polycaprolactone substrates for enhanced potentials in bone tissue engineering. Tissue Eng. 2006;12:2521. doi: 10.1089/ten.2006.12.2521. [DOI] [PubMed] [Google Scholar]
- 15.Wan Y. Wang Y. Liu Z. Qu X. Han B. Bei J. Wang S. Adhesion and proliferation of OCT-1 osteoblast-like cells on micro- and nano-scale topography structure poly(L-lactide) Biomaterials. 2005;26:4453. doi: 10.1016/j.biomaterials.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Honda MJ. Yada T. Ueda M. Kimata K. Cartilage formation by serial passaged cultured chondrocytes in a new scaffold: hybrid 75:25 poly(L-lactide-epsilon-caprolactone) sponge. J Oral Maxillofac Surg. 2004;62:1510. doi: 10.1016/j.joms.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann A. Konrad L. Gotzen L. Printz H. Ramaswamy A. Hofmann C. Bioengineering human bone tissue using autogenous osteoblasts cultured on different biomatrices. J Biomed Mater Res A. 2003;67:191. doi: 10.1002/jbm.a.10594. [DOI] [PubMed] [Google Scholar]
- 18.Silva MMCG. Cyster LA. Barry JJA. Yang XB. Oreffo ROC. Grant DM. Scotchford CA. Howdle SM. Shakesheff KM. Rose FRAJ. The effect of anisotrophic architecture on cell and tissue infiltration into tissue engineering scaffolds. Biomaterials. 2006;27:5909. doi: 10.1016/j.biomaterials.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Roh JD. Nelson GN. Udelsman BV. Brennan MP. Lockhart B. Fong PM. Lopez-Soler RI. Saltzman WM. Breuer CK. Centrifugal seeding increases seeding efficiency and cellular distribution of bone marrow stromal cells in porous biodegradable scaffolds. Tissue Eng. 2007;13:2743. doi: 10.1089/ten.2007.0171. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y. Uemura T. Dong J. Kojima H. Tanaka J. Tateishi T. Application of perfusion culture system improves in vitro and in vivo osteogenesis of bone marrow-derived osteoblastic cells in porous ceramic materials. Tissue Eng. 2003;9:1205. doi: 10.1089/10763270360728116. [DOI] [PubMed] [Google Scholar]
- 21.Solchaga LA. Tognana E. Penick K. Baskaran H. Goldberg VM. Caplan AI. Welter JF. A rapid seeding technique for the assembly of large cell/scaffold composite constructs. Tissue Eng. 2006;12:1851. doi: 10.1089/ten.2006.12.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uematsu K. Hattori K. Ishimoto Y. Yamauchi J. Habata T. Takakura Y. Ohgushi H. Fukuchi T. Sato M. Cartilage regeneration using mesechymal stem cells and a three-dimensional poly-lactic-glycolic acid (PLGA) scaffold. Biomaterials. 2005;26:4273. doi: 10.1016/j.biomaterials.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 23.Shen H. Hu X. Yang F. Bei J. Wang S. Combining oxygen plasma treatment with anchorage of cationized gelatin for enhancing cell affinity of poly(lactide-co-glycolide) Biomaterials. 2007;28:4219. doi: 10.1016/j.biomaterials.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y. Chan-Park MB. Density quantification of collagen grafted on biodegradable polyester: its application to esophageal smooth muscle cell. Anal Biochem. 2007;363:119. doi: 10.1016/j.ab.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Sahoo S. Ouyang H. Goh JC. Tay TE. Toh SL. Characterization of a novel polymeric scaffold for potential application in tendon/ligament tissue engineering. Tissue Eng. 2006;12:91. doi: 10.1089/ten.2006.12.91. [DOI] [PubMed] [Google Scholar]
- 26.Koegler WS. Griffith LG. Osteoblast response to PLGA tissue engineering scaffolds with PEO modified surface chemistries and demonstration of patterned cell response. Biomaterials. 2004;25:2819. doi: 10.1016/j.biomaterials.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 27.Spalazzi JP. Doty SB. Moffat KL. Levine WN. Lu HH. Development of controlled matrix heterogeneity on a triphasic scaffold for orthopedic interface tissue engineering. Tissue Eng. 2006;12:3497. doi: 10.1089/ten.2006.12.3497. [DOI] [PubMed] [Google Scholar]
- 28.Vunjak-Novakovic G. Kaplan DL. Tissue engineering: the next generation. Tissue Eng. 2006;12:3261. doi: 10.1089/ten.2006.12.3261. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y. Kim UJ. Blasioli DJ. Kim HJ. Kaplan DL. In vitro cartilage tissue engineering with 3D porous aqueous-derived silk scaffolds and mesenchymal stem cells. Biomaterials. 2005;26:7082. doi: 10.1016/j.biomaterials.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Opitza F. Schenke-Layland K. Cohnert TU. Stock UA. Phenotypical plasticity of vascular smooth muscle cells—effect of in vitro and in vivo shear stress for tissue engineered blood vessels. Tissue Eng. 2007;13:2505. doi: 10.1089/ten.2006.0424. [DOI] [PubMed] [Google Scholar]
- 31.Doroski D. Tittman BR. Miyasaka C. Study of biomedical specimens using scanning acoustic microscopy. Acoustical Imaging. 2007;28:13. [Google Scholar]
- 32.Flynn L. Prestwich GD. Semple JL. Woohouse KA. Adipose tissue engineering with naturally derived scaffolds and adipose-derived stem cells. Biomaterials. 2007;28:3834. doi: 10.1016/j.biomaterials.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Thurner P. Muller R. Raeber G. Sennhauser U. Hubbell JA. 3D morphology of cell cultures: a qualitative approach using micrometer synchrotron light tomography. Microsc Res Tech. 2005;66:289. doi: 10.1002/jemt.20170. [DOI] [PubMed] [Google Scholar]
- 34.Heymer A. Haddad D. Weber M. Gbureck U. Jakob PM. Eulert J. Noth U. Iron oxide labeling of human mesenchymal stem cells in collagen hydrogels for articular cartilage repair. Biomaterials. 2008;29:1473. doi: 10.1016/j.biomaterials.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Yang J. Shi G. Bei J. Wang S. Cao Y. Shang Q. Yang G. Wang W. Fabrication and surface modification of macroporous poly(L-lactic acid) and poly(L-lactic-co-glycolic acid) (70/30) cell scaffolds for human skin fibroblast cell culture. J Biomed Mater Res. 2002;62:438. doi: 10.1002/jbm.10318. [DOI] [PubMed] [Google Scholar]
- 36.Sander EA. Alb AM. Nauman EA. Reed WF. Dee KC. Solvent effects on the microstructure and porosity of 75/25 poly (D,L-lactide-co-glycolide) tissue scaffolds. J Biomed Mater Res A. 2004;70:506. doi: 10.1002/jbm.a.30109. [DOI] [PubMed] [Google Scholar]
- 37.Neimeyer P. Krause U. Fellenberg J. Kasten P. Seckinger A. Ho AD. Simank HG. Evaluation of mineralized cartilage and alpha-tricalcium phosphate as scaffolds for tissue engineering of bone using human mesenchymal stem cells. Cells Tissues Organs. 2004;177:68. doi: 10.1159/000079182. [DOI] [PubMed] [Google Scholar]
- 38.Blum JS. Temenoff JS. Park H. Jansen JA. Mikos AG. Barry MA. Development and characterization of green fluorescent protein and luciferase expressing cell line for non-destructive evaluation of tissue engineering constructs. Biomaterials. 2004;25:5809. doi: 10.1016/j.biomaterials.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien FJ. Harley BA. Yannas IV. Gibson LJ. The effect of pore size on cell adhesion in collagen-GAG scaffolds. Biomaterials. 2005;26:433. doi: 10.1016/j.biomaterials.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 40.Almarza AJ. Athanasiou KA. Seeding techniques and scaffolding choice for tissue engineering of temporomandibular joint disk. Tissue Eng. 2004;10:1787. doi: 10.1089/ten.2004.10.1787. [DOI] [PubMed] [Google Scholar]
- 41.Yasuda K. Inoue S. Tabata Y. Influence of culture method on the proliferation and osteogenic differentiation of human adipo-stromal cells in nonwoven fabrics. Tissue Eng. 2004;10:1587. doi: 10.1089/ten.2004.10.1587. [DOI] [PubMed] [Google Scholar]
- 42.Godbey WT. Hindy SB. Sherman ME. Atala A. A novel use of centrifugal force for cell seeding into porous scaffolds. Biomaterials. 2004;25:2799. doi: 10.1016/j.biomaterials.2003.09.056. [DOI] [PubMed] [Google Scholar]
- 43.Abramoff MD. Magelhaes PJ. Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36. [Google Scholar]
- 44.Ozawa T. Mickle DA. Weisel RD. Koyama N. Wong H. Ozawa S. Li RK. Histological changes of nonbiodegradable and biodegradable biomaterials used to repair right ventricular heart defects in rats. J Thorac Cardiovasc Surg. 2002;124:1157. doi: 10.1067/mtc.2002.127449. [DOI] [PubMed] [Google Scholar]
- 45.Robinson KA. Matheny RG. Myocardial tissue replacement with extracellular matrix scaffolds. Heart Surg Forum. 2002;6:8. [Google Scholar]
- 46.Kofidis T. Bruin JL. Hoyt G. Ho Y. Tanaka M. Yamane T. Lebl DR. Swijnenburg RJ. Chang CP. Quertermous T. Robbins RC. Myocardial restoration with embryonic stem cell bioartificial tissue transplantation. J Heart Lung Transplant. 2005;24:737. doi: 10.1016/j.healun.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Lutolf MP. Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 48.Takezawa T. A strategy for the development of tissue engineering scaffolds that regulate cell behavior. Biomaterials. 2003;24:2267. doi: 10.1016/s0142-9612(03)00038-3. [DOI] [PubMed] [Google Scholar]
- 49.Sodian R. Lemke T. Fritsche C. Hoerstrup SP. Fu P. Potapov EV. Hausmann H. Hetzer R. Tissue-engineering bioreactors: a new combined cell seeding and perfusion system for vascular tissue engineering. Tissue Eng. 2002;8:863. doi: 10.1089/10763270260424222. [DOI] [PubMed] [Google Scholar]
- 50.Tan EP. Lim CT. Characterization of bulk properties of nanofibrous scaffolds from nanomechanical properties of single nanofibers. J Biomed Mater Res A. 2006;77:526. doi: 10.1002/jbm.a.30646. [DOI] [PubMed] [Google Scholar]
- 51.Burg KJ. Delnomdedieu M. Beiler RJ. Culberson CR. Greene KG. Halberstadt CR. Holder WJ., Jr Loebsack AB. Roland WD. Johnson GA. Application of magnetic resonance microscopy to tissue engineering: a polylactide model. J Biomed Mater Res. 2002;61:380. doi: 10.1002/jbm.10146. [DOI] [PubMed] [Google Scholar]
- 52.Gong Y. He L. Li J. Zhou Q. Ma Z. Gao C. Shen J. Hydrogel-filled polylactide porous scaffolds for cartilage tissue engineering. J Biomed Mater Res B Appl Biomater. 2007;82:192. doi: 10.1002/jbm.b.30721. [DOI] [PubMed] [Google Scholar]
- 53.Nam YS. Yoon JJ. Park TG. A novel fabrication method of macroporous biodegradable polymer scaffolds using gas foaming salt as a porogen. J Biomed Mater Res. 2000;53:1. doi: 10.1002/(sici)1097-4636(2000)53:1<1::aid-jbm1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 54.Chen G. Sato T. Ushida T. Ochiai N. Tateishi T. Tissue engineering of cartilage using a hybrid scaffold of synthetic polymer and collagen. Tissue Eng. 2004;10:323. doi: 10.1089/107632704323061681. [DOI] [PubMed] [Google Scholar]
- 55.Ren T. Ren J. Jia X. Pan K. The bone formation in vivo and mandibular defect repair using PLGA porous scaffolds. J Biomed Mater Res A. 2005;74:562. doi: 10.1002/jbm.a.30324. [DOI] [PubMed] [Google Scholar]
- 56.Lin HR. Kuo CJ. Yang CY. Shaw SY. Wu YJ. Preparation of macroporous biodegradable PLGA scaffolds for cell attachement with the use of mixed salts as porogen additives. J Biomed Mater Res. 2002;63:271. doi: 10.1002/jbm.10183. [DOI] [PubMed] [Google Scholar]
- 57.Holy CE. Shoichet MS. Davies JE. Engineering three-dimensional bone tissues in vitro using biodegradable scaffolds: investigating initial cell-seeding density and culture period. J Biomed Mater Res. 2000;51:376. doi: 10.1002/1097-4636(20000905)51:3<376::aid-jbm11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 58.Chong AK. Ang AD. Goh JC. Hui JH. Lim AY. Lee EH. Lim BH. Bone marrow-derived mesenchymal stem cells influence early tendon-healing in a rabbit Achilles tendon model. J Bone Joint Surg Am. 2007;89:74. doi: 10.2106/JBJS.E.01396. [DOI] [PubMed] [Google Scholar]
- 59.Nieponice A. Soletti L. Guan J. Deasy BM. Huard J. Wagner WR. Vorp DA. Development of a tissue-engineered vascular graft combining a biodegradable scaffolds, muscle-derived stem cells and a rotational vacuum seeding technique. Biomaterials. 2008;29:825. doi: 10.1016/j.biomaterials.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauney JR. Blumberg J. Pirun M. Volloch V. Vunjak-Novakovic G. Kaplan DL. Osteogenic differentiation of human bone marrow stromal cells on partially demineralized bone scaffolds in vitro. Tissue Eng. 2004;10:81. doi: 10.1089/107632704322791727. [DOI] [PubMed] [Google Scholar]
- 61.Dvir-Ginzberg M. Gamlieli-Bonshtein I. Agbaria R. Cohen S. Live tissue engineering with alginate scaffolds: effects of cell-seeding density on hepatocyte viability, morphology, and function. Tissue Eng. 2003;9:757. doi: 10.1089/107632703768247430. [DOI] [PubMed] [Google Scholar]
- 62.Vacanti CA. Bonassar LJ. Vacanti MP. Shufflebarger J. Replacement of an avulsed phalanx with tissue-engineered bone. N Engl J Med. 2001;344:1511. doi: 10.1056/NEJM200105173442004. [DOI] [PubMed] [Google Scholar]
- 63.Gonen-Wadmany M. Gepstein L. Seliktar D. Controlling the cellular organization of tissue-engineered cardiac constructs. Ann NY Acad Sci. 2004;1015:299. doi: 10.1196/annals.1302.025. [DOI] [PubMed] [Google Scholar]
- 64.Boland ED. Matthews JA. Pawlowski KJ. Simpson DG. Wnek GE. Bowlin GL. Electrospinning collagen and elastin: Preliminary vascular tissue engineering. Front Biosci. 2004;9:1422. doi: 10.2741/1313. [DOI] [PubMed] [Google Scholar]
- 65.Liu H. Fan H. Wang Y. Toh SL. Goh JCH. The interaction between a combined knitted silk scaffolds and microporous silk sponge with human mesenchymal stem cells for ligament tissue engineering. Biomaterials. 2008;29:662. doi: 10.1016/j.biomaterials.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 66.Kim HJ. Kim UJ. Leisk GG. Bayan C. Georgakoudi I. Kaplan DL. Bone regeneration on aqueous-derived silk 3D scaffolds. Macromol Biosci. 2007;7:643. doi: 10.1002/mabi.200700030. [DOI] [PubMed] [Google Scholar]
- 67.Kim BS. Putnam AJ. Kulik TJ. Mooney DJ. Optimizing seeding and culture methods to engineering smooth muscle tissue on biodegradable polymer matrices. Biotechnol Bioeng. 1998;57:46. [PubMed] [Google Scholar]
- 68.Takahashi Y. Yamamoto M. Tabata Y. Osteogenic differentiation of mesenchymal stem cells in biodegradable sponges composed of gelatin and beta-tricalcium phosphate. Biomaterials. 2005;26:3587. doi: 10.1016/j.biomaterials.2004.09.046. [DOI] [PubMed] [Google Scholar]