Abstract
Background
Autism Spectrum Disorders (ASD) are phenotypically heterogeneous, characterized by impairments in the development of communication and social behaviour and the presence of repetitive behaviour and restricted interests. Dissecting the genetic complexity of ASD may require phenotypic data reflecting more detail than is offered by a categorical clinical diagnosis. Such data are available from the Social Responsiveness Scale (SRS) which is a continuous, quantitative measure of social ability giving scores that range from significant impairment to above average ability.
Methods
We present genome-wide results for 64 multiplex and extended families ranging from two to nine generations. SRS scores were available from 518 genotyped pedigree subjects, including affected and unaffected relatives. Genotypes from the Illumina 6 k single nucleotide polymorphism panel were provided by the Center for Inherited Disease Research. Quantitative and qualitative analyses were done using MCLINK, a software package that uses Markov chain Monte Carlo (MCMC) methods to perform multilocus linkage analysis on large extended pedigrees.
Results
When analysed as a qualitative trait, linkage occurred in the same locations as in our previous affected-only genome scan of these families, with findings on chromosomes 7q31.1-q32.3 [heterogeneity logarithm of the odds (HLOD) = 2.91], 15q13.3 (HLOD = 3.64), and 13q12.3 (HLOD = 2.23). Additional positive qualitative results were seen on chromosomes 6 and 10 in regions that may be of interest for other neuropsychiatric disorders. When analysed as a quantitative trait, results replicated a peak found in an independent sample using quantitative SRS scores on chromosome 11p15.1-p15.4 (HLOD = 2.77). Additional positive quantitative results were seen on chromosomes 7, 9, and 19.
Conclusions
The SRS linkage peaks reported here substantially overlap with peaks found in our previous affected-only genome scan of clinical diagnosis. In addition, we replicated a previous SRS peak in an independent sample. These results suggest the SRS is a robust and useful phenotype measure for genetic linkage studies of ASD. Finally, analyses of SRS scores revealed linkage peaks overlapping with evidence from other studies of neuropsychiatric diseases. The information available from the SRS itself may, therefore, reveal locations for autism susceptibility genes that would not otherwise be detected.
Background
Compelling evidence exists to suggest that autism spectrum disorder (ASD) has a complex heterogeneous genetic aetiology [1]. However, the specific nature of the genetic aetiology is not well understood. This genetic heterogeneity is, perhaps, not surprising given the phenotypic complexity and range of expression of ASD. Examining autism-related characteristics along a continuum of expression may yield more insight into the particular roles played by autism susceptibility genes. In addition, if ASD characteristics are viewed on a continuum, then clinically unaffected family members can also contribute information to family genetic studies.
Relatives of individuals with ASD have subclinical traits related to autism more frequently than the relatives of controls [2-8]. These subclinical traits can vary in intensity and have been collectively termed the broader autism phenotype (BAP). In addition, characteristics of clinically affected subjects - such as language delay, language deficits, social deficits, repetitive behaviours and insistence on sameness - have been successfully used to refine and clarify genetic studies of autism [9-12]. While categorical diagnostic status has been used successfully to identify autism linkage regions [1], quantitative measures that include the BAP provide additional information from clinically unaffected family members, and also provide more detailed phenotypic information on affected subjects.
The social, behavioural and communication characteristics of ASD are common and may be viewed as on a continuum with possible cutoffs for affected individuals [13,14]. The social responsiveness scale (SRS) is a quantitative measure of autistic traits with comparable child and adult versions, ranging continuously from significantly impaired to above average [13,15,16]. Several studies have reported significant familial correlations of subclinical features of autism as measured by the SRS [17-19], consistent with previous evidence of significant heritability of the BAP [7]. One previous genome-wide linkage scan has been done using SRS scores obtained from children in 99 nuclear families from the Autism Genetic Resource Exchange [20]. That study included 408 microsatellite markers and two suggestive genetic loci were identified on chromosomes 11 and 17, with other possible regions identified on chromosomes 4, 8, and 10.
We report genome-wide SRS linkage results using single nucleotide polymorphism (SNP) data genotyped on a unique resource of large extended and smaller multiplex ASD families from Utah. We have previously reported linkage evidence in these families using clinical diagnostic status and only including information from affected pedigree members. The current study has two advantages. First, we include additional phenotype information above and beyond our previous affected-only genome scan by using the quantitative SRS measure: clinically unaffected relatives can contribute information to this analysis. The SRS phenotype therefore adds to our previous study in our extended pedigrees. Second, we add to other previous studies of the SRS which focus on nuclear families. The likely heterogeneity of ASD increases the value of studies that include extended pedigrees [21,22]. Collections of small families may include cases with many different susceptibility loci. Subjects affected with ASD who are members of a large extended family may be more likely to share the same genetic causes through their common ancestors. Particularly for rare disease-causing variation, the type of variation most likely underlying autism spectrum disorders, extended pedigrees have substantially more power to reveal true findings that designs with single cases or small families [23]. Once variation has been found, pedigree members can reveal penetrance of the variant and specific associations with related phenotypes. The study of the SRS in pedigrees may be of particular interest, as recent evidence suggests differences in the SRS between simplex and multiplex families [24].
Methods
Subjects
Subjects were initially ascertained from 70 pedigrees that had at least two family members with an ASD. Six families did not have sufficient SRS phenotype information (defined as at least two family members with SRS scores) and were therefore dropped from this analysis. A total of 629 genotyped subjects from 64 families were used here, 181 of whom were defined as having either a strictly defined autistic disorder (AD) or a more broadly defined ASD. Table 1 gives the characteristics of these families which include 19 large extended pedigrees (6-9 generations), six families of moderate size (4-5 generations) and 39 smaller families (2-3 generations). The 19 extended pedigrees were identified or confirmed using the Utah Population Database (UPDB), a computerized genealogy database that contains family history information for over 6.5 million individuals who are, for the most part, descendants of the nineteenth century Utah pioneers http://www.hci.utah.edu/groups/ppr/. Previous studies have shown low rates of inbreeding within the UPDB [25,26]. Using the UPDB, we were able to identify many distant family relationships between the individuals with ASD that were not known to the subjects or their families. These relationships were kept confidential. This study has ongoing approval from the University of Utah Institutional Review Board (IRB). All adults participating in the research signed IRB-approved informed consent documents. All subjects under the age of 18 signed approved assent documents and their parents or guardians signed approved parental permission documents. Subjects in this study are independent from the Utah subjects who were contributed to the Autism Genome Project and studied by Liu et al. [27]. In addition, none of the families in the current analysis were included in the data set studied by Duvall et al. [20].
Table 1.
Description of the Utah autism spectrum disorder (ASD) families.
| Type of family | No. of families | Average No. of generations; standard deviation (SD; range) | Total subjects | Average No. of subjects per family; SD (range) | Total ASD subjects | Average No. of ASD subjects per family; SD (range) | Total No. of subjects with SRS | Average No. of subjects per family with SRS (range) |
|---|---|---|---|---|---|---|---|---|
| Large (6-9 generations) | 19 | 7.89; 0.66 (6 to 9) | 328 | 17.26; 12.92 (5 to 50) | 81 | 5.22; 2.54 (2 to 9) | 254 | 13.37; 10.32 (4 to 40) |
| Moderate (4 generations) | 6 | 4; 0.00 (4) | 85 | 14.17; 11.34 (6 to 32) | 21 | 4.00; 3.39 (2 to 9) | 70 | 11.67; 10.21 (3 to 27) |
| Small (2-3 generations) | 39 | 2.28; 0.46 (2 to 3) |
216 | 5.54; 2.40 (2 to 11) |
79 | 2.05; 0.57 (1 to 3) | 194 | 4.97; 2.11 (2 to 11) |
| Full sample | 64 | 629 | 181 | 518 | ||||
SRS, Social Responsiveness Scale.
Phenotyping
Families interested in participating were asked to give questionnaire consent, to give initial information regarding possible exclusion criteria and to complete the Social Communication Questionnaire (SCQ) [28]. The SCQ was developed as a parent report measure based on the Autism Diagnostic Interview-Revised [29]. A cutoff score of 15 on the SCQ has been shown to have good sensitivity (0.85) and specificity (0.75) for discriminating ASD from other diagnoses [30]. Subjects were contacted for possible inclusion if their SCQ score was above 15 or if they had a previous diagnosis of ASD. Subjects were excluded if they reported medical conditions known to be associated with autism (tuberous sclerosis, fragile X, neurofibromatosis, congenital rubella or phenylketonia) or evidence of brain injury. To the best of our knowledge, given currently available data, subjects did not have chromosomal anomalies that could confound our findings. If subjects were eligible for the study, they were asked to sign informed consent for DNA and additional assessments. When possible, all subjects with a suspected ASD were then given both the ADI-R and the Autism Diagnostic Observation Schedule-Generic [31] and study diagnoses were made using these assessments. For cases where assessments could not be obtained, the diagnoses were made according to Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV criteria by a psychologist trained in autism assessment (JSM) using all available information (clinical records, other behavioural data, other questionnaire and interview information).
All available subjects were given the SRS. Completed questionnaires were obtained from a total of 523 family members who were genotyped. Completed questionnaires were obtained for 92.1% of index cases, 83.9% of siblings (affected and unaffected), 87.2% of fathers, 78.8% of mothers and 73.7% of more extended relatives (grandparents, great-grandparents). The child version of the SRS was completed for 205 subjects and the adult version was completed by 318 subjects. The child version was filled out by a parent for almost all cases (203/205; 99.02%). The adult version was usually filled out by a spouse (236/318; 74.21%) but parents were raters for adult siblings over age 18 (59/318; 18.6%). Other informants filled out the remainder of the adult SRS forms (23/318; 7.2%). No children were rated by a parent with ASD. However, after surveys were returned, five adult raters were found to have ASD. Scores from these five raters (fathers) on their spouses (mothers) were not used for subsequent analyses, leaving 518 subjects with scores. There were an additional four children whose parent was coded as a poor rater on the ADI for under-reporting of behaviours. While no data regarding rater quality were available for the SRS directly, the similarity of content across the ADI and SRS made ratings done by these parents was suspect. Scores for these subjects were therefore not used in the quantitative analyses of data. For qualitative SRS analysis, data on these subjects were not deleted because possible under-reporting on the SRS did not impact upon the SRS category.
Genotyping
Genotyping services were provided by the Center for Inherited Disease Research (CIDR) using the 6 K single nucleotide polymorphism (SNP) Illumina Linkage Panel 12, which includes 6090 SNP markers, with an average genetic coverage of 0.65 cM. There were 46 SNPs who were dropped due to poor performance, leaving 6044 SNPs. Data from 653 pedigree subjects passed stringent quality control criteria for initial inclusion in the study [32]. Of the 629 genotyped subjects in 64 families used in this study, 518 of had SRS data.
Eliminating linkage disequilibrium (LD) between markers in linkage studies has been strongly recommended, as false positive results can occur in the presence of LD [33-36]. As these studies have shown, increases in type I error are most dramatic when LD is high and when there are missing genotypes for parents and other connecting relatives. Recommended thresholds of acceptable LD vary, but a pair-wise r2 value of 0.05 between SNPs has now been supported with extensive simulation studies [34]. Therefore, prior to linkage analysis, we screened all SNPs for LD using the PLINK software package [37], which recursively removes SNPs in LD within a sliding window. We set a window size of 50 SNPs, shifted the window by five SNPs at each step, and used a variance inflation factor of 1.5, which is equivalent to a multiple correlation coefficient R2 of 0.33 regressed simultaneously over all SNPs in the selected window. This R2 considers not only the correlations between SNPs but also between linear combinations of SNPs [37] and corresponds in our data to a pair-wise r2 of approximately 0.05. This screening for LD removed 1207 SNPs. Also, as part of the validation procedure, we removed 115 SNPs with a minor allele frequency less than 0.10 and 4 SNPs out of Hardy-Weinberg equilibrium. The total number of SNPs left after this phase was 4718.
Analyses
We used the genetic map provided by CIDR based on the deCODE genetic map [38]. Base pair positions were obtained from the March 2006 human reference sequence (hg18) assembly. The SRS was first treated as a qualitative trait. Published cutoff scores [15] were used to delineate three groups: SRS unaffected (raw score below 54), SRS spectrum (raw score between 54 and 87) and SRS affected (raw score 87 or above). Subjects who were missing SRS scores were coded as unknown.
This qualitative phenotype was tested using MCLINK, a Markov chain Monte Carlo (MCMC) method that allows for multilocus linkage analysis on large extended pedigrees [39]. Using blocked Gibbs sampling, MCLINK generates inheritance matrices for haplotypes of the markers being analysed and estimates the log-likelihood function linkage statistics. This method results in the reconstruction of haplotype configuration for the entire pedigree structure, allowing for analysis of the full genetic information available in the pedigree. MCMC methods therefore maximize the utility of extended pedigree data, as genetic information increases with the number of meioses in a family [40]. Further, the multipoint analysis maximizes the genetic information available from the entire map of SNP genotypes. In multipoint analyses, genetic information increases with the density of the genotypes [41-43] but is not decreased, with exclusion of markers in high linkage disequilibrium [42]. Internally, MCLINK runs the analysis five times to ensure a consistent solution. Results from MCLINK have shown a high degree of similarity to other MCMC linkage methods [44] and also to exact linkage methods and variance components linkage methods as applied to extended pedigrees [45]. MCLINK has been previously used to identify candidate genomic regions for a number of complex diseases in extended pedigrees [44,46-48]. Allele frequencies for the MCLINK analysis were estimated using all of the observed data.
We performed a non-parametric analysis and a general parametric model-based analysis using simple dominant and recessive model parameters that reproduced the reported population frequency of ASD [49]. For the recessive model, the disease allele frequency was set at 0.05; a penetrance of 0.01, 0.01, 0.5 was assigned to those with SRS scores in the spectrum category and 0.0014, 0.0014, 0.8 was assigned to those with SRS scores in the SRS affected category. For the dominant model, the disease allele frequency was set at 0.0025; penetrances were 0.01, 0.5, and 0.5 for those with SRS scores in the spectrum category and 0.0014, 0.8, 0.8 for those with SRS scores in the SRS affected category. Nonparametric methods provide an analytic approach independent of model assumptions. However, for the analysis of extended pedigrees, the parametric approach may be preferable. Parametric analysis provides assumptions about the genotype-phenotype relationship, simplifying the parameter space and allowing for more powerful and efficient analyses without increasing the false positive rate [22,50,51]. The multipoint heterogeneity LOD score (HLOD) allows for a proportion of unlinked pedigrees (1-alpha) and variation in the recombination fraction. The HLOD provided by MCLINK is robust to model misspecification and may reflect the true position of linkage peaks more accurately under conditions of appreciable heterogeneity [47,50,52]. HLOD scores have been shown to be more powerful than homogeneity LOD scores or model-free methods under these conditions [52,53]. The HLOD has been shown to produce scores consistent with other published methods [47,54]. P-values from the non-parametric qualitative analyses were converted to LOD scores for ease of interpretation and comparison with the parametric results. We present results in the commonly used framework of significance thresholds proposed by Lander and Kruglyak [55]: LOD score ≥ 1.88 (P = 0.0017) represents suggestive evidence and LOD score ≥ 3.30 (P = 0.000048) represents significant evidence. We recognize that our study design does not match perfectly with the design that generated these thresholds but they do provide reasonable benchmarks which enable us to organize and present our data. All P-values are theoretical, not empirical.
In order to analyse the SRS as a quantitative trait, we first applied a square root transform the SRS data to adjust for non-normality in the distribution, as has been done with previous SRS analyses [17]. The effects of age and gender were partialled out using regression. MCLINK was used to perform parametric quantitative trait analysis. In a quantitative trait analysis there is no concept of 'affected' or 'unaffected'; all individuals with a trait value are included in the analysis, other individuals are considered unknown. The MCLINK nonparametric analysis considers quantitative similarity among relatives and has been previously described [56]. For the parametric quantitative analysis, the penetrance vector for an individual is based on their observed trait value and three specified trait distributions for the three genotypes at the disease locus (no risk allele, one risk allele, two risk alleles). The three trait distributions are unknown, but can be estimated from the observed trait data. The SRS score cutoffs provide theoretical categories of unaffected, spectrum and affected subjects. Here we considered these three categories as a representing a hypothetical genetic model of: no risk allele, one risk allele and two risk alleles, and means and standard deviations of adjusted SRS scores were estimated. The three distributions for zero, one or two risk alleles were specified to be normal distributions with means of -1.552, 1.184, and 2.558 and standard deviations of 2.238, 1.581, and 1.356, respectively. The penetrance vector for an individual is simply the relative probabilities of the trait being observed from the three distributions. Only the relative probability between the three penetrance values is important as the linkage statistic is a likelihood ratio. For efficient calculation of parametric quantitative LOD scores, the continuous penetrance function is discretized into multiple classes (here, 40) and all individuals with trait values residing in a class are assigned penetrance vectors at the mid-point of the class. Classes were defined starting -3SD below the mean of the zero risk alleles distribution and adding equal increments to reach +3SD above the mean of the two risk alleles distribution.
In summary, we note that use of the SRS results in three important differences between this analysis and our previous affected-only analysis using clinical diagnosis on these families. First, this analysis includes information from an additional 337 clinically unaffected relatives. Second, for the qualitative analysis, affection status was defined using SRS data rather than clinical measures. Third, a quantitative analysis uses a full range of scores on autism-related traits in all measured pedigree members.
Results
Table 2 describes the diagnostic information for affected subjects. Of the 181 total affected subjects included in this analysis, 165 had data on both the ADI-R and ADOS-G; 16 other cases were assigned a diagnosis based on criteria specified in Diagnostic and Statistical Manual of Mental Disorders (DSM-IV-TR criteria). One hundred and thirty-five affected subjects met criteria for strictly defined autistic disorder (AD) and 46 met criteria for an ASD. There was a 7.4:1 male:female ratio among the subjects with strictly defined autism, which fell to 3.6:1 among the subjects with an ASD. For all subjects combined, the male:female ratio was 5.96:1. As expected, ADI-R scores were significantly higher for the autism group compared to the ASD group (t = 8.19, P < 0.0001 for social; t = 6.95, P < 0.0001 for verbal; t = 2.95, P = 0.004 for restricted interests/repetitive behaviours). The nonverbal total cannot be compared because no ASD subjects were nonverbal. Quantitative scores on the ADOS are not compared because of differences between modules but significantly more individuals in the autism group were administered Modules 1 and 2 compared to the ASD group.
Table 2.
Descriptive information for pedigree subjects.
| Mean ADI domain scores (SD; N) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnostic group | N | Male: female | IQ > 70 (%) | Mean age (SD) | Mean SRS score (SD; N; % child version) | Social | Verbal | Non-verbal |
Restr/ Repet |
| Autism* | 135 | 119:16 (7.44:1) | 69/125 (55.2) | 13.87 (11.44) | 107.0 (31.4; N = 126; 75.4%) | 21.8 (6.4) | 17.2 (4.7) | 12.3 (2.3; N = 26) | 6.7 (2.6) |
| ASD† | 46 | 36:10 (3.60:1) | 37/43 (86.1) | 16.98 (15.25) | 86.4 (29.1; N = 41; 75.6%) | 12.7 (5.9) | 10.5 (6.4) | (N = 0) | 5.3 (2.9) |
| All affected subjects | 181 | 155:26 (5.96:1) | 106/168 (63.1) | 14.64 (12.51) | 101.9 (33.5; N = 167; 75.5%) | 19.5 (7.5) | 15.2 (6.1) | 12.3 (2.3; N = 26) | 6.3 (2.7) |
| All relatives with no ASD diagnosis | 448 | 211:237(0.9:1) | 37.30 (20.49) | 28.7 (24.0; N = 351; 22.5%) | |||||
* 11 cases were missing an Autism Diagnostic Interview (ADI); five cases were missing an Autism Diagnostic Observation Schedule.
† Three cases were missing an ADI; no cases were missing an ADOS.
SD, standard deviation; SRS, Social Responsiveness Scale; Restr/Repet, Restricted Interests/Repetitive Behaviors Domain Score
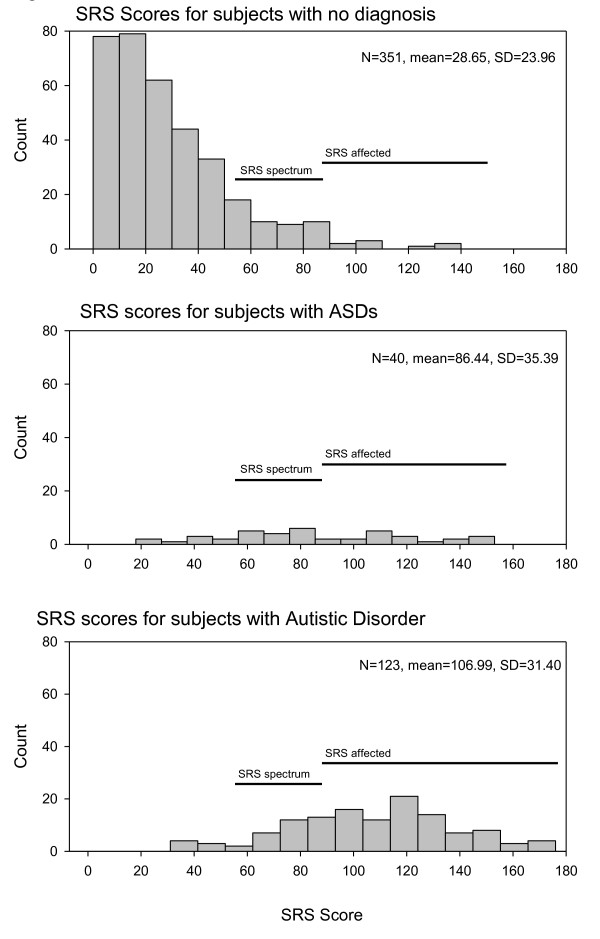
Regression adjustment of the SRS for age and gender effects resulted in parameter estimates of -0.074 for age and 1.6 for gender (males have higher scores). Figure 1 gives the distribution of SRS scores by diagnostic category. SRS scores were significantly different between autism and ASD (t = 3.71, P = 0.0003) and between ASD and other relatives (t = 10.57, P < 0.0001). Among affected subjects, substantial correlations were observed between ADI domain scores and the SRS total score. The highest correlations were observed with the ADI social domain score, with a correlation of 0.60 among ASD subjects and a correlation of 0.46 among subjects with AD. For ASD subjects, 17.1% scored in the normal range on the SRS, 39.0% scored in the spectrum range and 43.9% scored in the affected range. Among subjects with a clinical diagnosis of autism, 5.6% scored in the normal range, 19.1% scored in the spectrum range and 75.4% scored in the affected range. The 95 genotyped spouse pairs showed a significant spouse correlation of 0.33 (P = 0.001) using the square root transformed adjusted scores.
Figure 1.
Histograms of Social Responsiveness Scale scores for subjects in Utah autism pedigrees.
The genome-wide analyses of the data are summarized in Table 3 and in Figures 2 and 3. Note that MCLINK does not provide NPL scores for the X chromosome. Qualitative analyses show substantial overlap with our previous affected-only genome-wide study of these same families [32], in addition to revealing new positive results. We also saw positive findings from our analysis of the quantitative SRS data and replicated the chromosome 11 SRS finding reported by Duvall et al. [20].
Table 3.
Linkage results with P-values ≤ 0.0017 (suggestive evidence) for at least one method of analysis from quantitative and qualitative analyses of the total Social Responsiveness Score (SRS).
| Chromosome | Location1 | Score (alpha2; P-value) | Phenotype, model |
|---|---|---|---|
| 6p25.3 | 133,969 | 1.93 (0.40; 0.0014) | Qualitative, recessive |
| 6p22.1 | 26,261,314 | 2.36 (0.31; 0.00049) | Qualitative, recessive |
| 7q31.1-q32.3 | 113,934,453 | 2.07 (0.54; 0.001) | Quantitative, parametric |
| 129,451,548 | 2.91 (--; 0.00013) | Quantitative, NPL | |
| 130,291,151 | 2.55 (0.37; 0.00031) | Qualitative, recessive | |
| 9p24.33 | 593,192 | 2.66 (0.75; 0.00023) | Quantitative, parametric |
| 10p12.1 | 27,770,267 | 2.03 (0.74; 0.0011) | Qualitative, dominant |
| 10q22.1-q22.2 | 76,287,863 | 1.88 (0.65; 0.0016) | Qualitative, dominant |
| 11p15.1-p15.44 | 12,519,296 | 2.77 (0.44; 0.00018) | Quantitative, parametric |
| 13q12.3 | 30,070,594 | 2.23 (0.60; 0.00068) | Qualitative, recessive |
| 13q32.1 | 94,452,168 | 2.14 (0.85; 0.00085) | Qualitative, recessive |
| 15q13.3 | 31,177,959 | 3.64 (--; 0.000021) | Qualitative, NPL |
| 31,177,959 | 3.07 (0.70; 0.000085) | Qualitative, recessive | |
| 19q13.43 | 59,338,102 | 2.14 (0.71; 0.00085) | Quantitative, parametric |
Bold font indicates significant evidence (P ≤ 0.000048).
Note: All nonparametric linkage (NPL) scores have been converted to equivalent logarithm of the odds (LOD) scores. Chromosomal locations indicated in italics overlap our affected-only genome scan using clinical diagnosis [32].
1 Location at the maximum of the peak.
2 Alpha, the estimated proportion of linked families for the MCLINK heterogeneity LOD score, is given for parametric analyses.
3 The peak at 9p24.3 is in the same location as a peak reported in a single family from our sample [35].
4 The peak at 11p15.1-p15.4 replicates previous genome scan results of quantitative SRS scores [20].
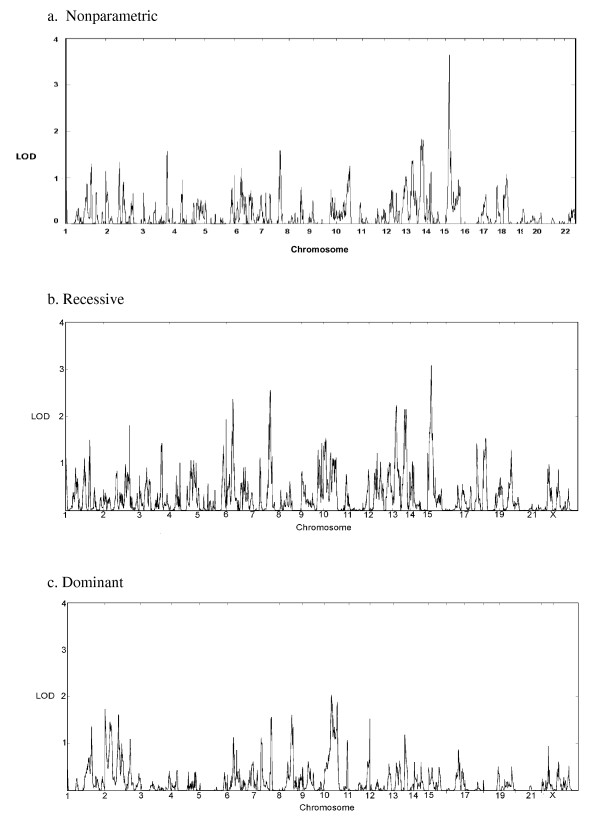
Figure 2.
Genome scan results of Social Responsiveness Scale scores treated as qualitative categories (unaffected, spectrum, and affected) using MCLINK. (a) The nonparametric analysis does not assume a genetic model; scores for this analysis have been converted to logarithm of odds (LOD) scores for ease of comparison. Analyses were also performed using basic recessive (b) and dominant (c) models with parameters consistent with the prevalence of autism spectrum disorder.
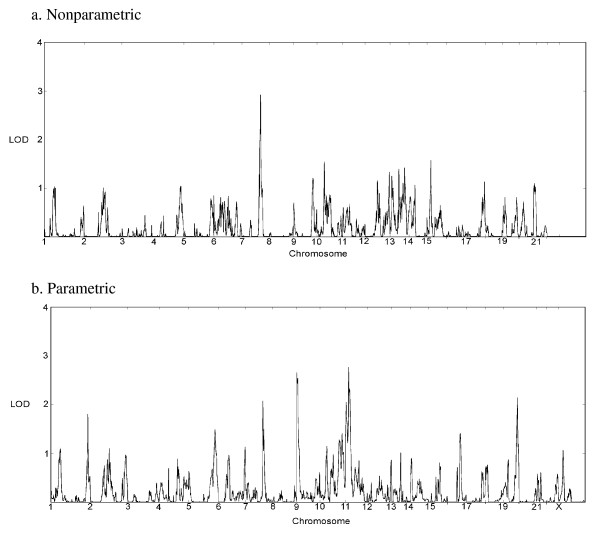
Figure 3.
Genome scan results of quantitative Social Responsiveness Scale (SRS) scores using MCLINK. The nonparametric analysis (a) does not assume a genetic model, and scores were converted to logarithm of odds (LOD) scores for ease of comparison. The parametric analysis (b), assumes the penetrance vector for an individual is based on their observed trait value and three specified trait distributions for the three genotypes at the disease locus (no risk allele, one risk allele, two risk alleles). These trait distributions are assumed to map onto the SRS score cutoffs (unaffected, spectrum, and affected). The penetrance vector for an individual is the relative probabilities of the trait being observed from the three distributions.
Figure 4 shows results for individual chromosomes where peaks overlap previous findings. The most significant peak was found in the MCLINK qualitative nonparametric analysis, with a genome-wide significant LOD score of 3.64 on chromosome 15q13.3 at rs1026752 (31,177,959 bp). Note that we converted all nonparametric linkage (NPL) scores to LOD scores for ease of comparison. The MCLINK recessive model maximized at the same location with a suggestive score of 3.07. This peak extended from about rs1719013 to rs580839 (27,940,000 bp - 32,780,000 bp). This peak was in the same location as the most significant linkage peak from our affected-only genome scan using clinical affection status in these pedigrees [32].
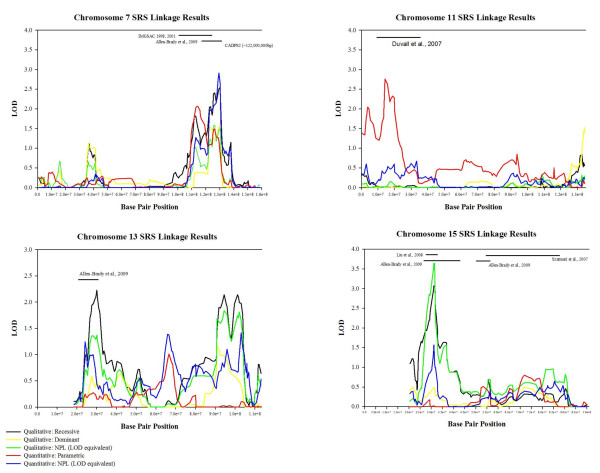
Figure 4.
Detail of levels of detail (LOD) score results on chromosomes 7, 11, 13, and 15. Scores of nonparametric linkage (NPL) analyses have been converted to LOD scores in order to facilitate comparisons.
We also saw evidence for linkage in other locations (see Table 3 and Figures 3 and 4). Of these peaks, those on chromosomes 7q31.1-q32.3 and 13q12.3 were also found in our previous genome scan using clinical affection status [32]. The peak at 7q31.1-q32.3 (125,450,000 bp - 130,300,000 bp) showed consistent evidence between quantitative and qualitative analyses. The 13q12.3 peak also showed overlap with our previous clinical diagnosis scan. In the current study, this peak extended from 26,500,000 bp to 33,000,000 bp and was supported by the qualitative recessive analyses. A second apparently distinct peak was found on chromosome 13q32.1 at 94,452,168 bp, also in the qualitative recessive analyses.
Our most significant quantitative peak was on chromosome 7q (LOD = 2.91) in the same region as the evidence from the qualitative analyses. Our next most significant peak was on chromosome 11p15.1-p15.4 spanning a region from 7,300,000 bp to 22,400,000 bp, with a maximum LOD of 2.77 at two adjacent SNPs (rs214101 and rs4757574) from a parametric analysis. The base pair location of our peak (17,246,930 bp - 17,682,950 bp, or 25.59cM - 26.84 cM) is well within one of the most significant peaks previously reported by Duvall et al. [20] in their quantitative SRS scan. Other quantitative peaks were found on chromosomes 9p24.3 and 19q13.43. The 9p24.3 peak is in a region previously identified in a clinical diagnosis scan done in our first extended family [44,57].
Discussion
The present genome-wide linkage scan of SRS scores was performed using 518 relatives from a unique resource of large extended and smaller multiplex ASD families from Utah. We found genome-wide significant linkage on chromosome 15q13.3, and several other suggestive findings. Peaks on chromosomes 7q, 13q and 15q were also found in our previous genome scan of affected-only subjects in these families based on clinical diagnosis [32]. SRS quantitative analyses also replicated a finding on chromosome 11 p reported in a previous genome scan of the SRS [20], providing some evidence for genetic consistency of this measure used as a quantitative trait across studies.
The SRS was developed as a continuous measure of autistic traits, with variation intended to extend across affected subjects and unaffected relatives. Among affected subjects, our data suggest substantial correlations between clinical measures of autism from the ADI and SRS total scores, particularly for the social behavioural domain. ADI and SRS are expected to differ to some degree because ADI scores are based on information from the developmental period between ages 4-5 and also on the most severe presentation through the individual's life, while the SRS focuses on current behaviour. In addition, most clinically affected subjects scored in the spectrum or affected ranges on the SRS, though as expected, we observed a substantial range of scores. Most of the clinically unaffected relatives scored in the normal range on the SRS (87.4%), but considerable variation also occurred in these scores.
In our previous genome-wide scan, all relatives with no clinical diagnosis were considered unknown, so results were based on phenotypic information from 192 genotyped affected cases. Using the SRS in the present study, we added information for 337 clinically unaffected relatives, though 11 clinically affected subjects from the previous scan were missing SRS scores and were not included here. A small percentage of clinically diagnosed relatives (N = 14) scored within the normal range on the SRS and therefore also contributed different information to the current scan. Our families also replicated previous evidence for significant SRS spouse resemblance (r = 0.33, P = 0.001) [17].
The new information provided by the SRS in these families revealed additional findings beyond our previous affected-only clinical diagnosis scan [32]. Not surprisingly, given the overlap in classification of affected subjects based on the SRS or the ADI/ADOS, the best overlap in findings between the current study and our previous study resulted when the SRS was treated as a qualitative trait. The use of a SRS-defined spectrum category and the addition of information from other clinically unaffected relatives provide the primary difference in findings between this study and our previous clinical diagnosis scan. The investigation of the whole range of quantitative information produced additional results, which again is perhaps not surprising. We used parametric quantitative methods in an effort to increase the similarity between qualitative and quantitative analyses. We realize that multiple tests may have increased our chances of a false positive result. If we conservatively assume that models are uncorrelated, our significance thresholds would be adjusted by log10(5) = 0.7 LOD score units [58] for these five tests: dominant, recessive, NPL qualitative tests and parametric and NPL quantitative tests.
Our genome-wide significant result on chromosome 15q13.3 was supported by the qualitative NPL and recessive models and some lesser evidence also by the quantitative analysis (see Figure 4). In our previous genome scan our best results also occurred on chromosome 15q in three locations that may represent a single peak (29,459,872 bp, 36,837,208 bp, and 55,629,733 bp) [32]. Liu et al. [27] have also reported evidence for linkage in this region (LOD = 4.01 at 15q13.3-q14).
The region on chromosome 7q31.3-32.3 also occurred in our previous clinical diagnosis scan, and was supported consistently across analysis models. This peak (125,450,000 bp - 130,300,000 bp) has been implicated previously by other studies [59-63]. Our findings suggest that SRS information strengthened this peak over the 7q peak in previous clinical diagnosis scan (LOD = 1.97), though results here are still only suggestive (LOD = 2.91). We note that the CNTNAP2 gene of recent interest in autism [64] is at 145,444,386 bp, downstream of our peak. The final peak showing overlap with our clinical diagnosis scan was on 13q12.3. A second downstream peak at 13q32.1 appeared in the current SRS study that was not evident in the clinical diagnosis scan. While some studies have reported positive findings on chromosome 13 for autism, attention has focused on the neurobeachin gene (NBEAL2) [65], which lies in between our two regions and is, therefore, not supported by our data.
Other peaks were observed on chromosomes 6 and 10. The regions on chromosome 6p25.3 and 6p22.1 have appeared in schizophrenia studies [66-68] and also in bipolar disorder [69,70]. The region also harbors a copy number variation (CNV) at 27,827,354-28,119,631 implicated in autism [71]. Interestingly, chromosome 10p12.1 and 10q22.1-q22.2 also has been reported for schizophrenia and bipolar disorder in many of the same samples that showed chromosome 6 findings [69,72-74]. Somewhat strikingly, two genome-wide associations with attention deficit and hyperactivity disorder (ADHD) have also been found on chromosome 10p12.1 at 27,672,044-27,694,523 and 10q22.1 at 72,126,813-72,120,519 [75]; both findings are quite close to our linkage peaks. Additional evidence exists in two other samples of an association between ADHD and autism using the SRS [76,77].
A quantitative analysis also revealed findings of interest. While results using the quantitative range of the SRS often paralleled the qualitative findings, there were interesting exceptions. We note that while we used the same genetic analysis software, the variation in the trait was treated quite differently, so direct overlap of findings would not necessarily be expected. For the quantitative analysis, we also note that adjusting for age and gender may have been a conservative adjustment, as clinical affection status is associated with age and gender. As a result of this adjustment, our quantitative results are unlikely to be due to behaviours that change with age or behaviour specific to gender. Our highest quantitative peak was on chromosome 7 in the same region as described above. Our next highest quantitative peak was on chromosome 11p15.5-p15.4, spanning a region from 7,300,000 bp to 22,400,000 bp (12.11 - 37.63 cM). This peak replicates a previously reported SRS peak from 15-45 cM found in an analysis of quantitative SRS scores [20]. Liu et al. [27] also found linkage in this region, primarily using a phenotype of delayed phrase speech, but with supporting evidence also from other phenotypes. A previous genome scan of clinical diagnosis in 1181 multiplex families also identified this region [78]. On chromosome 19q13.4, our quantitative evidence spanned a region from 57,150,000 bp to 63,780,000 bp. This region overlaps with previous autism linkage evidence [79] and also has several CNVs associated with autism [71].
Finally, we observed peaks in two other regions previously identified in our families in scans using affection status. We observed a quantitative peak on chromosome 9p24.3, from the telomere to about 4,330,000 bp. This peak has been previously reported for obsessive compulsive disorder (OCD) [80,81]. The glutamate transporter genet SLC1A1 is near this region (4,480,000 bp - 4,580,000 bp), and has shown some association with OCD [82,83].
While it is important to seek replication of these findings in independent samples, we intend first to pursue these findings within our sample families. It is possible that findings may be unique to our family sample; indeed, a rare genetic finding for autism would be our expectation. However, if we can find genetic variation responsible for these statistical findings, the results may still provide relevant evidence regarding biological mechanisms and genetic pathways implicated in autism. Our family data will allow us to investigate segregation of the genetic variants responsible for these peaks and to determine more detailed associations with phenotypes related to autism. The extended family design allows a more powerful tool to investigate rare genetic variants in relation to phenotype than the genome-wide association study design.
Conclusions
In summary, our results highlight three important points. First, using a quantitative analysis, we replicated a previously reported SRS peak on chromosome 11 peak found in the AGRE family sample [20]. This replication supports the SRS as a robust measure, as replication occurred across independent samples, and in spite of differences in study design and measurement. Our sample included large extended pedigrees and more than half the SRS scores in our sample were on adults; the Duvall et al. study [20] included only measurements on children. Second, our genome scan of SRS scores revealed several peaks that were the same as those found in our previous affected-only genome scan of clinical diagnosis [32]. These peaks in our Utah families are therefore robust to the inclusion of data from clinically unaffected relatives and to the analysis of autism measured in a manner quite different from the traditional structured interview and observation methods. This study supports our previous ASD linkage findings and suggests the SRS may be a reasonable surrogate measure for genetic analysis of ASD, though the SRS does not provide the depth of information used in a clinical setting. Finally, both qualitative and quantitative analyses of SRS scores revealed linkage peaks not observed using clinical affection status in our own previous pedigree studies, but often overlapping with other evidence from autism, ADHD, OCD, schizophrenia and/or bipolar studies. These results indicate that the additional information available from clinically unaffected relatives and the information in the SRS itself may reveal locations for autism susceptibility genes that would not otherwise be detected.
Abbreviations
AD: autistic disorder; ADHD: attention deficit and hyperactivity disorder; ADI-R: Autism Diagnostic Interview-Revised; ADOS-G: Autism Diagnostic Observation Schedule-Generic; ASD: autism spectrum disorder; BAP: broader autism phenotype; CIDR: Center for Inherited Disease Research; CNV: copy number variation; HLOD: heterogeneity logarithm of the odds (LOD); IRB: Institutional Review Board; LD: linkage disequilibrium; MCMC: Markov chain Monte Carlo; NPL: nonparametric linkage; OCD: obsessive compulsive disorder; SCQ: Social Communication Questionnaire; SNP: single nucleotide polymorphism; SRS: Social Responsiveness Scale; UPDB: Utah population database.
Competing interests
HC, WMM, and JSM received partial salary support from Lineagen Inc. (www.lineagen.com) from 12/1/07 to 12/31/08. This salary support is not ongoing.
Authors' contributions
HC conceived the study, participated in its design, directed the statistical analyses and drafted the manuscript. MEV verified the accuracy of the phenotype data and made contributions to the manuscript. RJR assisted with the interpretation of the results and helped draft the manuscript. NJC, DSC, and KAB helped with the statistical analyses and helped draft the manuscript. JSM confirmed the research diagnoses of ASD, supervised the collection of all phenotype data and made significant contributions to the interpretation of results. WMM participated in the design of the study and helped draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Hilary Coon, Email: hilary.coon@hsc.utah.edu.
Michele E Villalobos, Email: michele.villalobos@hsc.utah.edu.
Reid J Robison, Email: reid.robison@hsc.utah.edu.
Nicola J Camp, Email: nicola.camp@utah.edu.
Dale S Cannon, Email: dale.cannon@hsc.utah.edu.
Kristina Allen-Brady, Email: kristina.allen@hsc.utah.edu.
Judith S Miller, Email: judith.miller@hsc.utah.edu.
William M McMahon, Email: william.mcmahon@hsc.utah.edu.
Acknowledgements
This work was supported by R01 MH069359, Autism Speaks Mentor-based Fellowship 1592, the Utah Autism Foundation, the Carmen B Pingree School for Children with Autism and GCRC M01-RR025764 from the National Center for Research Resources. Partial support for all datasets within the Utah Population Database (UPDB) was provided by the University of Utah Huntsman Cancer Institute. We thank our staff whose countless hours of work have made this study possible. We also greatly appreciate the time and effort given by the family members who participated in this study.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265(5596):726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Fisman S, Tuff L, Bartolucci G, Mahoney WJ, Bryson SE. Parents and collateral relatives of children with pervasive developmental disorders: a family history study. Am J Med Genetics. 1995;60(4):282–289. doi: 10.1002/ajmg.1320600405. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychological Med. 1995;25(1):63–77. doi: 10.1017/S0033291700028099. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Holden J, Bryson S, Mahoney W, Tuff L, MacLean J, White B, Bartolucci G, Schutz C, Robinson P, Hoult L. High phenotypic correlations among siblings with autism and pervasive developmental disorders. Am J Med Genetics. 1996;67(4):354–360. doi: 10.1002/(SICI)1096-8628(19960726)67:4<354::AID-AJMG7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Piven J, Palmer P. Cognitive deficits in parents from multiple-incidence autism families. J Child Psychology Psychiatry. 1997;38(8):1011–21. doi: 10.1111/j.1469-7610.1997.tb01618.x. [DOI] [PubMed] [Google Scholar]
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry. 1997;154(2):185–190. doi: 10.1176/ajp.154.2.185. [DOI] [PubMed] [Google Scholar]
- Piven J, Palmer P, Landa R, Santangelo S, Jacobi D, Childress D. Personality and language characteristics in parents from multiple-incidence autism families. Am J Med Genetics. 1997;74(4):398–411. doi: 10.1002/(SICI)1096-8628(19970725)74:4<398::AID-AJMG11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Bradford Y. Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genetics. 2001;105(8):539–547. doi: 10.1002/ajmg.1497. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet. 2001;68(6):1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, Abramson RK, Wright HH, DeLong RG, Gilbert JR, Cuccaro ML, Pericak-Vance MA. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Med Genetics. 2002;70(4):1058–1061. doi: 10.1086/339765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Cuccaro ML, Hauser ER, Raiford KL, Menold MM, Wolpert CM, Ravan SA, Elston L, Decena K, Donnelly SL, Abramson RK, Wright HH, DeLong GR, Gilbert JR, Pericak-Vance MA. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet. 2003;72(3):539–548. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, Metzger LM, Shoushtari CS, Splinter R, Reich W. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33(4):427–433. doi: 10.1023/A:1025014929212. [DOI] [PubMed] [Google Scholar]
- Bolte S, Poustka F, Constantino JN. Assessing autistic traits: cross-cultural validation of the social responsiveness scale (SRS) Autism Res. 2008;1(6):354–363. doi: 10.1002/aur.49. [DOI] [PubMed] [Google Scholar]
- Constantino JN. The Social Responsiveness Scale Los Angeles. California: Western Psychological Services; 2002. [Google Scholar]
- Constantino JN, Gruber CP, Davis S, Hayes S, Passanante N, Przybeck T. The factor structure of autistic traits. J Child Adolesc Psychiat. 2004;45:719–726. doi: 10.1111/j.1469-7610.2004.00266.x. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Todd RD. Intergenerational transmission of subthreshold autistic traits in the general population. Biological Psychiatry. 2005;57(6):655–660. doi: 10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Lajonchere C, Lutz M, Gray T, Abbacchi A, McKenna K, Singh D, Todd RD. Autistic social impairment in the siblings of children with pervasive developmental disorders. Am J Psychiat. 2006;163:294–296. doi: 10.1176/appi.ajp.163.2.294. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Abbacchi AM, Lavesser RD, Reed H, Givens L, CHiang L, Gray T, Gross M, Zhang Y, Todd RD. Developmental course of autistic social impairment in males. Dev Psychopathol. 2009;21:127–138. doi: 10.1017/S095457940900008X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH. A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am J Psychiatry. 2007;164(4):656–662. doi: 10.1176/appi.ajp.164.4.656. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1:1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Goring HH. Gene mapping in the 20th and 21st centuries: statistical methods, data analysis, and experimental design. Hum Biol. 2000;72(1):63–132. [PubMed] [Google Scholar]
- Thomas A, Camp NJ, Farnham JM, Allen-Brady K, Cannon-Albright LA. Shared genomic segment analysis, mapping disease predisposition genes in extended pedigrees using SNP genotyping assays. Ann Hum Genet. 2008;72:279–287. doi: 10.1111/j.1469-1809.2007.00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkud YV, Todd RD, Abbacchi AM, Zhang Y, Constantino JN. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Neuropsychiat Genet. 2009;105B:328–334. doi: 10.1002/ajmg.b.30810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB. Inbreeding in the Utah Mormons: an evaluation of estimates based on pedigrees, isonymy, and migration matrices. Ann Hum Genet. 1989;53(Pt 4):339–355. doi: 10.1111/j.1469-1809.1989.tb01803.x. [DOI] [PubMed] [Google Scholar]
- Jorde LB. Consanguinity and prereproductive mortality in the Utah Mormon population. Hum Hered. 2001;52(2):61–65. doi: 10.1159/000053356. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P. Autism Genome Project Consortium. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol Psychiatry. 2008;64(7):561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berument S, Rutter M, Lord C, Pickles A, Bailey A. Autism Screening Questionniare. CA.: Western Psychological Services; 1999. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Baranek G, Bodfish J, Gordon A, Houser M, Poe M. Concurrent validity of the ADI-R and SCQ in high functioning autism. Abstract presented at Collaborative Programs of Excellence in Autism/Studies to Advance Autism Research and Treatment Annual Scientific Meeting, Bethesda MD, May 2004.
- Lord C, Risi S, Lambrecht L, Cook EH Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Developmental Disorders. 2000;30(3):205–223. doi: 10.1023/A:1005592401947. [DOI] [PubMed] [Google Scholar]
- Allen-Brady K, Miller J, Matsunami N, Stevens J, Block H, Farley M, Krasny L, Pingree C, Lainhart J, Leppert M, McMahon WM, Coon H. A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol Psychiatry. 2009;14:590–600. doi: 10.1038/mp.2008.14. [DOI] [PubMed] [Google Scholar]
- Huang Q, Shete S, Amos C. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analyses. Am J Hum Genet. 2004;75:1106–1112. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Holmans P. The effect of linkage disequilibrium on linkage analysis of incomplete pedigrees. BMC Genetics. 2005;6(Suppl):S6. doi: 10.1186/1471-2156-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Brady K, Horne BD, Malhotra A, Teerlink C, Camp NJ, Thomas A. Analysis of high-density single-nucleotide polymorphism data: three novel methods that control for linkage disequilibrium between markers in a linkage analysis. BMC Proc. 2007;1(Suppl 1):S160. doi: 10.1186/1753-6561-1-s1-s160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Amos CI, Etzel CJ, Shete S, Gregersen PK. Comparison of genome-wide single-nucleotide polymorphism linkage analyses in Caucasian and Hispanic NARAC families. BMC Proceedings. 2007;1(Suppl 1):S97. doi: 10.1186/1753-6561-1-s1-s97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Thomas A, Gutin A, Abkevich V, Bansal A. Multipoint linkage analysis by blocked Gibbs sampling. Statistics Computing. 2000. pp. 259–269. [DOI]
- Hossjer O. Information and effective number of meioses in linkage analysis. J Math Biol. 2005;50:208–232. doi: 10.1007/s00285-004-0289-z. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility loci. Am J Hum Genet. 2004;75:948–965. doi: 10.1086/425870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal P, Gillanders EM, Mathias R, Ibay GP, Klein AP, Baffoe-Bonnie AB, Ou L, Dusenberry IP, Tsai Y-Y, Chines PS, Doan BQ, Bailey-Wilson JE. Identification of tag single-nucleotide polymorphisms in regions with varying linkage disequilibrium. BMC Genet. 2005;6(Suppl2):S73. doi: 10.1186/1471-2156-6-S1-S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jacobs K, Kerstann KF, Bergen AW, Goldstein AM, Goldin LR. Linkage analysis of the GAW14 simulated dataset with microsatellite and single-nucleotide polymorphism markers in large pedigrees. BMC Genetics. 2005;6(Suppl 1):S14. doi: 10.1186/1471-2156-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon H, Matsunami N, Stevens J, Miller J, Pingree C, Camp NJ, Thomas A, Krasny L, Lainhart J, Leppert MF, McMahon WM. Evidence for linkage on chromosome 3q25-27 in a large autism extended pedigree. Hum Hered. 2005;60(4):220–226. doi: 10.1159/000090546. [DOI] [PubMed] [Google Scholar]
- Horne BD, Malhotra A, Camp NJ. Framingham Heart Study. Comparison of linkage analysis methods for genome-wide scanning of extended pedigrees, with application to the TG/HDL-C ratio in the Framingham Heart Study. BMC Genet. 2003;4(Suppl 1):S93. doi: 10.1186/1471-2156-4-S1-S93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GB, Camp NJ, Farnham JM, Cannon-Albright LA. Genome-wide linkage analysis for aggressive prostate cancer in Utah high-risk pedigrees. Prostate. 2007;67(6):605–613. doi: 10.1002/pros.20554. [DOI] [PubMed] [Google Scholar]
- Hunt SC, Abkevich V, Hensel CH, Gutin A, Neff CD, Russell DL, Tran T, Hong X, Jammulapati S, Riley R, Weaver-Feldhaus J, Macalma T, Richards MM, Gress R, Francis M, Thomas A, Frech GC, Adams TD, Shattuck D, Stone S. Linkage of body mass index to chromosome 20 in Utah pedigrees. Hum Genet. 2001;109(3):279–285. doi: 10.1007/s004390100581. [DOI] [PubMed] [Google Scholar]
- Teerlink CC, Camp NJ, Bansal A, Crapo R, Hughes D, Kort E, Rowe K, Cannon-Albright LA. Significant evidence for linkage to chromosome 5q13 in a genome-wide scan for asthma in an extended pedigree resource. Eur J Hum Genet. 2009;17:636–643. doi: 10.1038/ejhg.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADDMN. Prevalence of autism spectrum disorders--autism and developmental disabilities monitoring network, 14 sites, United States, 2002. MMWR Surveill Summ. 2007;56(1):12–28. [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD. Linkage analysis in the presence of errors I: complex-valued recombination fractions and complex phenotypes. Am J Hum Genet. 2000;66(3):1095–1106. doi: 10.1086/302797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE. The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet. 1998;63(3):870–879. doi: 10.1086/301997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin LR. Detection of linkage under heterogeneity: comparison of the two-locus vs. admixture models. Genet Epidemiol. 1992;9(1):61–66. doi: 10.1002/gepi.1370090107. [DOI] [PubMed] [Google Scholar]
- Abreu PC, Greenberg DA, Hodge SE. Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet. 1999;65(3):847–857. doi: 10.1086/302536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GB, Cannon-Albright LA, Thomas A, Camp NJ. Extracting disease risk profiles from expression data for linkage analysis: application to prostate cancer. BMC Proc 1. 2007. p. S82. [DOI] [PMC free article] [PubMed]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Camp NJ, Gutin A, Abkevich V, Farnham JM, Cannon-Albright L, Thomas A. A new nonparametric linkage statistic for mapping both qualitative and quantitative trait loci. Genet Epidemiol. 2001;21:S461–466. doi: 10.1002/gepi.2001.21.s1.s461. [DOI] [PubMed] [Google Scholar]
- Allen-Brady K, Robison R, Cannon D, Varvil T, Villalobos ME, Pingree C, Leppert MF, Miller J, McMahon WM, Coon H. Genome-wide linkage in Utah autism pedigrees. Molecular Psychiatry. 2009. in press . [DOI] [PMC free article] [PubMed]
- Ott J. Analysis of Human Genetic Linkage. 3. Baltimore: The Johns Hopkins University Press; 1999. [Google Scholar]
- IMGSAC. A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet. 1998;7(3):571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- IMGSAC. A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16 p. International Molecular Genetic Study of Autism Consortium. Am J Hum Genet. 2001;69(3):570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMGSAC. An autosomal genomic screen for autism. International Molecular Genetic Study of Autism Consortium. Am J Med Genet. 2001;105(8):609–615. [PubMed] [Google Scholar]
- Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, Rosenthal E, Rothstein J, Flodman P, Smith M, Coon H, Leong L, Yu CE, Stodgell C, Rodier PM, Spence MA, Minshew N, McMahon WM, Wijsman EM. Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatry. 2006;11(11):1049–1060. doi: 10.1038/sj.mp.4001874. 979. [DOI] [PubMed] [Google Scholar]
- Trikalinos TA, Karvouni A, Zintzaras E, Ylisaukko-oja T, Peltonen L, Jarvela I, Ioannidis JP. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11(1):29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bromar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castermans D, Wilquet V, Parthoens E, Huysmans C, Steyaert J, Swinnen L, Fryns JP, Ven W Van de, Devriendt K. The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet. 2003;40(5):352–356. doi: 10.1136/jmg.40.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, Trixler M, Falkai P, Rietschel M, Maier W, Wildenauer DB. A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10 p and 6. Mol Psychiatry. 2000;5(6):638–649. doi: 10.1038/sj.mp.4000791. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, Oord E van den, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13(6):570–84. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ, Barci BM, Torrey EF. Meta-analysis of 12 genomic studies in bipolar disorder. J Mol Neurosci. 2007;31(3):221–243. doi: 10.1385/jmn:31:03:221. [DOI] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13(6):558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7(6):542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Hwu HG, Liu CM, Chen WJ, Tsuang MM, Liu SK, Shieh MH, Hwang TJ, Ou-Yang WC, Chen CY, Chen CC, Lin JJ, Chou FH, Chueh CM, Liu WM, Hall MH, Su J, Van Eerdewegh P, Tsuang MT. Genome scan of Han Chinese schizophrenia families from Taiwan: confirmation of linkage to 10q22.3. Am J Psychiatry. 2006;163(10):1760–1766. doi: 10.1176/appi.ajp.163.10.1760. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Lasky-Su J, Glatt SJ, Van Eerdewegh P, Tsuang MT. Early onset bipolar disorder: possible linkage to chromosome 9q34. Bipolar Disord. 2006;8(2):144–151. doi: 10.1111/j.1399-5618.2006.00289.x. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sham PC, Purcell S, Banaschewski T, Buitelaar J, Franke B, Sonuga-Barke E, Ebstein R, Eisenberg J, Mulligan A, Gill M, Manor I, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Chen W, Zhou K, Asherson P, Faraone SV. Population differences in the International Multi-Centre ADHD Gene Project. Genet Epidemiol. 2008;32(2):98–107. doi: 10.1002/gepi.20265. [DOI] [PubMed] [Google Scholar]
- Reiersen AM, Constantino JN, Grimmer M, Martin NG, Todd RD. Evidence for shared genetic influences on self-reported ADHD and autistic symptoms in young adult Australian twins. Twin Res Hum Genet. 2008;11(6):579–585. doi: 10.1375/twin.11.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiersen AM, Constantino JN, Volk HE, Todd RD. Autistic traits in a population-based ADHD twin sample. J Child Psychol Psychiat. 2007;48(5):464–472. doi: 10.1111/j.1469-7610.2006.01720.x. [DOI] [PubMed] [Google Scholar]
- Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC. Autism Genetic Resource Exchange Consortium. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69(2):327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Kim SJ, Gonen D, Hanna GL, Leventhal BL, Cook EH Jr. Genomic organization of the SLC1A1/EAAC1 gene and mutation screening in early-onset obsessive-compulsive disorder. Mol Psychiatry. 2001;6(2):160–167. doi: 10.1038/sj.mp.4000806. [DOI] [PubMed] [Google Scholar]
- Willour VL, Yao Shugart Y, Samuels J, Grados M, Cullen B, Bienvenu OJ III, Wang Y, Liang KY, Valle D, Hoehn-Saric R, Riddle M, Nestadt G. Replication study supports evidence for linkage to 9p24 in obsessive-compulsive disorder. Am J Hum Genet. 2004;75(3):508–513. doi: 10.1086/423899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold PD, Sicard T, Burroughs E, Richter MA, Kennedy JL. Glutamate transporter gene SLC1A1 associated with obsessive-compulsive disorder. Arch Gen Psychiatry. 2006;63(7):769–776. doi: 10.1001/archpsyc.63.7.769. [DOI] [PubMed] [Google Scholar]
- Liang KY, Wang Y, Shugart YY, Grados M, Fyer AJ, Rauch S, Murphy D, McCracken J, Rasmussen S, Cullen B, Hoehn-Saric R, Greenberg B, Pinto A, Knowles J, Piacentini J, Pauls D, Bienvenu O, Riddle M, Samuels J, Nestadt G. Evidence for potential relationship between SLC1A1 and a putative genetic linkage region on chromosome 14q to obsessive-compulsive disorder with compulsive hoarding. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):1000–1002. doi: 10.1002/ajmg.b.30713. [DOI] [PubMed] [Google Scholar]