Abstract
Peroxisome proliferator-activated receptors (PPARs) regulate lipid and glucose homeostasis, are targets of pharmaceuticals, and are also activated by environmental contaminants. Almost nothing is known about expression of PPARs during human fetal development. This study examines expression of PPARα, β, and γ mRNA and protein in human fetal tissues. With increasing fetal age, mRNA expression of PPARα and β increased in liver, but PPARβ decreased in heart and intestine, and PPARγ decreased in adrenal. Adult and fetal mean expression of PPARα, β, and γ mRNA did not differ in intestine, but expression was lower in fetal stomach and heart. PPARα and β mRNA in kidney and spleen, and PPARγ mRNA in lung and adrenal were lower in fetal versus adult. PPARγ in liver and PPARβ mRNA in thymus were higher in fetal versus adult. PPARα protein increased with fetal age in intestine and decreased in lung, kidney, and adrenal. PPARβ protein in adrenal and PPARγ in kidney decreased with fetal age. This study provides new information on expression of PPAR subtypes during human development and will be important in evaluating the potential for the developing human to respond to PPAR environmental or pharmaceutical agonists.
1. Introduction
Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear hormone receptor superfamily and there are three primary subtypes (α, β/δ, and γ) [1]. These receptors play important roles in embryonic and fetal development as well as placental function [2, 3], regulating many cellular and metabolic processes [4]. PPARs control energy homeostasis, are important regulators of adipogenesis, lipid metabolism, inflammatory responses, and hematopoiesis, and are implicated in chronic diseases such as diabetes, obesity and atherosclerosis [5–8]. PPARβ and γ have roles in early embryonic survival and implantation [9, 10]. PPARs regulate gene expression by binding to specific DNA sequences, peroxisome proliferator response elements (PPREs), in the promoter regions of target genes. Prior to DNA binding PPAR forms a heterodimer with the retinoid X receptor (RXR) [11, 12]. A number of endogenous ligands have been identified for each PPAR subtype, including long-chain fatty acids, polyunsaturated fatty acids such as linoleic and arachidonic acids, saturated fatty acids, and eicosanoids [1]. A variety of synthetic ligands have been developed for pharmaceutical purposes to treat chronic diseases such as hyperlipidemia, diabetes, and metabolic syndrome. In addition, some chemicals and environmental contaminants activate PPARs, for example, phthalates, tri- and dichloroacetic acids, trichloroethylene, and the perfluorinated alkyl and sulfonyl acid compounds (PFAAs) [11–14].
PFAAs, including perfluorooctanoic acid (PFOA), perfluorononanoic acid (PFNA), and perfluorooctane sulfonate (PFOS), are highly stable molecules with chemical properties that make them excellent surfactants [15]. For many years these chemicals were widely used in industrial applications and are now found as persistent environmental contaminants that are also present in the tissues and serum of wildlife and humans [15–17]. In laboratory studies, prenatal exposure of rodents to these compounds produces dose-related effects on pre- and postnatal survival, developmental delay, and deficits in postnatal growth [18–23]. PFAAs activate PPARα, and the developmental toxicity of PFOA and PFNA in the mouse was shown to be dependent on expression of PPARα in the fetus (effects of prenatal exposure to PFOA or PFNA that occur in wild-type mice were not observed in PPARα knockout offspring) [24, 25]. PPARs are expressed in the mouse embryo and fetus (reviewed in [26]), and prenatal exposure to PFAAs was shown to change gene expression in the pre- and postnatal livers in a pattern indicative of activation of PPAR as well as the CAR nuclear receptor [27–29].
Almost nothing is known about expression of PPAR during human development. At the present time, a search of the literature revealed only one paper that described the expression of PPARs in the human fetus, and that paper described expression in the gastrointestinal (GI) tract [30]. This gap in scientific knowledge of PPAR expression during human development requires attention as PFAAs, which activate PPAR, alter gene expression, and have developmental toxicity in the rodent, are pervasive in the environment and have been found in serum and blood samples of populations around the world, including samples from infants, children, and in umbilical cord blood and milk (indicating prenatal and postnatal exposure of infants) [17, 31–36]. Thus, in order to evaluate the potential for these environmental compounds, and others, to affect human fetal development, it is important to have information regarding the expression of PPARs in the developing human fetus. The present study reports mRNA and protein expression for PPARα, β, and γ in embryonic day (ED) 54 to 125 human fetal liver, heart, lung, kidney, stomach, intestine, adrenal, spleen, and thymus.
2. Methods and Materials
2.1. Human Fetal and Adult Samples
Human fetal tissues ranging in age from embryonic day (ED) 54 to 125 were obtained from the Birth Defects Research Laboratory at the University of Washington, Seattle. The collection of tissue specimens from clinically aborted fetuses by the Birth Defects Research Laboratory (including informed consent for the donation and all procedures) was conducted with Human Subjects Institutional Review Board (IRB) approval. At the EPA, the study was reviewed by the Office of Human Research Ethics, UNC Biomedical IRB, and approved by the National Health Effects and Environmental Research Laboratory (NHEERL) Human Research Protocol Office (HRPO).
Tissues were snap frozen as soon as possible after collection and stored at −80°C until shipped on dry ice. On arrival at EPA, samples were stored at −80°C until processed for total RNA and protein. The nine tissues analyzed included liver, heart, lung, kidney, stomach, intestine, adrenal, spleen, and thymus. Prior to processing the fetal tissues to prepare RNA and protein, samples were weighed and smaller samples were designated for RNA preparation only, while larger samples were divided for both RNA and protein preparation, and any excess sample was returned (still frozen) for storage at −80°C. Handling during the weighing and division of samples was done over dry ice to the extent possible to minimize thawing. Adult total RNA for the 9 tissues examined in the study was obtained from FirstChoice Human Total RNA Survey Panel, Ambion, Inc, (each adult sample consisted of pooled total RNA from 3 individuals). In addition, tissue samples from 23 adult human livers were available for comparison of PPAR mRNA expression in adult and fetal liver. These samples were obtained from CellzDirect, Inc. (Durham, NC). Total RNA was prepared from the frozen adult liver tissue samples and qPCR performed, as described for the fetal samples.
2.2. qPCR Experimental Design and Procedures
Each tissue was run in separate qPCR experiments (e.g., liver samples were not run with those of any other tissue). In the qPCR experiments, expression of PPARα, β, γ and an internal control gene were examined on each plate, and samples on the plate included 2 replicates of each fetal sample and of the appropriate pooled adult tissue (FirstChoice Human Total RNA). In cases where there were too many samples of a tissue to run all of the reactions on one plate, the samples were run across 2 plates such that each age was represented as equally as possible on each plate. The actual number of samples examined for each tissue is stated in the results section, but the number available ranged from 23 to 46 specimens, except for thymus which had 11 specimens. PPAR gene expression was expressed relative to an internal control gene. The fetal samples of each tissue were examined for expression of β-actin, β2-microglobulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as potential internal control genes. Regression analysis of cycle threshold (Ct) was performed for each potential control gene to detect any changes in expression with age. Based on favorable regression outcomes in all of the tissues (no significant change with age), β2-microglobulin (B2M) was selected as the internal control gene (data not shown).
Tissue was homogenized and extracted in TRI Reagent (Sigma Chemical, St. Louis, MO) according to the manufacturer's directions, and RNA pellets were stored in 70% ethanol at −80°C until further use. Following resuspension in nuclease-free water (Promega Corporation, Madison, WI), the RNA was quantified and evaluated for purity (260 nm/280 nm and 260 nm/230 nm ratio) using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies,Wilmington, DE). Prior to qPCR, 2 μg total RNA was digested using 2 units of DNaseI (Promega Corporation, Madison, WI) for 30 min at 37°C followed by 10 min at 65°C in a buffer containing 40 mM Tris (pH 8.0), 10 mM MgSO4, and 1 mM CaCl2. The DNase-treated RNA was then quantified using a Quant-iT RiboGreen RNA assay kit according to the manufacturer's protocol (Invitrogen Corporation, Carlsbad, CA). Approximately 1 μg of the DNase-treated RNA was reverse transcribed using a High-Capacity cDNA Archive Kit according to the provided protocol (Applied Biosystems). Amplification was performed on an Applied Biosystems model 7900HT Fast Real-Time PCR System in duplicate using 25 ng cDNA and TaqMan Universal PCR Master Mix (Applied Biosystems) in a total volume of 12 μl. The following TaqMan assays (Applied Biosystems) were included in the study: PPARα (Hs00947539_m1), PPARβ (Hs00602622_m1), PPARγ (Hs00234592_m1), β-actin (Hs99999903_m1), GAPDH (Hs99999905_m1), β-2 microglobulin (Hs99999907_m1).
PPAR mRNA Ct values, calculated by Applied Biosystems SDS2.2.2 software, were normalized by subtraction of the Ct for the internal control, B2M, generating δCt values. The mean δCt for each sample was calculated from the 2 replicates and then analyzed to evaluate changes in expression with fetal age (regression analysis). Differences in expression between subtypes were determined using ANOVA of all mean δCt values (without regard to age), with Bonferroni's post-test applied for pairwise comparisons (Prism 4.0, GraphPad Software, San Diego, CA). Data are graphed as a log plot of 2−δCt. For all tissues except liver, a comparison of the fetal samples with the single adult pooled sample was performed using Ct values and a t-distribution test (sample size less than 30) or the Z-distribution test (sample size equal to or greater than 30) to determine the probability that an adult value of this extreme or more extreme would be found in the distribution of fetal values (using probability calculators available on-line at http://faculty.vassar.edu/lowry/tabs.html or http://davidmlane.com/hyperstat/z_table.html). As the internal control gene expression was not the same in adult and fetal tissues, Ct values, and not δCt values, were used for this comparison. Data comparing fetal (mean Ct of 2 replicates) and adult Ct (each replicate shown in the plot) are graphed as a vertical scatter plot of Ct. For the liver, there were 23 adult liver specimens available for comparison with the fetal liver samples. The adult samples were from both males and females and ranged in age from 21 to 86 years, but analysis of either Ct or dCt showed no effect of either sex or age on the expression of PPARα, β, or γ or B2M (data not shown). The adult and fetal liver samples had comparable levels of the internal control gene, B2M, and thus it was possible to compare the normalized Ct values (dCt) using ANOVA with Bonferroni's post-test applied for pairwise comparisons (Prism 4.0, GraphPad Software, San Diego, CA).
2.3. Western Blot Experimental Design and Procedures
Samples of each tissue were run in separate Western blot experiments (e.g., liver samples were not run with those of any other tissue). In general, most of the tissues required 2-3 Western blots to accommodate all of the samples (only 12 sample lanes were available per gel), and the samples were blocked across blots such that the age range was represented as uniformly as possible on each blot. The actual number of samples examined for each tissue is stated in the results section, but the number available ranged from 5 to 36 specimens, and only thymus and spleen had fewer than 22 specimens. Each blot was examined for expression of one of the PPAR subtypes and for the internal control protein. GAPDH was selected as the internal control protein as expression did not change with age (based on regression analyses of GAPDH protein expression across age in each of the 9 tissues, data not shown). A positive control for antibody detection of PPARα, β, or γ was also run on each blot. Positive controls were Hep G2 whole cell extract (Santa Cruz, SC-2227), Jurkat cell nuclear extract (Santa Cruz, SC-2132), and U937 whole cell extract (Santa Cruz, SC-2239), for expression of PPARα, β, or γ, respectively. After all tissues were examined in this manner, additional Western blots were run in which all 9 tissues were represented on the blot, with most of the tissues from the same 91-day-old fetus (to the extent possible, as not all tissues were available from any single fetus, and thymus was not available from a 91-day fetus). Three “across-tissue” blots were run for each PPAR subtype (i.e., n = 3 fetuses per tissue examined in the “across-tissue” survey).
Western blots were run with 25 μg of total cell lysate or positive control per lane. All gels were 7.5% acrylamide precast gels (Biorad, Hercules, CA) and were run for 90 min at 125 V. Protein transfer to nitrocellulose membrane (Biorad) was done for 40 min using the Biorad semidry transfer system. Membranes were blocked for 1 hr in 5% milk and incubated overnight in primary antibody in 5% milk. Primary antibodies for PPARα (SC-9000) and PPARγ (SC-7196) were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA) and used at a dilution of 1:200. PPARβ (Abcam 21209) antibody was obtained from Abcam, Inc, (Cambridge, MA) and used at 1 : 750 dilution. Antibody for GAPDH (SC-25778) was from Santa Cruz Biotechnologies and was diluted at 1 : 10,000. After overnight incubation with primary antibody, blots were probed 1 hr with a horseradish peroxidase-conjugated secondary antibody in 5% milk. Secondary antibodies were diluted 1 : 5000 and included goat anti-rabbit Jax 111-035-144 (West Grove, PA), goat anti-rabbit KPL 074-1506 and rabbit anti-goat KPL 14-13-06 (KPL, Gaithersburg, MD). Chemiluminescence was imaged using a Biorad Fluor-S machine with 2 or 5 min exposures. Biorad Quantity One software was used to perform volume rectification densitometry with background subtraction on the chemiluminescence images, generating data for both the PPAR and GAPDH bands. PPAR protein expression was expressed relative to the internal control gene, GAPDH, and regression analysis of the relative values was performed to detect any significant change in slope with age (Prism 4.0, GraphPad Software, San Diego, CA).
3. Results
The expression of PPARs is presented for each tissue, reporting any change in expression of protein or mRNA with gestational age, comparing the relative level of mRNA expression of the isotypes, and comparing mRNA expression in the fetal organ to that observed in the human adult sample of that tissue. Protein and mRNA for all PPAR isotypes were detected in all of the 9 tissues and the results are summarized in Table 1, which also lists the tissues from highest to lowest expression of mRNA for each PPAR. Table 2 summarizes the relative expression of the isotypes within each tissue. The data is shown using the same presentation format for each tissue. A full narrative presentation is given for the first tissue presented (liver) and, for the sake of brevity, the results for other tissues omit repetitive explanations which apply to all the data sets.
Table 1.
Relative RNA1 expression for each subtype (listed from highest to lowest mean expression), characteristics2 of RNA and protein expression across age, and fetal mRNA expression relative to adult.
| RNA abundance: high to low | RNA change with age | Fetal versus adult mRNA | Protein change with age |
|---|---|---|---|
| PPARα | |||
|
| |||
| Intestine | NS | NS | Increase |
| Liver | Increase | NS | NS |
| Lung | NS | NS | Decrease |
| Heart | NS | Lower | NS |
| Kidney | NS | Lower | Decrease |
| Adrenal | NS | NS | Decrease |
| Thymus | NS | NS | NS |
| Stomach | NS | Lower | NS |
| Spleen | NS | Lower | NS |
| PPARβ | |||
|
| |||
| Intestine | Decrease | NS | NS |
| Adrenal | NS | NS | Decrease |
| Lung | NS | NS | NS |
| Kidney | NS | Lower | NS |
| Thymus | NS | Higher | NS |
| Liver | Increase | NS | NS |
| Heart | Decrease | Lower | NS |
| Spleen | NS | Lower | NS |
| Stomach | NS | Lower | NS |
| PPARγ | |||
|
| |||
| Thymus | NS | NS | NS |
| Intestine | NS | NS | NS |
| Spleen | NS | NS | NS |
| Liver | NS | Higher | NS |
| Kidney | NS | NS | Decrease |
| Lung | NS | Lower | NS |
| Stomach | NS | Lower | NS |
| Heart | NS | Lower | NS |
| Adrenal | Decrease | Lower | NS |
1Relative RNA expression based on mean Ct for all samples across all ages for each tissue, listed from highest to lowest mean expression for each subtype. 2Change in RNA and protein expression with age shown as increased, decreased, or not significant (NS) and fetal mRNA expression compared to adult expression shown as higher, lower, or not significantly (NS) different from adult.
Table 2.
Relative RNA expression of PPAR isotypes within each tissue.
| Intestine | α = β = γ |
| Liver | α = γ > β |
| Lung | α > β = γ |
| Heart | α > β > γ |
| Adrenal | α = β > γ |
|
| |
| Thymus | γ > β > α |
| Spleen | γ > α = β |
| Kidney | γ > α > β |
| Stomach | γ > α = β |
Relative expression based on mean Ct across ages for all samples of each tissue.
3.1. Liver
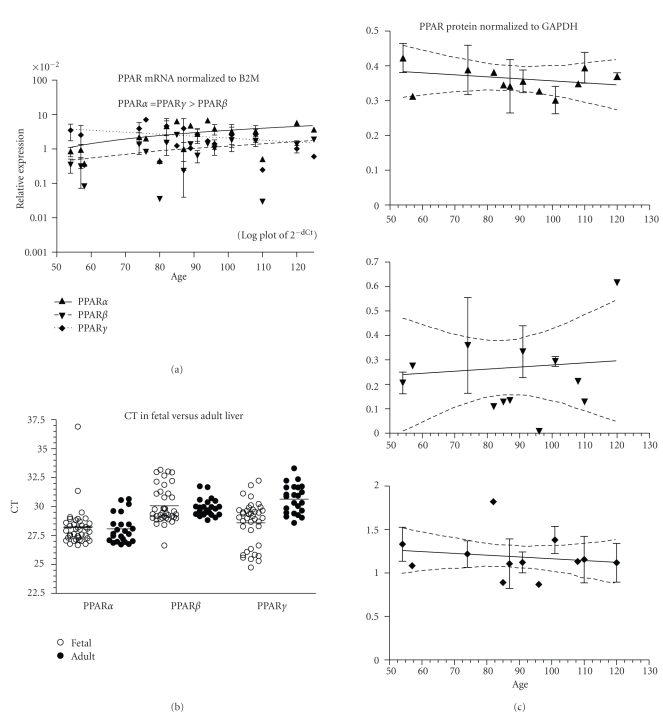
Human fetal liver expressed PPARα, β, and γ mRNA from ED54–125 (data acquired from 39 fetuses). Expression of PPARα and PPARβ increased significantly while PPARγ remained unchanged across the age range (Figure 1(a), P < .01 and .05, resp.).Expression levels of PPARα and γ across all ages were not significantly different and both were more highly expressed than PPARβ (P < .001). β2M mRNA expression was considerably higher than any of the PPAR subtypes (mean ± SEM Ct across all ages, note that a lower Ct signifies more abundant mRNA than a high Ct: β2M = 22.4 ± 0.3, PPARα = 28.2 ± 0.3, PPARβ = 30.1 ± 0.3, PPARγ = 28.6 ± 0.3). The expression of PPAR mRNA in the fetal liver was compared to that in 23 adult human liver samples. The adult samples were from both males and females and ranged in age from 21 to 86 years, but analysis showed no effect of either sex or age on the expression of PPARα, β, γ, or B2M (data not shown). The expression of B2M, the internal control gene, in adult and fetal liver samples was not significantly different (21.9 ± 0.2, 22.4 ± 0.3, mean ± SEM, resp.), and thus it was possible to analyze PPAR values normalized to B2M (dCt). Analysis of unadjusted Ct or dCt gave the same outcomes, and Figure 1(b) shows plots of the Ct values for adult and fetal livers. The human fetal and adult livers expressed PPARα and β mRNA at levels that were not significantly different (Figure 1(b)), but PPARγ was significantly higher in the fetal liver (P < .001, lower mean Ct indicates higher mRNA present in a sample). The overall outcome for PPARα and β was the same from the pooled total RNA from 3 donors (Ambion FirstChoice liver sample, data not shown) as that from the 23 individuals; adult and fetal liver expression did not significantly differ. The Ambion FirstChoice pooled adult liver RNA indicated that PPARγ did not differ between adult and fetal livers, but the data from the 23 individuals showed a significantly higher expression in the fetus, and the larger “n” of that assay would lend support to the validity of that outcome. In the fetal liver, PPARα, β, and γ protein expression did not change with fetal age (Figure 1(c); data from 22 fetuses, ED54–120).
Figure 1.
Liver. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult liver. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) and adult (filled circles) sample (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density is normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
3.2. Heart
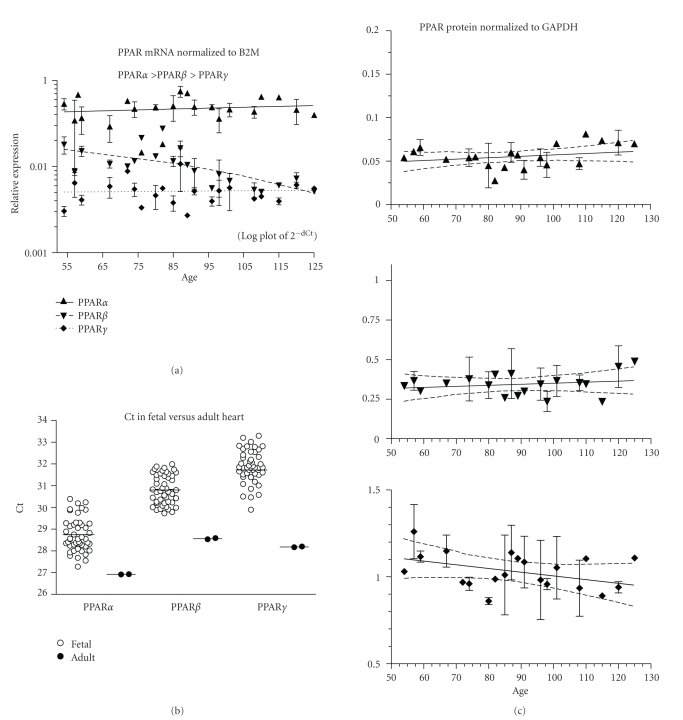
PPARα and γ expression did not change, but PPARβ expression decreased (P < .0001) with fetal age (Figure 2(a), ED54–125, n = 46 fetuses).PPARα, PPARβ, and PPARγ relative expressions are shown in Table 2 and differences between isotypes were significant at P < .001. β2M mRNA expression was higher than PPAR (mean Ct ± SEM: β2M = 24.2 ± 0.8, PPARα = 28.7 ± 0.1, PPARβ = 30.8 ± 0.1, and PPARγ = 31.9 ± 0.1). Fetal PPARα, β, and γ mRNA expression was lower than that of the adult sample (Figure 2(b); P < .05,.001,.001, resp.). PPAR protein expression did not change with fetal age (Figure 2(c); 36 fetuses, ED54–125).
Figure 2.
Heart. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult heart. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
3.3. Lung
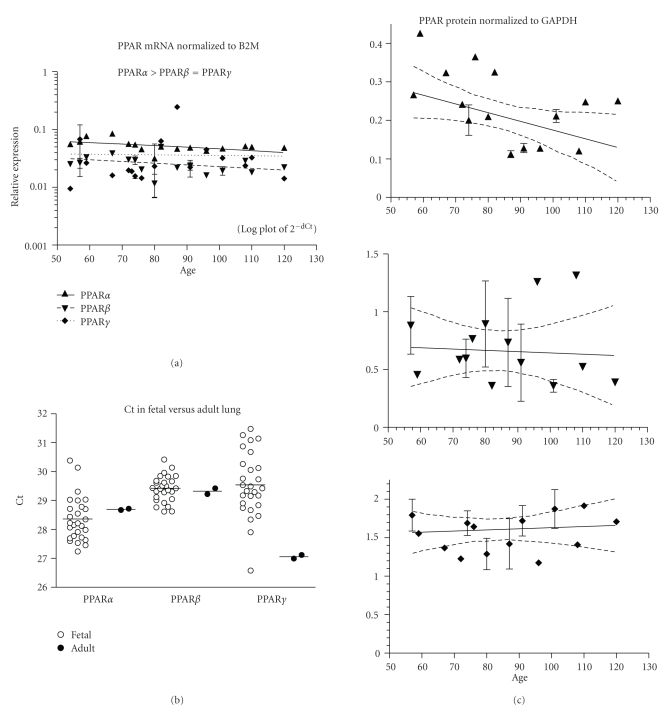
PPAR mRNA expression in fetal lung did not change with age (Figure 3(a), ED54–120, n = 27 fetuses). PPARα was the most highly expressed isotype (P < .001, Table 2). β2M mRNA expression was higher than PPAR, (mean Ct ± SEM: β2M = 24.0 ± 0.2, PPARα = 28.4 ± 0.1, PPARβ = 29.4 ± 0.1, and PPARγ = 29.5 ± 0.2). Fetal and adult PPARα and β mRNA expressions were not different, but fetal PPARγ was lower (Figure 3(b); P < .05). PPARα protein levels decreased (P < .05), but PPARβ and γ did not change with fetal age (Figure 3(c), ED57 to 120, n = 27 fetuses).
Figure 3.
Lung. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult lung. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
3.4. Kidney
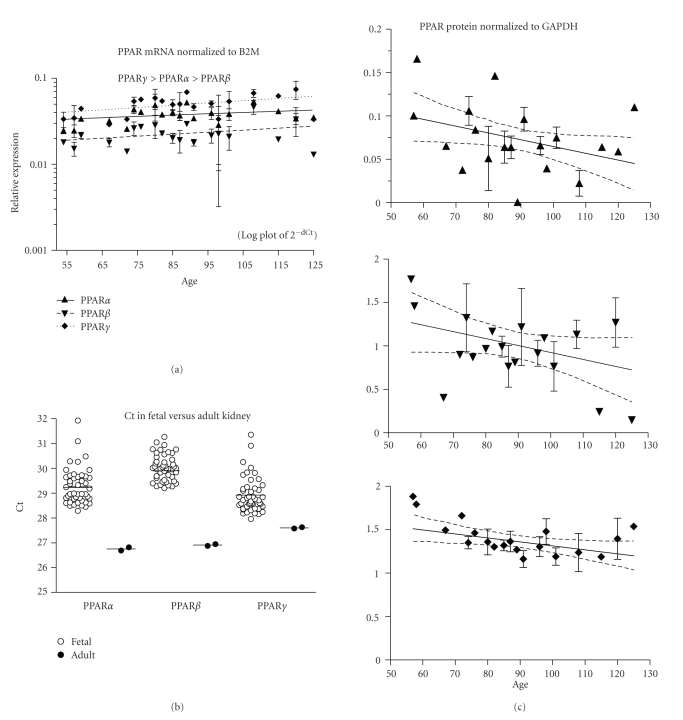
PPAR mRNA expression did not change with age (Figure 4(a), 46 fetuses, ED54–125). PPARγ expression was higher than PPARα and PPARβ (P < .05, P < .001, resp.), and PPARα was higher than PPARβ (P < .001). β2M mRNA expression was higher than PPAR (mean Ct ± SEM: β2M = 24.5 ± 0.1, PPARα = 29.3 ± 0.1, PPARβ = 30.0 ± 0.1, PPARγ = 28.9 ± 0.1). PPARα and β fetal mRNAs were lower than in the adult (Figure 4(b); P < .01, <.0001, respectively), but PPARγ was similar (P = .07). PPARβ protein expression did not change with fetal age (P = .09), but PPARα and γ decreased (P < .05, Figure 4(c); 36 fetuses, ED57–125).
Figure 4.
Kidney. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult kidney. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
3.5. Stomach
PPAR mRNA expression did not change with age (Figure 5(a), 35 fetuses, ED54–120). PPARγ was the most highly expressed isotype (P < .001, Table 2). β2M mRNA expression was higher than PPAR (mean Ct ± SEM: β2M = 28.0 ± 0.3, PPARα = 32.4 ± 0.3, PPARβ = 33.3 ± 0.3, and PPARγ = 29.8 ± 0.4). PPARα, β, and γ mRNA expression was lower in fetal than in adult stomach (Figure 5(b); P < .01, <.0001, <.05, resp.). PPAR protein expression did not change with fetal age (Figure 5(c); 26 fetuses, ED59–120).
Figure 5.
Stomach. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult stomach. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
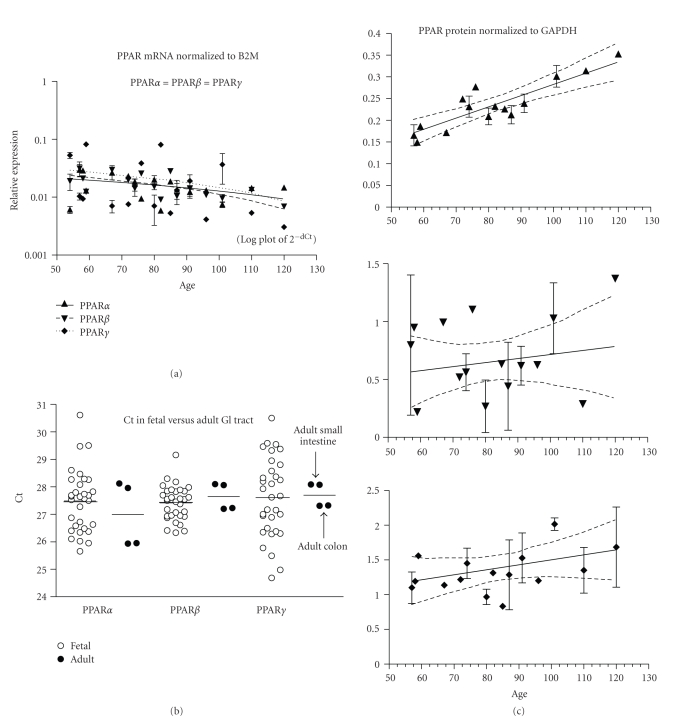
3.6. Intestine
PPARα and γ mRNA expression did not change with age, but PPARβ decreased (P < .001, Figure 6(a), 32 fetuses, ED54–120). PPARα, β, and γ were expressed at similar levels (Table 2). β2M mRNA expression was higher than PPAR (mean Ct ± SEM: β2M = 21.4 ± 0.2, PPARα = 27.5 ± 0.2, PPARβ = 27.4 ± 0.1, and PPARγ = 27.6 ± 0.3). Fetal intestinal PPAR mRNA was not significantly different from either the adult small intestine or the adult colon (Figure 6(b)). PPARα protein expression increased (P < .001), while PPARβ and γ proteins did not change with fetal age (Figure 6(c); 29 fetuses, ED57–120).
Figure 6.
Intestine. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult small intestine and colon (filled circles show the 2 replicates of small intestine above and 2 replicates of colon below the line indicating the mean of the combined tissue values). Each open symbol represents the mean Ct value of 2 replicates for each fetal sample (overall mean is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density is normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
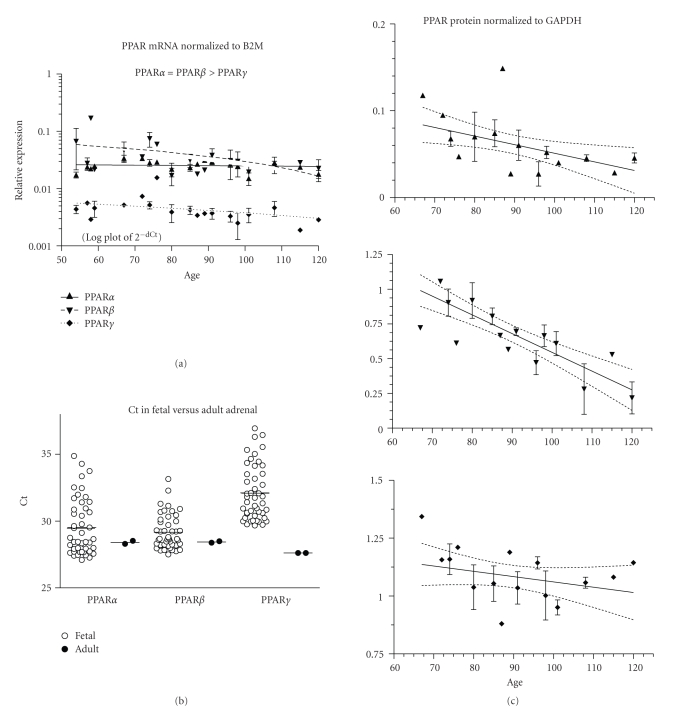
3.7. Adrenal
PPARγ mRNA decreased with age (P < .05), while PPARα and β remained unchanged (although P = .0503 for PPARβ; Figure 7(a), 46 fetuses, ED54–120). PPARα and β mRNAs were more highly expressed than PPARγ (P < .001, Table 2). β2M mRNA expression was higher than PPAR, (mean Ct ± SEM: β2M = 24.2 ± 0.3, PPARα = 29.5 ± 0.3, PPARβ = 29.2 ± 0.2, and PPARγ = 32.1 ± 0.3). Fetal and adult PPARα and β mRNAs were not different, but PPARγ was lower in fetal adrenal (P < .05; Figure 7(b)). PPARα and β protein expression decreased with fetal age (P < .05, P < .001, resp.; Figure 7(c); 36 fetuses, ED67–120).
Figure 7.
Adrenal. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult adrenal. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density is normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
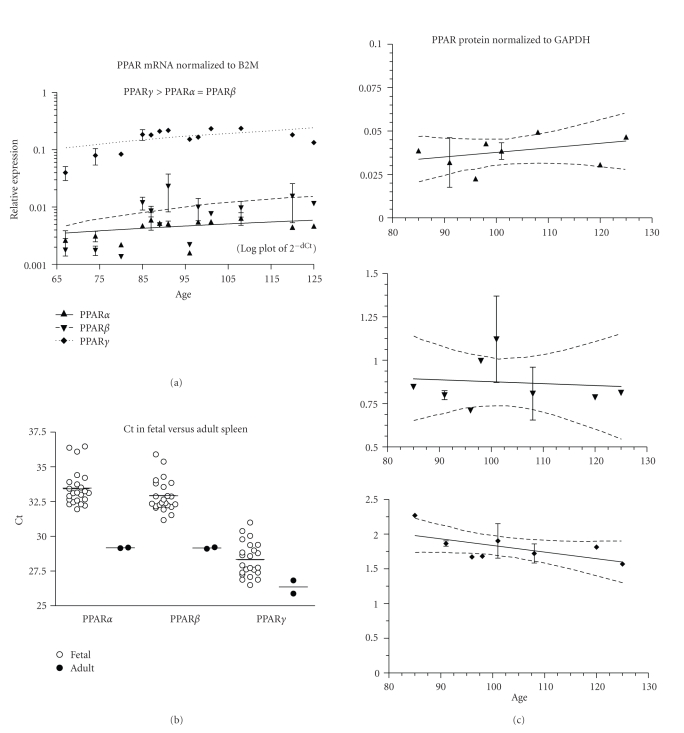
3.8. Spleen
PPAR mRNA expression did not change with age (Figure 8(a), 23 fetuses, ED67–125). PPARγ was the most highly expressed isotype (P < .001, Table 2). β2M mRNA expression was higher than PPAR (mean Ct ± SEM: β2M = 25.6 ± 0.2, and PPARα = 33.5 ± 0.3, PPARβ = 32.9 ± 0.2, PPARγ = 28.3 ± 0.3). Fetal PPARα and β mRNAs were lower than in the adult (P < .01, Figure 8(b)). PPARα, β, and γ protein expression did not change with fetal age (Figure 8(c); 11 fetuses, ED85–125).
Figure 8.
Spleen. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult spleen. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density is normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, and down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
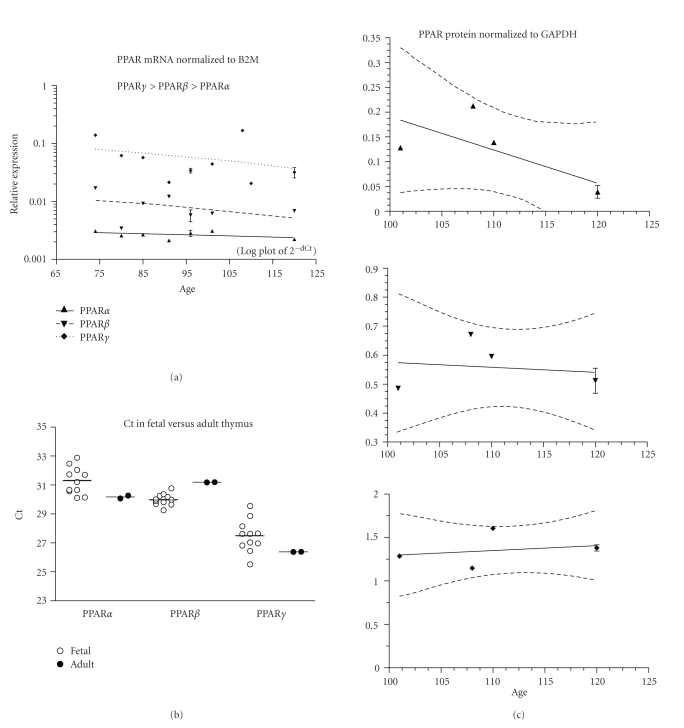
3.9. Thymus
PPAR mRNA expression did not change with age (Figure 9(a), 11 fetuses, ED74–120). PPARγ mRNA expression was higher than PPARα or β (P < .001), and that of PPARβ was higher than PPARα (P < .01). β2M mRNA expression was higher than PPAR (mean Ct ± SEM Ct: β2M = 23.1 ± 0.4, PPARα = 31.3 ± 0.3, PPARβ = 30.0 ± 0.1, and PPARγ = 27.5 ± 0.3). PPARβ fetal mRNA expression was higher than in the adult (P < .05, Figure 9(b)). PPAR protein expression did not change with fetal age (Figure 9(c); 5 fetuses, ED101–120).
Figure 9.
Thymus. (a) The expression of PPARα, β, and γ mRNA is shown across the fetal age range. Log plot of mean ± SEM Ct is normalized to β-2-microglobulin (B2M). (b) The fetal expression of PPARα, β, and γ is shown relative to expression in adult thymus. Each symbol represents the mean Ct value of 2 replicates for each fetal (open circles) sample and adult (filled circles) individual replicates are shown (overall mean for each group is shown as a horizontal line). (c) PPARα, β, and γ protein expression is shown across the fetal age range. Western blot density is normalized to glyceraldehyde-3-phophate dehydrogenase (GAPDH). Up arrowhead indicates PPARα, and down arrowhead PPARβ, and diamond PPARγ. If only one sample was available for a particular age, then an error term could not be calculated and no SEM bar is shown. Regression analysis evaluated change with age. Dashed lines in graphs of C are the 95% confidence interval.
3.10. Comparison of PPARα, β, γ Expression Levels in Different Tissues
Table 1 lists the tissues in an order based on the level of RNA expression in fetal tissues such that the first tissue listed for each subtype has the highest and the last in the list has the lowest expression. The ranking for RNA expression is based on the mean Ct across all ages for each tissue. Expression of PPARα mRNA is the highest in the intestine, liver, and lung and is relatively low in stomach and spleen. PPARβ was the highest in intestine, adrenal, and lung, while expression in spleen and stomach was relatively low. PPARγ was the highest in thymus, intestine, and spleen, but was poorly expressed in heart and adrenal. Among all the tissues, intestine was unique in having high expression of all three subtypes, although lung expressed high levels of both PPARα and β. Stomach poorly expressed all three subtypes relative to the other tissues. Spleen showed weak expression of both PPARα and β, while expression of PPARβ and γ was weak in the heart.
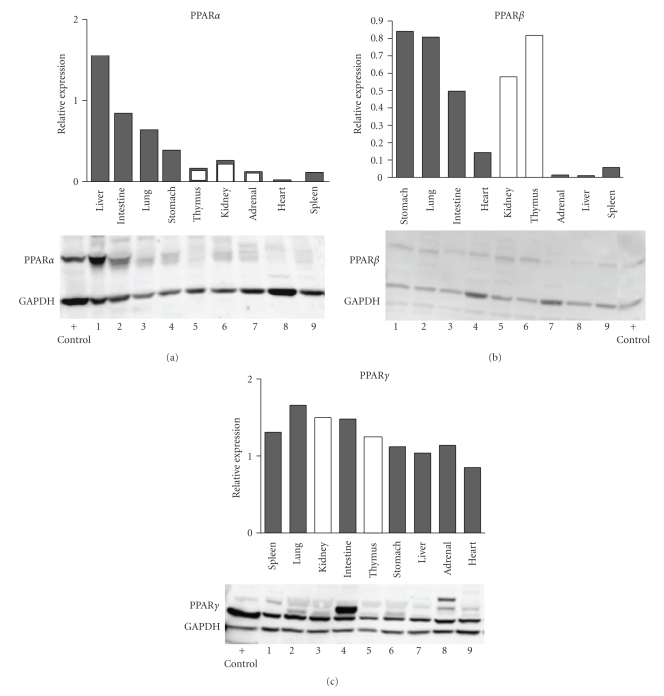
Comparison of the relative levels of protein expression for each PPAR subtype is not presented. The expression of GAPDH did not change with age, making it a suitable loading control for normalization of the PPAR densitometry values from the Western blots for each tissue across age (data not shown); however, GAPDH expression was substantially different between the tissues and that makes it inappropriate to compare or rank the normalized expression between the different tissues. Even considering that the levels of GAPDH were not uniformly expressed in the different tissues, it is clear that there were different levels of PPARα, β, and γ proteins in the various tissues. This can be seen in Figure 10 which illustrates the expression of PPAR proteins in all 9 tissues on single blots for each subtype. In these assays, all 9 tissues were present on each blot and, to the extent possible, the tissues on each blot were from the same 91-day old fetus. Thymus was only available from 101-, 108-, and 110-day old fetuses. Three of the cross-tissue assays (each using tissues from different fetuses) were run for each PPAR subtype and examples of the multiple tissue blots are shown in Figure 10.
Figure 10.
Western blots are shown in which all 9 tissues are present on each blot. On the PPARα blot, all tissues shown by dark bars were from a single 91-day-old fetus, and adrenal and kidney (white bars) were from different 91-day-old fetuses, and thymus was from a 101-day-old fetus. The blots for PPARβ and PPARγ used samples from a 91-day-old fetus (dark bars, the same set of samples for both PPARβ and γ) with kidney and thymus samples (white bars) from different fetuses (91 and 108 days, resp.). Blot images are labeled to show the location of the PPAR band, the GAPDH band, and lane containing the positive control (Hep G2 whole cell extract, Jurkat cell nuclear extract, and U937 whole cell extract, for expression of PPARα, β, or γ, resp.). The densitometry data (PPAR expression normalized to GAPDH) for each gel is shown above the blot image. Lanes 1–9 contain the samples listed on the x-axis of the bar graphs.
4. Discussion
This study provides new information regarding the expression of PPAR subtypes during human fetal development. PPARα, β and γ are expressed in the human fetus from embryonic days 54 to 125. Protein and mRNA for all three PPAR subtypes were detected in the 9 tissues examined in this study. In some organs, the expression of mRNA or protein changed during the developmental period examined. Relative levels of mRNA expression of the PPAR subtypes varied by tissue. In some organs, the level of mRNA expressed was comparable to or higher than that of the adult tissue.
Human fetal expression of PPAR subtypes can be considered similar to the expression patterns reported for the laboratory rodent, reviewed in [26]. In mouse and rat, PPAR mRNA and/or protein was detected during prenatal and postnatal development for liver, kidney, heart, lung, adrenal, spleen, vertebra, tissues of the central nervous system (CNS), brain, adipose, fat, muscle, and skin. The patterns of expression varied by tissue and were dependent on developmental stage. It is difficult to make specific comparisons between developmental patterns of PPAR expression in the laboratory animal and the human fetal tissues of this study as comparisons between comparable developmental stages become complicated following the end of organogenesis [26]. In the present study, the period of human fetal development ranged from about 8 to 18 weeks, a period following organogenesis and encompassing the fetal stage of rapid growth, differentiation, and functional maturation of the organ systems. The end of organogenesis and beginning of the fetal period are generally considered to occur at the end of the eighth week of gestation [37] and a landmark of the entry to the fetal stage is the fusion of the secondary palate. Palatal fusion in human fetuses begins around embryonic day 54 and is generally complete in the 56-57-day-old fetus [37, 38]. In the mouse and rat, palatal fusion occurs on ED14-15 and 16-17, respectively, although this can vary by a day or two depending on the strain. Thus, it may be reasonable to consider the ED14 mouse, ED16 rat, and the ED54–56 human fetal tissues to be at comparable developmental stages for purposes of comparison of PPAR expression. Restricting the discussion to that specific developmental period (end of organogenesis marked by palatal fusion), the comparisons of human and rodent PPAR expression are somewhat limited. Overall, as discussed below, there are similarities, and also some differences, in the expression of PPAR in rodent and human fetuses at the end of organogenesis.
In the ED15.5 rat liver, moderate levels of mRNA for PPARα and β were found and PPARβ protein was reported in ED15 mouse liver and PPARγ2 protein was detected at a slightly earlier stage (ED13) in mouse liver [39, 40]. In the present study, PPARα was highly expressed in the human fetal liver and relatively abundant compared to other tissues (only intestine was higher). When evaluated across all ages, PPARα and γ were more abundant than β in liver.
Rat heart and lung expressed PPARα and β, and PPARβ protein expression is reported for mouse heart and lung [40, 41]. In the present study, human fetal lung and heart had high expression of PPARα and lung strongly expressed PPARα and β relative to the other organs. In human fetal heart and lung, PPARα was more abundant than β or γ, and in heart γ was the subtype with the least expression.
The ED15.5 rat and 14.5 mouse kidneys expressed PPARα mRNA. PPARβ mRNA was found in rat and PPARβ and γ were weakly detected in the mouse kidney [41, 42]. In human fetal kidney, PPARγ was expressed at higher levels than α or β, and β was the least abundant subtype in kidney.
We are not aware of any published data regarding expression of PPAR in thymus or spleen of the developing rodent. In the human fetal spleen, PPARα and β were expressed at low but equivalent levels and PPARγ was the most abundant subtype with relatively high expression (only those of thymus and intestine were higher). In human fetal thymus, PPARγ mRNA was very abundant (higher than in any other tissue) and PPARβ and α were detected at lower levels than PPARγ.
In rat GI tract, mRNAs for PPARα and β, but not γ (reported as not detected), were expressed, and PPARβ protein was reported in mouse GI tissue [40, 41]. The present study found high expression in intestine for all PPAR subtypes relative to the other organs, and PPARα, β, and γ mRNAs were at equivalent levels. Stomach, which was examined separately, had lower expression of all subtypes relative to intestine, and PPARγ was the most highly expressed subtype in stomach. Huin et al. [30] examined PPAR protein expression in the fetal human digestive tract (aged 7 to 22 weeks) using immunohistochemistry and found spatial and temporal patterns of expression in esophagus, stomach, jejunum, ilium, and colon. In the present study, using qPCR and Western blotting methods, no change with age was detected in stomach for expression of PPARα, β, or γ. Huin's report found slightly less PPARα at 19 weeks compared to 12 and 15 weeks of age, while PPARβ and γ were slightly higher at 15 and 16 weeks, respectively, than at 12 or 19 weeks of gestation. The 19-week observations of Huin's study are just outside the age range of the present study, but the slight changes in protein reported by Huin differ from our observations of mRNA and protein at the earlier ages. The intestinal expression of PPAR reported for the various regions observed in Huin's study is similar as an overall pattern to that found in the present study; however, in the present study it was not possible to separate regions of the intestinal tract. Huin reported increasing PPARα in the ileum from 12 to 22 weeks of age, similar to the increase with age in PPARα protein observed from 8 to 18 weeks in the present study. Similarly, Huin reported that PPARβ and γ protein expression in the jejunum and ilium was similar across time (7–16 and 12–22 weeks, resp.), and the present study also found no significant change in protein expression of PPARβ or γ with age.
An important finding of the present study is that fetal tissues can express PPAR at levels equivalent to those of the adult tissues (or higher in the case of PPARγ in liver and PPARβ in thymus). However, some caution is needed as the adult data for each tissue (with the exception of liver) is based on a pooled total RNA sample from 3 donors and it is not known whether a similar outcome would be derived from a larger number of adult donors. However, in the case of liver, the data from 23 individuals supported the data from the pooled sample, that is, expressions of PPARα and β were not significantly different in adult and fetal livers. However, the pooled sample did not detect the increased expression of PPARγ in fetal liver relative to the adult, as observed in the 23 individual liver samples. Thus, the adult versus fetal comparison provides data that were previously not available and represent the only information for this endpoint. However, it is important to recognize that comparisons of these data with additional analyses from larger adult tissue sets would be desirable.
In summary, this study is unique in providing substantial information on the expression of PPARα, β, and γ during human fetal development. Among the strengths of the study are the acquisition of both protein and mRNA data from the same samples, the inclusion of multiple tissues from most fetuses, and the large number of individuals represented in the sample set. Representation of tissues across a range of ages supported an evaluation of whether PPAR expression changed as development progressed. The qPCR approach supported estimation of the relative expression of subtypes within a tissue as well as supporting comparisons of expression of each subtype across the different tissues. As mentioned in the introduction, an important aspect of this study was to provide information for use in assessing the potential for the human fetus to respond to PPAR agonists. Studies in human fetal tissues of responses to PPAR agonists are generally not feasible; thus, it is important to at least have information on the developmental expression of PPAR and how that compares to adult expression. This study contributes to our knowledge regarding the expression of PPAR during development and compares fetal and adult PPAR expression. An important finding of the study is that fetal tissues can have expression levels equivalent to those of the adult tissues (or higher in the case of PPARγ in liver and PPARβ in thymus). The role of PPAR subtypes in the developing fetus remains unclear, but it is likely that these nuclear receptors have roles similar to those described for adult tissues, including regulation of energy homeostasis as well as lipid and glucose utilization. During the fetal stages examined in this study, the organs undergo rapid growth, differentiation, and acquisition of functionality. Exogenous agents that alter PPAR signaling in the adult, such as environmental agents, chemicals, or drugs, are capable of affecting lipid and glucose utilization, cholesterol biosynthesis, and other metabolic pathways, and these attributes make PPAR signaling an attractive target for pharmaceuticals directed at management of disease states (diabetes, metabolic syndrome, hyperlipidemia) [4, 40]. This study showed that PPAR subtypes are expressed during human fetal development in many organs and it is likely that PPAR expression and function during development are tightly regulated. It is not clear whether specific agents perturb PPAR expression or function in the fetus, whether such perturbations will have consequences or whether effects might emerge at or persist through much later life stages. However, the demonstration of expression of PPARα, β, and γ in nine major organs during human fetal development renders consideration of such issues highly relevant.
Acknowledgments
Drs. Michael Devito and Chris Corton (US EPA) generously made available samples of adult human liver, which they obtained from CellzDirect, Inc. (Durham, NC). The authors also appreciate the assistance of Cynthia J. Wolf and Jason Stanko with the animal work in this study.
References
- 1.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Reviews. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 2.Keller H, Devchand PR, Perroud M, Wahli W. PPARα structure-function relationships derived from species-specific differences in responsiveness to hypolipidemic agents. Biological Chemistry. 1997;378(7):651–655. doi: 10.1515/bchm.1997.378.7.651. [DOI] [PubMed] [Google Scholar]
- 3.Michalik L, Desvergne B, Dreyer C, Gavillet M, Laurini RN, Wahli W. PPAR expression and function during vertebrate development. International Journal of Developmental Biology. 2002;46(1):105–114. [PubMed] [Google Scholar]
- 4.Escher P, Wahli W. Peroxisome proliferator-activated receptors: insight into multiple cellular functions. Mutation Research. 2000;448(2):121–138. doi: 10.1016/s0027-5107(99)00231-6. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y, Kim S. Genetic manipulations of PPARs: effects on obesity and metabolic disease. PPAR Research. 2007;2007:12 pages. doi: 10.1155/2007/12781. Article ID 12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurnell M. ’Striking the right balance’ in targeting PPARγ in the metabolic syndrome: novel insights from human genetic studies. PPAR Research. 2007;2007:14 pages. doi: 10.1155/2007/83593. Article ID 83593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees WD, McNeil CJ, Maloney CA. The roles of PPARs in the fetal origins of metabolic health and disease. PPAR Research. 2008;2008:8 pages. doi: 10.1155/2008/459030. Article ID 459030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semple RK, Chatterjee VKK, O’Rahilly S. PPARγ and human metabolic disease. Journal of Clinical Investigation. 2006;116(3):581–589. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barak Y, Liao D, He W, et al. Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barak Y, Nelson MC, Ong ES, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 11.Tan NS, Michalik L, Desvergne B, Wahli W. Multiple expression control mechanisms of peroxisome proliferator-activated receptors and their target genes. Journal of Steroid Biochemistry and Molecular Biology. 2005;93(2–5):99–105. doi: 10.1016/j.jsbmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 12.Goya K, Sumitani S, Otsuki M, et al. The thiazolidinedione drug troglitazone up-regulates nitric oxide synthase expression in vascular endothelial cells. Journal of Diabetes and Its Complications. 2006;20(5):336–342. doi: 10.1016/j.jdiacomp.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Klaunig JE, Babich MA, Baetcke KP, et al. PPARα agonist-induced rodent tumors: modes of action and human relevance. Critical Reviews in Toxicology. 2003;33(6):655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- 14.Zoete V, Grosdidier A, Michielin O. Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. Biochimica et Biophysica Acta. 2007;1771(8):915–925. doi: 10.1016/j.bbalip.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Andersen ME, Butenhoff JL, Chang S-C, et al. Perfluoroalkyl acids and related chemistries—toxicokinetics and modes of action. Toxicological Sciences. 2008;102(1):3–14. doi: 10.1093/toxsci/kfm270. [DOI] [PubMed] [Google Scholar]
- 16.Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, Seed J. Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicological Sciences. 2007;99(2):366–394. doi: 10.1093/toxsci/kfm128. [DOI] [PubMed] [Google Scholar]
- 17.Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, Seed J. Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicological Sciences. 2007;99(2):366–394. doi: 10.1093/toxsci/kfm128. [DOI] [PubMed] [Google Scholar]
- 18.Wolf CJ, Fenton SE, Schmid JE, et al. Developmental toxicity of perfluorooctanoic acid in the CD-1 mouse after cross-foster and restricted gestational exposures. Toxicological Sciences. 2007;95(2):462–473. doi: 10.1093/toxsci/kfl159. [DOI] [PubMed] [Google Scholar]
- 19.Lau C, Thibodeaux JR, Hanson RG, et al. Effects of perfluorooctanoic acid exposure during pregnancy in the mouse. Toxicological Sciences. 2006;90(2):510–518. doi: 10.1093/toxsci/kfj105. [DOI] [PubMed] [Google Scholar]
- 20.Lau C, Thibodeaux JR, Hanson RG, et al. Exposure to perfluorooctane sulfonate during pregnancy in rat and mouse. II: postnatal evaluation. Toxicological Sciences. 2003;74(2):382–392. doi: 10.1093/toxsci/kfg122. [DOI] [PubMed] [Google Scholar]
- 21.Lau C, Butenhoff JL, Rogers JM. The developmental toxicity of perfluoroalkyl acids and their derivatives. Toxicology and Applied Pharmacology. 2004;198(2):231–241. doi: 10.1016/j.taap.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 22.Luebker DJ, Case MT, York RG, Moore JA, Hansen KJ, Butenhoff JL. Two-generation reproduction and cross-foster studies of perfluorooctanesulfonate (PFOS) in rats. Toxicology. 2005;215(1-2):126–148. doi: 10.1016/j.tox.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Luebker DJ, York RG, Hansen KJ, Moore JA, Butenhoff JL. Neonatal mortality from in utero exposure to perfluorooctanesulfonate (PFOS) in Sprague-Dawley rats: dose-response, and biochemical and pharamacokinetic parameters. Toxicology. 2005;215(1-2):149–169. doi: 10.1016/j.tox.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Abbott BD, Wolf CJ, Schmid JE, et al. Perfluorooctanoic acid-induced developmental toxicity in the mouse is dependent on expression of peroxisome proliferator-activated receptor-alpha. Toxicological Sciences. 2007;98(2):571–581. doi: 10.1093/toxsci/kfm110. [DOI] [PubMed] [Google Scholar]
- 25.Wolf CJ, Lau C, Abbott BD. Developmental effects of perfluorononanoic acid are dependent on peroxisome proliferator-activated receptor-alpha. The Toxicologist. 2009;108(1):p. 353. doi: 10.1155/2010/282896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbott BD. Review of the expression of peroxisome proliferator-activated receptors alpha (PPARα), beta (PPARβ), and gamma (PPARγ) in rodent and human development. Reproductive Toxicology. 2009;27(3-4):246–257. doi: 10.1016/j.reprotox.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Rosen MB, Abbott BD, Wolf DC, et al. Gene profiling in the livers of wild-type and PPARα-null mice exposed to perfluorooctanoic acid. Toxicologic Pathology. 2008;36(4):592–607. doi: 10.1177/0192623308318208. [DOI] [PubMed] [Google Scholar]
- 28.Rosen MB, Schmid JE, Das KP, Wood CR, Zehr RD, Lau C. Gene expression profiling in the liver and lung of perfluorooctane sulfonate-exposed mouse fetuses: comparison to changes induced by exposure to perfluorooctanoic acid. Reproductive Toxicology. 2009;27(3-4):278–288. doi: 10.1016/j.reprotox.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Rosen MB, Thibodeaux JR, Wood CR, Zehr RD, Schmid JE, Lau C. Gene expression profiling in the lung and liver of PFOA-exposed mouse fetuses. Toxicology. 2007;239(1-2):15–33. doi: 10.1016/j.tox.2007.06.095. [DOI] [PubMed] [Google Scholar]
- 30.Huin C, Corriveau L, Bianchi A, et al. Differential expression of peroxisome proliferator-activated receptors (PPARs) in the developing human fetal digestive tract. Journal of Histochemistry and Cytochemistry. 2000;48(5):603–611. doi: 10.1177/002215540004800504. [DOI] [PubMed] [Google Scholar]
- 31.Apelberg BJ, Witter FR, Herbstman JB, et al. Cord serum concentrations of perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) in relation to weight and size at birth. Environmental Health Perspectives. 2007;115(11):1670–1676. doi: 10.1289/ehp.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada K, Saito N, Inoue K, et al. The influence of time, sex and geographic factors on levels of perfluorooctane sulfonate and perfluorooctanoate in human serum over the last 25 years. Journal of Occupational Health. 2004;46(2):141–147. doi: 10.1539/joh.46.141. [DOI] [PubMed] [Google Scholar]
- 33.Karrman A, Ericson I, van Bavel B, et al. Exposure of perfluorinated chemicals through lactation: levels of matched human milk and serum and a temporal trend, 1996–2004, in Sweden. Environmental Health Perspectives. 2007;115(2):226–230. doi: 10.1289/ehp.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuklenyik Z, Reich JA, Tully JS, Needham LL, Calafat AM. Automated solid-phase extraction and measurement of perfluorinated organic acids and amides in human serum and milk. Environmental Science and Technology. 2004;38(13):3698–3704. doi: 10.1021/es040332u. [DOI] [PubMed] [Google Scholar]
- 35.Butenhoff JL, Gaylor DW, Moore JA, et al. Characterization of risk for general population exposure to perfluorooctanoate. Regulatory Toxicology and Pharmacology. 2004;39(3):363–380. doi: 10.1016/j.yrtph.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 36.So MK, Yamashita N, Taniyasu S, et al. Health risks in infants associated with exposure to perfluorinated compounds in human breast milk from Zhoushan, China. Environmental Science and Technology. 2006;40(9):2924–2929. doi: 10.1021/es060031f. [DOI] [PubMed] [Google Scholar]
- 37.Sadler TW. Langman's Medical Embryology. 8th edition. Baltimore, Md, USA: Lippincott Willians & Wilkins; 2000. [Google Scholar]
- 38.Abbott BD, Probst MR, Perdew GH, Buckalew AR. AH receptor, ARNT, glucocorticoid receptor, EGF receptor, EGF, TGFα, TGFβ1, TGFβ2, and TGFβ3 expression in human embryonic palate, and effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Teratology. 1998;58(2):30–43. doi: 10.1002/(SICI)1096-9926(199808)58:2<30::AID-TERA4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Beck F, Plummer S, Senior PV, Byrne S, Green S, Brammar WJ. The ontogeny of peroxisome-proliferator-activated receptor gene expression in the mouse and rat. Proceedings of the Royal Society B. 1992;247(1319):83–87. doi: 10.1098/rspb.1992.0012. [DOI] [PubMed] [Google Scholar]
- 40.Keller JM, Collet P, Bianchi A, et al. Implications of peroxisome proliferator-activated receptors (PPARS) in development, cell life status and disease. International Journal of Developmental Biology. 2000;44(5):429–442. [PubMed] [Google Scholar]
- 41.Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor- α, -β, and -γ during rat embryonic development. Endocrinology. 1998;139(6):2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 42.Yang T, Michele DE, Park J, et al. Expression of peroxisomal proliferator-activated receptors and retinoid X receptors in the kidney. American Journal of Physiology. 1999;277(6):F966–F973. doi: 10.1152/ajprenal.1999.277.6.F966. [DOI] [PubMed] [Google Scholar]