Abstract
Background
HOXB1 plays a major role in brainstem morphogenesis and could partly determine the cranial circumference in conjunction with HOXA1. In our sample, HOXA1 alleles significantly influence head growth rates both in autistic patients and in population controls. An initial report, suggesting that HOXB1 could confer autism vulnerability in interaction with HOXA1, was not confirmed by five small association studies.
Methods
Our sample includes 269 autistic individuals, belonging to 219 simplex and 28 multiplex families. A mutational analysis of the two exons and flanking intronic sequences of the HOXB1 gene was carried out in 84 autistic patients by denaturing high performance liquid chromatography, followed by DNA sequencing. Identified rare variants were then searched by a restriction analysis in 236 autistic patients and 325-345 controls. Case-control and family-based association studies were performed on two common variants in 169 Italian patients versus 184 Italian controls and in 247 trios.
Results
We identified three common polymorphisms, rs72338773 [c.82insACAGCGCCC (INS/nINS)], rs12939811 [c.309A>T (Q103H)], and rs7207109 [c.450G>A (A150A)] and three rare variants, namely IVS1+63G>A, rs35115415 [c.702G>A (V234V)] and c.872_873delinsAA (S291N). SNPs rs72338773 and rs12939811 were not associated with autism, using either a case-control (alleles, exact P = 0.13) or a family-based design [transmission/disequilibrium test (TDT)χ2 = 1.774, P = 0.183]. The rare variants, all inherited from one of the parents, were present in two Italian and in two Caucasian-American families. Autistic probands in two families surprisingly inherited a distinct rare variant from each parent. The IVS1+63A allele was present in 3/690 control chromosomes, whereas rare alleles at rs35115415 and c.872_873delinsAA (S291N) were not found in 662 and 650 control chromosomes, respectively. The INS-T309 allele influenced head size, but its effect appears more modest and shows no interaction with HOXA1 alleles. The INS-T309 allele is also associated with more severe stereotypic behaviours, according to ADI-R scores (N = 60 patients, P < 0.01).
Conclusions
HOXB1 mutations do not represent a common cause of autism, nor do HOXB1 common variants play important roles in autism vulnerability. HOXB1 provides minor, albeit detectable contributions to head circumference in autistic patients, with HOXA1 displaying more prominent effects. HOXB1 variants may modulate the clinical phenotype, especially in the area of stereotypic behaviours.
Background
Genetic contributions to autism have received strong support from family and twin studies [1,2]. A relatively small number of major loci was initially predicted to explain the disease in the majority of affected individuals [2]. However, the number of common genetic variants conferring vulnerability to autism has grown well beyond the initial expectations, in addition to several rare variants identified to this date, each explaining the disease in a very small number of patients [[3-5]. The genetic underpinnings of autism spectrum disorders have thus proven to be far more complex than expected, likely due to genetic heterogeneity, epistasis and gene-environment interactions [3-5].
Several lines of evidence have demonstrated altered prenatal neurodevelopment as central to autism pathogenesis. Abnormal neurodevelopment possibly occurring in the first trimester of pregnancy best accounts for the microscopic alterations shown by post-mortem neuroanatomical studies of the brains of autistic patients [6,7]. Phenotypic evidence also supports a prenatal aetiology, as fine motor abnormalities are detectable at 4-6 months of age or even at birth in children later developing autism [8]. Finally, a prenatal time window as early as days 20-25 post-fertilization appears crucial to autism's aetiology, since exposure to thalidomide during pregnancy leads to autism only if occurring within this limited developmental interval [9]. Genes encoding proteins involved in early neural development could thus encompass polymorphisms or mutations contributing to the disease process.
The HOXB1 gene, located on human chr 17q21.32, is a member of the HOX gene family of homeobox transcription factors and critically involved in the development of the brain stem. HOXB1 gene expression is limited to rhombomere 4 and to neural crest cells migrating out of this region, resulting in a gross reduction or complete loss of the facial motor nucleus in HOXB1 mutant mice [10,11]. A similar loss of facial motor neurons has been described in one autistic brain [12]. HOXB1 gene expression occurs very early in mouse development (E8.5-E9.5) [10,11], at a time, interestingly, overlapping with the window of maximal prenatal sensitivity in rodent models of autism [13]. HOXB1 gene expression is also strongly up-regulated by HOXA1 [14] and the two genes indeed synergize in patterning hindbrain structures, cranial nerves and pharyngeal arches, so that double-mutant mice display prominent malformations while single mutants suffer much milder abnormalities [14-16]. A gene × gene interaction between HOXB1 and HOXA1 gene variants was initially proposed to contribute to autism [17]. In our sample, the HOXA1 c.218A>G [His73Arg] polymorphism was significantly associated with autism, although we found an association with the A218 and not the G218 allele [18], in contrast to the original study [17]. Moreover, HOXA1 c.218A>G alleles were found to significantly influence head growth rates, and not final head size, both in autistic patients and in typically developing children [18,19]. The latter result is especially interesting, considering that approximately 20% of autistic patients consistently show macrocephaly and may represent an endophenotypic subgroup possibly sharing common pathophysiological underpinnings [20,21]. Following the initial positive study [17], five studies subsequently reported negative association findings using HOXB1 gene markers [22-26]. However, the sample sizes assessed in all of these studies were too small to detect moderate-size effects and rare variants of potential clinical relevance (see Discussion). Furthermore, only two of these studies apparently performed mutational searches by DNA sequencing [22,23]. The present study was thus undertaken to screen a larger sample of autistic patients, unaffected controls and nuclear families for HOXB1 gene mutations, rare variants and common polymorphisms, either causing the disorder, conferring autism vulnerability, influencing the clinical expression of the disease, or modulating cranial growth rates by themselves or in interaction with HOXA1 gene variants.
Methods
Subjects
Families were recruited for this study based on the presence of a proband diagnosed with primary autism spectrum disorder (idiopathic, non-syndromic ASD). The clinical sample includes 269 autistic individuals and 593 first-degree relatives, belonging to 219 simplex and 28 multiplex families. In reference to ethnicity, these families encompass 171 simplex and one multiplex Italian families, as well as 48 simplex and 27 multiplex Caucasian-American families, the latter including 15 simplex and 23 multiplex families obtained from the Autism Genetic Resource Exchange (AGRE) repository. Demographic and clinical characteristics of the autistic patients are summarized in Table 1. In addition, we also recruited 345 normal adult controls (M/F ratio = 2.43; mean age ± standard deviation = 33.1 ± 9.2 years, range 18-53), among blood donors at the Haematology Unit of 'Casa Sollievo della Sofferenza' hospital (S Giovanni Rotondo, FG, Italy) and self-reporting no history of major psychiatric disorders or psychotropic drug use for at least 6 months prior to blood donation. This clinical sample largely overlaps with the sample assessed in our previous works on the HOXA1 gene [18,19]. The size of subgroups of patients and families involved in the different studies described in the present report are summarized in Additional File 1: Table S1.
Table 1.
Demographic and clinical characteristics of the autistic patients (N = 269, unless otherwise specified).
| Mean/median | Standard deviation | Range | ||
|---|---|---|---|---|
| Mean Age (year) | 9.3 | 5.6 | 3-33 | |
| Median VABS scores (N = 149) | ||||
| Communication | 71 | 19-112 | ||
| Daily living skills | 65 | 19-107 | ||
| Socialization | 66 | 20-103 | ||
| Motor skills | 80 | 37-114 | ||
| Composite | 60 | 19-103 | ||
| N | (%) | |||
| Sex | ||||
| Male | 242 | (90.0) | ||
| Female | 27 | (10.0) | ||
| M/F ratio | 9.0:1 | |||
| Family type | ||||
| Simplex | 219 | (88.7) | ||
| Multiplex | 28 | (11.3) | ||
| DSM-IV diagnosis | ||||
| Autistic disorder | 208 | (77.3) | ||
| Asperger syndrome | 22 | (8.2) | ||
| PDD-NOS | 39 | (14.5) | ||
| Intellectual quotient (N = 99) | ||||
| >70 | 38 | (38.4) | ||
| ≤ 70 | 61 | (61.6) | ||
VABS, Vineland Adaptive Behavior Scales; DSM, Diagnostic and Statistical Manual of Mental Disorders; PDD-NOS, pervasive development disorder not otherwise specified.
Inclusion criteria and diagnostic screening methods have been previously reported [18,19,21] . Briefly, patients fulfilling the Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV diagnostic criteria for autistic disorder, Asperger's disorder or pervasive developmental disorder not otherwise specified (PDD-NOS) [27] were screened for non-syndromic autism using magnetic resonance imaging, electroencephalogram, audiometry, urinary aminoacid and organic acid measurements, cytogenetic and fragile-X testing. Patients with dysmorphic features were excluded, even in the absence of detectable cytogenetic alterations. Patients with sporadic seizures (< 1 every 6 months) were included; patients with frequent seizures or focal neurological deficits were excluded. Autistic behaviours were assessed using the official Italian version of the Autism Diagnostic Observation Schedule (ADOS) and the Autism Diagnostic Interview-Revised (ADI-R) [28,29], available since 2005; adaptive functioning was assessed using the Vineland Adaptive Behavior Scales; intellectual quotient was determined using either the Griffith Mental Developmental Scales, the Coloured Raven Matrices, the Bayley Developmental Scales or the Leiter International Performance Scale. Head circumference was measured in all ASD patients aged <16 years old and transformed into percentiles using the sex- and age-specific standard tables currently adopted in the vast majority of European countries and by the Italian Pediatric Association, as described [21]. This study, as well as the consent form signed by all parents for themselves and their children, was approved by the Institutional Review Board (IRB) of the University Campus Bio-Medico (Rome, Italy). Controls provided written informed consent to the use of their biomaterials for scientific research, using the consent form approved by the IRB of 'Casa Sollievo dalla Sofferenza' hospital (S Giovanni Rotondo, FG, Italy).
Primer design and polymerase chain reaction (PCR) amplification conditions
PCR primers were designed to amplify the two exons of the HOXB1 gene (Ensembl: ENST00000239174) and exon-intron boundaries, extending 30-100 bases into intronic sequences both 5' and 3' of each exon. Primers are described in Table 2. PCR amplifications were performed using a Gene Amp PCR System 9700 (Perkin Elmer, CA, USA) in a final reaction volume of 25 μL containing 100 ng of genomic DNA template, 250 μM dNTPs, 0.5 μM of each primers, 1.25 U Ampli taq GoldTM DNA polymerase (PE Applied Biosystems, CA, USA), in 1X reaction Buffer (10 mM Tris HCl pH 8.3, 50 mM KCl, 2.5 mM MgCl2). PCR cycling conditions consisted of an initial denaturation step at 95°C for 12 min, followed by 35 cycles of 95°C for 30 s, annealing at temperatures reported in Table 2 for 30 s and elongation at 72°C for 10 min.
Table 2.
Primer sequences for HOXB1 gene PCR amplification and DHPLC analysis.
| Exon | Primer name | Primer sequences (5'-3') | Amplicon size (bp) | PCR annealing temp (°C) | DHPLC oven temp (°C) |
|---|---|---|---|---|---|
| 1 | HOXB1-F1 for | CATACTGCCGAAAGGTTGTAG | 365 | 60 | 61.5, 65.6, 66.2 |
| HOXB1-F1 rev | TAGTACTGAGAAGGCCCGTA | ||||
| 1 | HOXB1-F2 for | GGTATGCTCCTGCCGCCTGCA | 227 | 58 | 63.3, 65.5 |
| HOXB1-F2 rev | ATCAGCATAGGCCGGTGCAA | ||||
| 1 | HOXB1-F3 for | AGCATCCCCCTTATGGGAA | 282 | 58 | 62.8 |
| HOXB1-F3 rev | CTTACCTGTGTCTACCAGAG | ||||
| 2 | HOXB1-F4 for | GAGAATTGACCTGGCCTTTC | 359 | 60 | 62.9, 63.4 |
| HOXB1-F4 rev | TGACAGAGCTGGGTGAGGCTT | ||||
| 2 | HOXB1-F5 for | TTTGGTTCCAGAACCGACGA | 300 | 60 | 64.0, 65.5 |
| HOXB1-F5 rev | GGCAGCTCTAAACTGGACTT | ||||
PCR, polymerase chain reaction; DHPLC, denaturing high performance liquid chromatography; temp, temperature.
Wave® System denaturing high performance liquid chromatography (DHPLC) analysis
PCR products from each patient and one normal control were mixed, denaturated for 5 min at 95°C and cooled slowly for 30 min down to 40°C in a thermal cycler, to enable the formation of heteroduplexes. Analytical conditions for each fragment were determined using the Transgenomic WaveMaker™ Software v.4.1.44 (Transgenomic Inc, NE, USA). Samples were run on the 3500HT Wave™ DNA Analysis System (Transgenomic). PCR amplicons were loaded (5 μL) on a C18 reverse-phase column based on nonporous poly (styrene/divinil-benzene) particles (DnaSep™ column; Transgenomic). Hetero and homoduplex analysis was carried out with an acetonitrile gradient formed by mixing buffer A (0.1 M TEAA) and buffer B (0.1 M TEAA, 25% acetonitrile). Flow-rate was 0.7 mL/min with an increase of buffer B of 2% per min for 4.5 min and DNA was detected at 260 nm. All samples showing an abnormal elution profile were sequenced.
Sequence analysis
PCR products of patients yielding abnormal chromatograms were purified using GFXTM PCR DNA and Gel Band Purification Kit (Amersham Biosciences, CA, USA). DNA sequencing was performed in a 10 μL final volume, with 3 pmol of primer, 4-6 ng of DNA template and 2 μL of Big Dye Terminator Ready Reaction mix v. 2.1 (PE-ABI, CA, USA), using an ABI PRISM 3100 Genetic Analyser, v.3.7 (PE Applied Biosystems, Foster City, CA). DNA sequence analysis was performed using Sequencing Analysis program v. 3.7 (PE Applied Biosystems).
Genotyping
SNP rs12939811 (Q103H) was genotyped by PCR amplification and DHPLC analysis (temperature melting 63.3°C). In order to discriminate homozygosity for the common allele (AA) from homozygosity for the rare allele (TT), the samples resulting in a single elution profile were mixed with a control sample carrying the AA genotype and were analysed again by DHPLC under at the same temperature conditions. All samples compatible with a TT genotype were then sequenced for confirmation. SNPs rs72338773 (c.82insACAGCGCCC), IVS1+63G>A, S291N and rs35115415 (V234V) were each genotyped by PCR amplification and allele-specific restriction analysis. Primer sequences for HOXB1 genotyping are listed in Table 3. Briefly, restriction analyses were performed, as follows: (a) rs72338773 (c.82insACAGCGCCC) was amplified using primers HOXB1-F1_for/HOXB1-F1_rev; the 365 bp fragment was digested with MspI, yielding 140, 121 and 113 bp fragments in the presence of the insertion; (b) the IVS1+63G>A SNP was amplified with primers IVS1+63G>A-mut_for/IVS1+63G>A_rev and the 118 bp amplicon was digested using HphI, producing two 83 and 35 bp fragments in the presence of the A allele; (c) the c.872_873delinsAA (S291N) SNP was amplified using primers S291N-mut_for/HOXB1-F5_rev and the 366 bp fragment was digested with MnlI, yielding 186, 112bp, 50bp and 18bp fragments with the N allele; and (d) rs35115415 (V234V) was amplified using primers S291N-mut_for/HOXB1-F5 rev and the 366 bp fragment was digested with FokI, yielding 335 and 31 bp in the presence of the A allele.
Table 3.
Primer sequence for HOXB1 SNP genotyping.
| Exon | Primer name | Primer sequences (5'-3') | Amplicon size (bp) | PCR annealing temp (°C) |
|---|---|---|---|---|
| 1 | HOXB1-F1 for | CATACTGCCGAAAGGTTGTAG | 365 | 60 |
| HOXB1-F1 rev | TAGTACTGAGAAGGCCCGTA | |||
| 1 | HOXB1-F2 for | GGTATGCTCCTGCCGCCTGCA | 227 | 58 |
| HOXB1-F2 rev | ATCAGCATAGGCCGGTGCAA | |||
| 1 | IVS1+63 G/A-mut_for | AAAGCATCTCTGCTTCCCCTGCGG | 118 | 58 |
| IVS1+63G/A_rev | GACCTCACCTGACCTGAGAC | |||
| 2 | S291N-mut_for | ACCTGAGCCGGGCCCGGAGGATG | 366 | 58 |
| HOXB1-F5 rev | GGCAGCTCTAAACTGGACTT | |||
PCR, polymerase chain reaction; temp, temperature.
Data analysis
Hardy-Weinberg analyses were performed using the Hardy-Weinberg equilibrium (HWE) and HWE2 programs [30]. Case-control allelic and genotypic distributions were contrasted applying the χ2 statistics after randomly selecting one patient per multiplex family. Single-marker and family-based association analyses were performed applying the transmission/disequilibrium test (TDT) [31] using the TDTPHASE software of the UNPHASED package [32]; only complete trios and one trio per multiplex family were included in these analyses. All family-based association analyses were carried out on Italian and Caucasian-American families merged together after population structure analyses provided no evidence of genetic dyshomogeneity in a subgroup of 179 autistic patients including 155 Italians and 24 Caucasian-Americans randomly chosen one per family, genotyped at 90 unlinked SNPs distributed genome-wide and analysed using the STRUCTURE program [33]. Since this stratification analysis did not include unaffected controls, case-control contrasts employed only individuals of Italian ancestry. Power analyses were performed using P2BAT [34]. The phylogenetic conservation of sequences encompassing HOXB1 polymorphisms was assessed by orthologue sequence alignments performed using the ClustalW2 [35] and VISTA [36] softwares. The distributions of cranial circumference percentiles in cases and controls were contrasted using the Mann-Whitney U test and the Moses test of extreme reactions, to analyse differences in central tendency and dispersion, respectively [37]. Data are expressed as mean ± standard error of mean, except for the head circumference which is expressed as median ± interquartilic range. Two-tail P values are reported throughout the manuscript. The outcome of analyses with phenotypic variables (items of behavioural scales, patient and family history variables and head circumference) underwent three levels of Bonferroni correction for multiple testing: (a) 'stringent', assuming each phenotypic variable as independent: P = 0.05/41 = 0.0012; (b) 'intermediate', counting as a single entity only those items which record clearly overlapping phenomena (for example, 'presence of motor stereotypes' in patient history and ADI-R item C): P = 0.05/28 = 0.0018; and (c) 'relaxed', considering all variables, except for the head circumference, as non-independent entities clustered into four principal components, in accordance with our recent work [38]: P = 0.05/5 = 0.01.
Results
HOXB1 gene variants
The mutational analysis performed by DHPLC and DNA sequencing in 84 autistic patients unveiled three common polymorphisms and three rare variants. The common polymorphisms, which were all previously described [17,22,39], are: (a) rs72338773 [c.82insACAGCGCCC (INS/nINS)], here also named c.82ins9; (b) rs12939811 [c.309A>T (Q103H)]; and (c) rs7207109 [c.450G>A (A150A)]. The 9-bp insertion c.82insACAGCGCCC was initially reported by Faiella et al. [39] and introduces into the amino acid sequence the tripeptide H-S-A. Two of the three common variants, namely rs72338773 and rs12939811, were genotyped in 169 ASD patients and 184 controls, all of Italian ethnicity. Genotypic and allelic distributions are presented in Table 4.
Table 4.
Case-control study for the HOXB1 polymorphisms rs72338773 [c.82insACAGCGCCC (INS/nINS)] and rs12939811 [c.309A>T (Q103H)].
| Genotypes | Italian patients (N = 169) | Italian controls (N = 184) | Alleles | Italian patients (N = 338) | Italian controls (N = 368) |
|---|---|---|---|---|---|
| nINS/nINS | 100 (59.2%) | 125 (67.9%) | nINS | 262 (0.7751) | 303 (0.8234) |
| nINS/INS | 62 (36.7%) | 53 (28.8%) | INS | 76 (0.2249) | 65 (0.1766) |
| INS/INS | 7 (4.1%) | 6 (3.3%) | |||
| χ2 = 2.93, 2 df, P = 0.23, ns | Exact 2-tail P = 0.13, ns | ||||
| A/A | 102 (60.4%) | 125 (67.9%) | A | 263 (0.7781) | 304 (0.8261) |
| A/T | 59 (34.9%) | 54 (29.3%) | T | 75 (0.2219) | 64 (0.1739) |
| T/T | 8 (4.7%) | 5 (2.8%) | |||
| χ2 = 2.61, 2 df, P = 0.27, ns | Exact two-tail P = 0.13, ns | ||||
INS, insertion; ns, not significant.
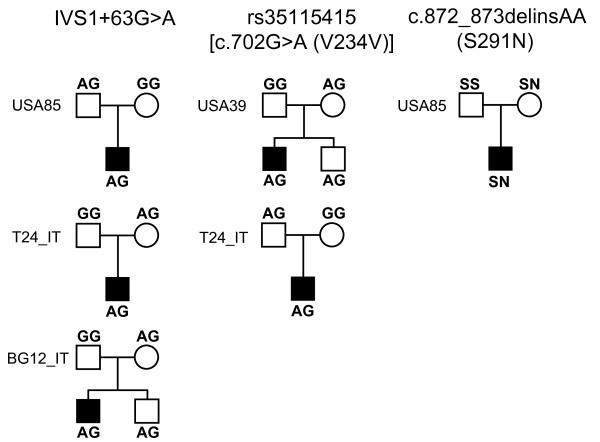
The three rare variants - including (a) IVS1+63G>A, (b) rs35115415 [c.702G>A (V234V)] and (c) c.872_873delinsAA (S291N) - were searched in a total of 236 autistic patients. In all cases, rare variants were inherited by the proband from one of the parents, as depicted in Figure 1. Allelic frequencies are as follows:
Figure 1.
Pedigrees of families carrying rare variants in the HOXB1gene.
(a) the IVS1+63G>A was found in 3/472 (0.64%) chromosomes from ASD patients and in 3/690 (0.43%) chromosomes belonging to 345 Italian controls (patients versus controls, P = 0.62)
(b) rs35115415 (c.702G>A) was found in one Italian and one Caucasian-American families (2/472 = 0.42%). This variant was not found in 662 chromosomes belonging to 331 Italian controls. This same variant is reported in dbSNP with an allelic frequency of A = 0.02, assessed in 21 Caucasian- and 20 African-Americans from the Coriell Cell Repository collection of apparently healthy individuals
(c) c.872_873delinsAA (S291N) involves the change of contiguous base pairs (TC>AA), present on the same chromosome inherited by the proband from the maternal side in one Caucasian-American family (1/472 = 0.21%). This variant was not found in 650 chromosomes belonging to 325 Italian controls.
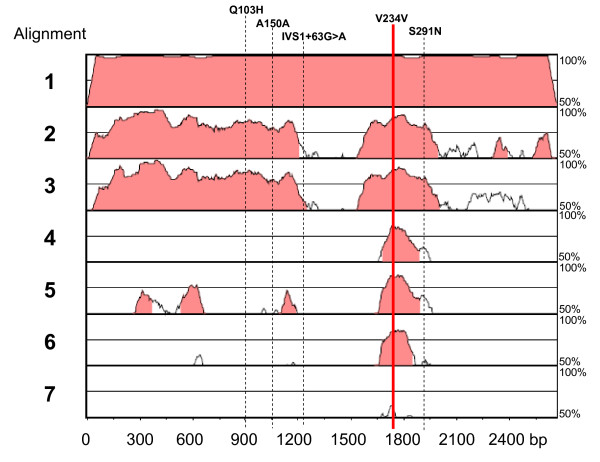
In two families, rare variants IVS1+63G>A and 702G>A (V234V) were transmitted also to an unaffected sibling (Figure 1). Phylogenetic analyses performed using VISTA (Figure 2) and ClustalW2 (Figure 3) demonstrate that especially rs35115415 [c.702G>A (V234V)] and, to some extent, also the S291N rare variants are located in evolutionarily conserved regions.
Figure 2.
Phylogenetic conservation analysis by sequence alignment: VISTA plot of the entire HOXB1 locus, highlighting the position of SNPs Q103H, A150A, IVS1+63G>A, V234V, and S291N. Species include Homo sapiens, aligned with (1) Pan troglodytes, (2) Mus musculus, (3) Rattus norvegicus, (4) Xenopus laevis, (5) Gallus gallus, (6) Tetraodon nigroviridis and (7) Caenorhabditis elegans, in this order.
Figure 3.
Phylogenetic conservation analysis by sequence alignment: ClustalW2 output focussed on the V234V (top) and S291N (bottom) SNPs.
Case-control and family based association studies
No significant deviation from HWE was detected by analysing HOXB1 common variants in our sample (Table 4). Case-control and family-based association studies were performed using rs72338773 (INS/nINS) and rs12939811 (Q103H); rs7207109 (A150A) was dropped, because it is a synonymous variant in complete linkage disequilibrium with rs12939811 (data not shown). The results of the case-control study are summarized in Table 4. Neither rs72338773 nor rs12939811 show a genotypic or allelic association with autism. Genotypic and allelic distributions in our control sample are very similar to those obtained in 56 CEU individuals (i.e., Utah residents with Northern and Western European ancestry from the CEPH collection) available in public databases [A/A = 41 (73.2%), A/T = 13 (23.2%), T/T = 2 (3.6%); allele A = 95 (0.848), allele T = 17 (0.152)] and also these do not differ significantly from genotypic and allelic distributions present in our autistic patients [genotype χ2 = 3.65, 2 df, P = 0.16; allelic exact P = 0.14]. Similarly, a TDT performed on 247 trios, including 172 Italian and 75 Caucasian-American trios, yields a non-significant trend toward the overtransmission of the T309 allele from heterozygous parents to affected offspring (transmitted:non-transmitted = 118:103; χ2 = 1.774, 1 df, P = 0.183). In accordance with case-control results, the 9-bp insertion displays an even smaller overtransmission for the INS allele (transmitted: non-transmitted = 127: 118; χ2 = 0.398, 1 df, P = 0.528). Collectively, our results do not support an association of large/moderate effect size between autism and HOXB1 gene variants marked by SNPs rs72338773 and rs12939811.
Phenotypic correlates of HOXB1 gene variants in autism
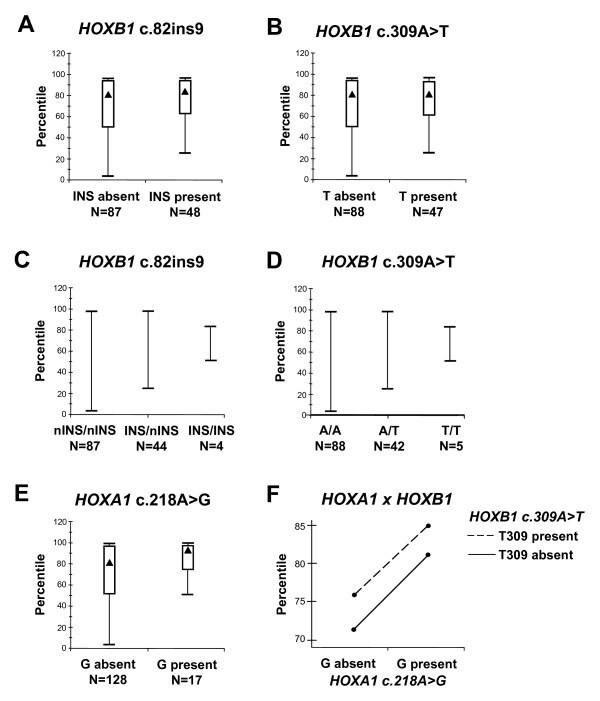
HOXB1 alleles exert a modest effect on cranial circumference in autistic patients (Figure 4). Median head sizes are practically superimposable in autistic patients with a presence/absence of INS and/or T309 alleles [INS present versus INS absent = 86.25 ± 17.5 versus 82.5 ± 23.75 percentile, Mann-Whitney (M-W) U = 2609.0, P = 0.31; T309 present versus T309 absent = 82.5 ± 17.5 versus 82.5 ± 23.75 percentile, M-W U = 2740.5, P = 0.66]. However, autistic patients carrying at least one copy of the INS or T309 alleles display significantly less dispersion in their head circumference distribution compared to patients carrying the nINS/nINS or AA genotypes (Moses test of extreme reactions, P < 0.01 for both) (Figure 4, panels A-D). Practically, the lower end of the head circumference range is at the 50th, 25th and 3rd percentile in ASD patients carrying the INS/INS, INS/nINS, and nINS/nINS (or TT, AT and AA) genotype, respectively, and all of the 7/88 (8.0%) ASD patients with head circumferences below the 25th percentile carry the nINS/nINS and AA genotypes (Figure 4, panels C and D). Importantly, these same patients display a much more prominent effect on head size by HOXA1 c.218A>G alleles, as previously reported [18,19] (G218 present versus G218 absent = 93.5 ± 16.9 versus 82.5 ± 23.75 percentile, M-W U = 2841.0, P = 0.08 and Moses test, P < 1 × 10-7; (Figure 4, panel E). Finally, we find no evidence of HOXA1 × HOXB1 interactions, as both appear to independently influence head circumference in autistic patients (Figure 4, panel F).
Figure 4.
Cranial circumference and allelic status at SNPs (A, C) HOXB1 rs72338773 [c.82insACAGCGCCC (INS/nINS)], (B, D) HOXB1 rs12939811 [c.309A>T (Q103H)], (E) HOXA1 rs10951154 [c.218A>G (H73R)], and (F) HOXA1 × HOXB1 interactions at SNPs c.218A>G and c.309A>T, respectively. Sample sizes are reported for each group below the X-axis. Data are expressed as median+interquartilic range (▲ = median; T = I.Q.R.; ▯= non-outliers).
Analyses exploring possible clinical correlates of HOXB1 allelic status in the subset of patients characterized using the ADOS and ADI-R unveiled a possible influence on stereotypic behaviours. Mean total C score at the ADI-R were 7.18 versus 5:30 for 'T309 present' versus 'T309 absent' patients (N = 17 and 43, respectively, Student t = 2.72, 58 df, P < 0.01). This analysis survives Bonferroni correction only when applying the 'relaxed' criterion (see Methods).
Discussion
The present study describes three rare (IVS1+63G>A, V234V and S291N) and three common (c.82ins9, Q103H and A150A) variants in the HOXB1 locus of a total sample of 269 ASD patients. No pathogenetic de novo mutation was found as all these gene variants were inherited from one of the parents and at least two are present in other population samples available in public databases. Also, HOXB1 common variants do not seem to play major roles in autism pathogenesis, although minor contributions cannot be excluded due to sample size limitations (see below). At the phenotypic level, HOXB1 alleles appear to modulate head growth rates to a much lesser extent compared to the HOXA1 c.218A>G SNP (Figure 4, compare panels E versus A and B) [18,19]. In reference to cranial circumference, we also find no evidence of gene-gene interaction, since both HOXA1 and HOXB1 influence head growth independently of each other (Figure 4F). Finally, preliminary analyses involving the subgroup of patients characterized also with the ADI-R suggest that HOXB1 alleles may influence stereotypic behaviours. Given the relatively large number of clinical variables assessed for association with HOXB1 alleles, this finding survives correction for repeated measures only applying the 'relaxed' criteria, namely those accounting for the non-independence of several variables which are significantly cross-correlated in our sample [38].
In interpreting these results, three limitations of our study design should be considered: (a) we have not specifically assessed for the presence of CNVs encompassing the HOXB1 locus in our sample - from a methodological standpoint, DHPLC is not able to distinguish true homozygosity from deletion of one allele; (b) despite employing a significantly larger sample size, compared to previously published reports [17,22-26], our study is still underpowered for common variants in the allelic frequency range of the INS and T309 alleles (see Table 5 at allelic frequencies of 0.2-0.3) and for highly penetrant de novo mutations with dominant effects and allelic frequency below 0.02 (Table 6 at an allelic frequency of 0.01); and (c) we cannot evaluate the possible differences in mutation burden at this locus between patients and controls because mutational analysis was performed only among our patient sample.
Table 5.
Power analysis referring to a family-based design for common variants with low penetrance under an additive model.
| Allele frequency | Penetrance | Power | ||
|---|---|---|---|---|
| AA | AB | BB | ||
| 0.2 | 0.0 | 0.2 | 0.4 | 0.483 |
| 0.3 | 0.0 | 0.2 | 0.4 | 0.689 |
| 0.4 | 0.0 | 0.2 | 0.4 | 0.800 |
| 0.5 | 0.0 | 0.2 | 0.4 | 0.855 |
| 0.6 | 0.0 | 0.2 | 0.4 | 0.899 |
| 0.7 | 0.0 | 0.2 | 0.4 | 0.902 |
| 0.2 | 0.0 | 0.15 | 0.3 | 0.351 |
| 0.3 | 0.0 | 0.15 | 0.3 | 0.511 |
| 0.4 | 0.0 | 0.15 | 0.3 | 0.637 |
| 0.5 | 0.0 | 0.15 | 0.3 | 0.730 |
| 0.6 | 0.0 | 0.15 | 0.3 | 0.795 |
| 0.7 | 0.0 | 0.15 | 0.3 | 0.735 |
| 0.2 | 0.0 | 0.1 | 0.2 | 0.202 |
| 0.3 | 0.0 | 0.1 | 0.2 | 0.277 |
| 0.4 | 0.0 | 0.1 | 0.2 | 0.396 |
| 0.5 | 0.0 | 0.1 | 0.2 | 0.477 |
| 0.6 | 0.0 | 0.1 | 0.2 | 0.488 |
| 0.7 | 0.0 | 0.1 | 0.2 | 0.505 |
Table 6.
Power analysis referring to a family-based design for rare variants with high penetrance and a dominant model.
| Allele frequency | Penetrance | Power | ||
|---|---|---|---|---|
| AA | AB | BB | ||
| 0.10 | 0.8 | 0.8 | 0.0 | 1.000 |
| 0.08 | 0.8 | 0.8 | 0.0 | 1.000 |
| 0.06 | 0.8 | 0.8 | 0.0 | 1.000 |
| 0.04 | 0.8 | 0.8 | 0.0 | 0.986 |
| 0.02 | 0.8 | 0.8 | 0.0 | 0.642 |
| 0.01 | 0.8 | 0.8 | 0.0 | 0.075 |
| 0.10 | 0.9 | 0.9 | 0.0 | 1.000 |
| 0.08 | 0.9 | 0.9 | 0.0 | 1.000 |
| 0.06 | 0.9 | 0.9 | 0.0 | 1.000 |
| 0.04 | 0.9 | 0.9 | 0.0 | 0.998 |
| 0.02 | 0.9 | 0.9 | 0.0 | 0.767 |
| 0.01 | 0.9 | 0.9 | 0.0 | 0.144 |
| 0.10 | 1.0 | 1.0 | 0.0 | 1.000 |
| 0.08 | 1.0 | 1.0 | 0.0 | 1.000 |
| 0.06 | 1.0 | 1.0 | 0.0 | 1.000 |
| 0.04 | 1.0 | 1.0 | 0.0 | 0.999 |
| 0.02 | 1.0 | 1.0 | 0.0 | 0.751 |
| 0.01 | 1.0 | 1.0 | 0.0 | 0.146 |
A novel and interesting finding is represented by the location and pattern of inheritance of rare variants. In particular, rs35115415 [c.702G>A (V234V)] and c.872_873delinsAA (S291N) are located in genomic regions which have been evolutionarily conserved from Tetraodon nigroviridis and Caenorhabditis elegans all the way up to Homo sapiens (Figures 2 and 3). This homeobox domain is probably under high selective pressure and disruptive mutations are probably incompatible with life. Mutations or chromosomal rearrangements involving HOX genes and compatible with life are in most cases highly disruptive in animals and in humans, resulting in abnormal limb formation, severe neurological syndromes, leukaemias, or solid tumours [40,41]. In reference to unaffected individuals, publically available databases report one large chorodial neovascularization (CNV; duplication/deletion) involving HOXB1, among many other genes, found in two of the 270 individuals of the HapMap collection (see http://projects.tcag.ca/variation/, variation 4040, landmarks chr17:43,957,672... 44,191,836) [42], indicating that the human genome may tolerate some deletions and duplications involving the entire HOXB1 locus. However, the HOXB1 gene segments hosting rs35115415 [c.702G>A (V234V)] and c.872_873delinsAA (S291N) may possibly accommodate only the less disruptive and non-pathogenic gene variants. Another interesting issue is the parental transmission to the same autistic proband of two distinct rare variants, one transmitted from each parent (Figure 1). Instances where rare variants (single nucleotides and/or CNVs) have no causal effect in heterozygous carriers but are pathogenic in a state of compound heterozygosity are increasingly recognized as a possible cause of autism [43]. However, the rare HOXB1 variants described in the present study do not appear pathogenic, as it is biologically implausible that in family T24_IT, for example, the convergence of the intronic variant IVS1+63G>A and the conserved variant 702G>A (V234V) in the same individual may bear dramatic functional consequences. Within the framework of the 'compound heterozygosity' hypothesis, a similar phenomenon should also be occurring in at least one other autism-causing locus. The a priori probability of the compound heterozygosity occurs by chance at the HoxB1 locus can be estimated at 1.8 × 10-5 and 9.0 × 10-6 for families T24_IT and USA85, respectively. The probability of this same phenomenon occurring by chance at two separate loci in the same family is extraordinarily low, unless boosted by a deficit in genome maintenance mechanisms. Some [44,45], though not all studies [46], support the hypothesis that a small, yet a sizable minority of autism families may display an increased degree of genomic instability. This hypothesis, which is clearly not addressed in the present report, will be the object of further investigation
Conclusions
In summary, our data indicate that: (a) HOXB1 gene variants, either rare or common, do not exert large or moderate effects on affection status in autism spectrum disorders, while minor contributions cannot be excluded due to sample size limitations; (b) modulatory effects on head growth rates are compatible with the developmental roles of this homeobox gene: and (c) HOXB1 could influence the clinical autistic phenotype in the area of stereotypic behaviours.
The peculiar coincidence of two distinct rare variants inherited by the autistic probands from both parents in two families raises interest in dysfunctional genome maintenance mechanisms in a subgroup of ASD patients.
Abbreviations
ADI-R: Autism Diagnostic Interview-Revised; ADOS: Autism Diagnostic Observation Schedule; AGRE: Autism Genetic Resource Exchange; ASD: autism spectrum disorders; CNV: chorodial neovascularization; DHPLC: denaturing high performance liquid chromatography; HWE: Hardy-Weinberg equilibrium; INS: insertion; M-W: Mann-Whitney; PCR: polymerase chain reaction; PDD-NOS: pervasive developmental disorder not otherwise specified; TDT: transmission/disequilibrium test.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LAM and VG, coordinated by LDA, performed the DHPLCs, genotyping, DNA sequencing and phylogenetic analyses and commented on the manuscript. RS updated the clinical and genetic databases and performed all statistical analyses. Patient recruitment, clinical assessment and sample collections were performed by PC, BM, RA and GG in Rome, RMi and CB in Naples, CL and MS in Milan, CS and RM in Phoenix. AMP contributed to study design, coordinated the recruitment and data collection and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1: Sample sizes involved in the different studies reported here, distinguished by cases/control status and by ethnicity.
Contributor Information
Lucia A Muscarella, Email: l.muscarella@operapadrepio.it.
Vito Guarnieri, Email: v.guarnieri@operapadrepio.it.
Roberto Sacco, Email: r.sacco@unicampus.it.
Paolo Curatolo, Email: curatolo@uniroma2.it.
Barbara Manzi, Email: bmanzi@libero.it.
Riccardo Alessandrelli, Email: riccalessandrelli@libero.it.
Grazia Giana, Email: graziagiana@hotmail.com.
Roberto Militerni, Email: roberto.militerni@unina2.it.
Carmela Bravaccio, Email: carmela.bravaccio@unina.it.
Carlo Lenti, Email: carlo.lenti@unimi.it.
Monica Saccani, Email: monica.saccani@ao-sanpaolo.it.
Cindy Schneider, Email: cschneider@center4autism.org.
Raun Melmed, Email: raun.melmed@melmedcenter.com.
Leonardo D'Agruma, Email: l.dagruma@operapadrepio.it.
Antonio M Persico, Email: a.persico@unicampus.it.
Acknowledgements
The authors gratefully acknowledge all the families who participated in this study, the resources provided by the AGRE Consortium and Roberto Rigardetto, Marina Gandione, Simona Trillo and Maria Paola Santangelo for contributing to the patient recruitment and data collection. This work was supported by the Italian Ministry for University, Scientific Research and Technology (PRIN No. 2006058195), the Italian Ministry of Health (RFPS-2007-5-640174 and RC2003), and the Autism Speaks Foundation (NJ, USA).
References
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–78. doi: 10.1017/S0033291700028099. [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, MacDonald H, Bailey A, Le Couteur A, Sim CH, Rutter M. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin Neurosci. 2009;11:35–43. doi: 10.31887/DCNS.2009.11.1/jdbuxbaum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintas C, Persico AM. Autistic phenotypes and genetic testing: state-of-the-art for the clinical geneticist. J Med Genet, J Med Genet. 2009;46:1–8. doi: 10.1136/jmg.2008.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P. A clinicopathological study of autism. Brain. 1998;121:889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005;23:183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Teitelbaum O, Benton T, Shah PK, Prince A, Kelly JL, Teitelbaum P. Eshkol-Wachman movement notation in diagnosis: the early detection of Asperger's syndrome. Proc Natl Acad Sci USA. 2004;101:11909–11914. doi: 10.1073/pnas.0403919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Stromland K, Ventura L, Johansson M, Bandim JM, Gillberg C. Autism associated with conditions characterized by developmental errors in early embryogenesis: a mini review. Int J Dev Neurosci. 2005;23:201–219. doi: 10.1016/j.ijdevneu.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Rossel M, Manley NR, Capecchi MR. Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development. 1996;122:3217–3228. doi: 10.1242/dev.122.10.3217. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. J Comp Neurol. 1996;370:247–261. doi: 10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Courchesne E. Brainstem, cerebellar and limbic neuroanatomical abnormalities in autism. Curr Opin Neurobiol. 1997;7:269–278. doi: 10.1016/S0959-4388(97)80016-5. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- Rossell M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–5040. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- Ingram JL, Stodgell CJ, Hyman SL, Figlewicz DA, Weitkamp LR, Rodier PM. Discovery of allelic variants of HOXA1 and HOXB1: genetic susceptibility to autism spectrum disorders. Teratology. 2000;62:393–405. doi: 10.1002/1096-9926(200012)62:6<393::AID-TERA6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Conciatori M, Stodgell CJ, Hyman SL, O'Bara M, Militerni R, Bravaccio C, Trillo S, Montecchi F, Schneider C, Melmed R, Elia M, Crawford L, Spence SJ, Muscarella L, Guarnieri V, D'Agruma L, Quattrone A, Zelante L, Rabinowitz D, Pascucci T, Puglisi-Allegra S, Reichelt KL, Rodier PM, Persico AM. Association between the HOXA1 A218G polymorphism and increased head circumference in patients with autism. Biol Psychiatry. 2004;55:413–419. doi: 10.1016/j.biopsych.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Muscarella LA, Guarnieri V, Sacco R, Militerni R, Bravaccio C, Trillo S, Schneider C, Melmed R, Elia M, Mascia ML, Rucci E, Piemontese MR, D'Agruma L, Persico AM. HOXA1 gene variants influence head growth rates in humans. Am J Med Genet (Neuropsychiatric Genet) 2007;144:388–390. doi: 10.1002/ajmg.b.30469. [DOI] [PubMed] [Google Scholar]
- Woodhouse W, Bailey A, Rutter M, Bolton P, Baird G, Le Couteur A. Head circumference in autism and other pervasive developmental disorders. J Child Psychol Psychiat. 1996;37:665–671. doi: 10.1111/j.1469-7610.1996.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Sacco R, Militerni R, Frolli A, Bravaccio C, Gritti A, Elia M, Curatolo P, Manzi B, Trillo S, Lenti C, Saccani M, Schneider C, Melmed R, Reichelt KL, Pascucci T, Puglisi-Allegra S, Persico AM. Clinical, morphological, and biochemical correlates of head circumference in autism. Biol Psychiatry. 2007;62:1038–1047. doi: 10.1016/j.biopsych.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Li J, Tabor HK, Nguyen L, Gleason C, Lotspeich LJ, Spiker D, Risch N, Myers RM. Lack of association between HoxA1 and HoxB1 gene variants and autism in 110 multiplex families. Am J Med Genet (Neuropsychiat Genet) 2002;114:24–30. doi: 10.1002/ajmg.1618. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Bittel DC, Miles JH, Takahashi N, Wang CH, Kibiryeva N, Butler MG. No association between HOXA1 and HOXB1 genes and autism spectrum disorders (ASD) J Med Genet. 2002;39:e70. doi: 10.1136/jmg.39.11.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano V, Calì F, Mirisola M, Gambino G, D'Anna R, Di Rosa P, Seidita G, Chiavetta V, Aiello F, Canziani F, De Leo G, Ayala GF, Elia M. Lack of association of HOXA1 and HOXB1 mutations and autism in Sicilian (Italian) patients. Mol Psychiatry. 2003;8:716–717. doi: 10.1038/sj.mp.4001285. [DOI] [PubMed] [Google Scholar]
- Gallagher L, Hawi Z, Kearney G, Fitzgerald M, Gill M. No association between allelic variants of HOXA1/HOXB1 and autism. Am J Med Genet B Neuropsychiatr Genet. 2004;124B:64–67. doi: 10.1002/ajmg.b.20094. [DOI] [PubMed] [Google Scholar]
- Sen B, Sinha S, Ahmed S, Ghosh S, Gangopadhyay PK, Usha R. Lack of association of HOXA1 and HOXB1 variants with autism in the Indian population. Psychiatr Genet. 2007;17:1. doi: 10.1097/YPG.0b013e328010de0d. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- Lord C, Rutter M, DiLavore PC, Risi S. In: ADOS, Autism Diagnostic Observation Schedule. Tancredi R, Saccani M, Persico AM, Parrini B, Igliozzi R, Faggioli R, editor. Los Angeles: Western Psychological Services; 2002. (Italian version, Firenze: Organizzazioni Speciali; 2005) [Google Scholar]
- Rutter M, Le Couter A, Lord C. In: ADI-R, Autism Diagnostic Interview - Revised. Faggioli R, Saccani M, Persico AM, Tancredi R, Parrini B, Igliozzi R, editor. Los Angeles: Western Psychological Services; 2003. (Italian version, Firenze: Organizzazioni Speciali, 2005) [Google Scholar]
- Statistical Genetics Utility programs. http://linkage.rockefeller.edu/ott/util.htm
- Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- UNPHASED software. http://portal.litbio.org/Registered/Option/unphased.html
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PBAT. http://www.biostat.harvard.edu/~clange/default.htm
- ClustalW2. http://www.ebi.ac.uk/Tools/clustalw2/index.html
- VISTA. http://genome.lbl.gov/vista/index.shtml
- Wayne WD. Applied Nonparametric Statistics. 2. Boston: PWS-KENT; 1990. pp. 82–143. [Google Scholar]
- Persico AM, Sacco R, Curatolo P, Manzi B, Lenti C, Saccani M, Militerni R, Bravaccio C, Elia M. Program No. 446.20, Neuroscience Meeting Planner. Washington, DC: Society for Neuroscience; 2008. Isolation of principal components in autistic disorder symptomatology and their association with biological endophenotypes [abstract]http://www.sfn.org [Google Scholar]
- Faiella A, Zortea M, Barbaria E, Albani F, Capra V, Cama A, Boncinelli E. A genetic polymorphism in the human HOXB1 homeobox gene implying a 9bp tandem repeat in the amino-terminal coding region. Mutations in brief no. 200. Online. Hum Mutat. 1998;12:363. [PubMed] [Google Scholar]
- Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- Hung YC, Ueda M, Terai Y, Kumagai K, Ueki K, Kanda K, Yamaguchi H, Akise D, Ueki M. Homeobox gene expression and mutation in cervical carcinoma cells. Cancer Sci. 2003;94:437–441. doi: 10.1111/j.1349-7006.2003.tb01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin Neurosci. 2009;11:35–43. doi: 10.31887/DCNS.2009.11.1/jdbuxbaum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont ML, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois MC, Prieur M, Vekemans M, Carter NP, Munnich A, Colleaux L, Philippe A. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Martin C, Lese-Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Sample sizes involved in the different studies reported here, distinguished by cases/control status and by ethnicity.