Summary
During mitosis, chromosomes must become aligned at the equator of the mitotic spindle before segregation. Recent work suggests that a kinesin-8 motor utilizes a unique combination of activities to regulate this process.
To properly segregate chromosomes during cell division, cells employ a system of proteins to capture, bi-orient and position chromosomes at the equator of the microtubule-based mitotic spindle. Chromosomes make relatively stable attachments to spindle microtubule plus-ends through specialized protein complexes called kinetochores. Throughout the process of chromosome alignment, referred to as “congression”, kinetochore microtubules elongate and shorten while maintaining attachment to the chromosomes. Detailed observations of vertebrate chromosome movements and micromanipulation studies have established that assembly (lengthening) and disassembly (shortening) of the 20–25 microtubules that bind each vertebrate kinetochore may contribute the bulk of the forces required for chromosome congression during early mitosis, as well as chromosome segregation during anaphase [1]. New evidence suggests that the human kinesin-8 motor, Kif18A, regulates kinetochore microtubule dynamics to promote chromosome congression [2].
Chromosomes attached to kinetochore microtubules in vertebrate systems typically move with a constant velocity (1–3 um/min) and make rapid directional changes from poleward to anti-poleward movement, where the direction of movement is described relative to the spindle pole that the chromosome is attached to. The direction of chromosome movement is complemented by kinetochore microtubule plus-end dynamics. The addition of αβ-tubulin dimers at kinetochore microtubule plus-ends correlates with anti-poleward movements and removal of tubulin dimers results in poleward movements. Once chromosomes reach the equator of the spindle, they continue to undergo oscillations of alternating poleward and anti-poleward movements until anaphase onset [1]. Interestingly, the sole parameter of movement that changes after chromosome alignment is the microtubule “switching” potential, the potential to switch from assembly to disassembly (catastrophe) or vice versa (rescue). The switching potential is lower prior to alignment resulting in directionally persistent chromosome movements [3]. Modeling studies in yeast have implicated a spatial gradient of either microtubule catastrophe or rescue as an essential component of chromosome positioning [4]
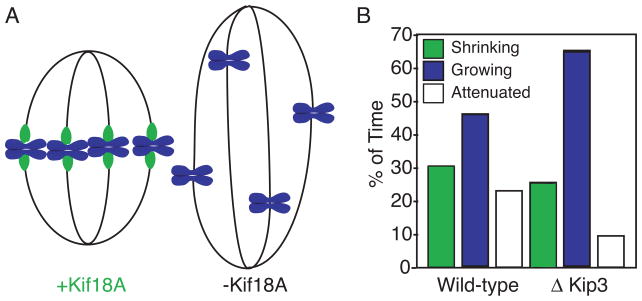
Molecules that modulate microtubule dynamics, especially with respect to switching parameters, are likely candidates to promote chromosome congression. Previous studies have implicated the kinesin-8 motors in chromosome congression, however the specific contribution that these motors make to chromosome movements remains unsolved [5–10]. A new report from Mayr and colleagues suggests that a human kinesin-8 motor, Kif18A, may use a combination of directional motility and microtubule depolymerization to regulate microtubule dynamics and control chromosome movements during mitosis [2]. The authors demonstrate that Kif18A depolymerizes stable microtubules in vitro and localizes to the plus-ends of kinetochore microtubules in mitotic cells. Depletion of Kif18A affects kinetochore movements and results in mitotic cells with abnormally long spindles that fail to congress chromosomes (Figure 1A).
Figure 1.
Effects of kinesin-8 depletion in live cells. (A) Depletion of Kif18A results in longer spindles and chromosomes that do not line up at the metaphase plate. (B) Loss of yeast Kip3 results in microtubules that spend comparatively more time growing than attenuated relative to wild-type cells. Data modified from Gupta et al., 2006 [11].
Interestingly, Kif18A, like its yeast ortholog Kip3p, translocates unidirectionally along microtubules and depolymerizes stable microtubule plus-ends in vitro [2, 11, 12]. Similar to the depolymerizing kinesin-like motors of the kinesin-13 family, the ATPase activity of kinesin-8 motors is stimulated by both microtubules and free-tubulin, suggesting that members of these two subfamilies depolymerize microtubules via a similar mechanism [2, 11, 12, 13 ]. However, unlike the well-studied kinesin-13 motor, MCAK, which does not translocate directionally and can depolymerize both ends of a microtubule, kinesin-8s translocate to and specifically act to destabilize plus-ends [11–13]. Furthermore, the rate of kinesin-8-dependent depolymerization is affected by the length of the microtubule substrate, i.e. kinesin-8 motors depolymerize longer microtubules faster than shorter ones in vitro [2, 12]. This length-dependent effect on the depolymerization rate is likely due to the accumulation of more motor at the tips of longer microtubules [12]. Such a combination of activities could allow kinesin-8 motors in the context of a cell to specifically target depolymerizing activity to the plus-ends of unusually long or stable microtubules, such as kinetochore microtubules. In this manner, microtubule length and by extension, chromosome position would be self-limiting.
The length-dependent model could explain why the loss of kinesin-8 function in various experimental systems results in failed chromosome congression, abnormally long mitotic spindles, and nuclear positioning defects as all of these processes require regulation of microtubule length [12]. Consistent with this idea, the evidence presented by Mayr et al., suggests that Kif18A exerts its effects on both spindle length and chromosome alignment by regulating the plus-end dynamics of kinetochore microtubules [2] (diagrammed in Figure 1A). Similarly, Kip3p regulates the plus-end dynamics of microtubules that interact with the cell cortex during nuclear positioning [11]. Taken together, these data suggest that kinesin-8 motors from yeast and human have similar biochemical activities that are utilized to directly regulate microtubule plus-end dynamics and microtubule length in vivo. Surprisingly, however, the specific effects of Kif18A and Kip3p on cellular microtubule dynamics are quite different.
Mayr et al. report that Kif18A is needed to achieve normal rates of chromosome movement, as chromosome velocities are slower in its absence. In addition, the loss of Kif18A function results in spindle microtubules that are resistant to depolymerization, which leads the authors to conclude that microtubule dynamics in cells lacking Kif18A are suppressed [2]. Together, these results suggest a function for Kif18A in depolymerizing kinetochore microtubules to directly drive chromosome movements. These findings indicate that Kif18A has an important function in regulating kinetochore microtubule dynamics. However, it is not entirely clear whether reduced chromosome velocity can account for the complete disruption of congression seen in Kif18A depleted cells. For example, tubulin mutations that suppress microtubule dynamics, and thus presumably reduce the rate of chromosome movement during mitosis, do not prevent proper chromosome positioning in yeast [14]. This raises a question about whether Kif18A might have additional functions during chromosome congression?
The measured effects of Kif18A on kinetochore microtubule dynamics correspond well with its ability to depolymerize stabilized microtubules in vitro, however, they differ from the effects of kip3 deletion on cytoplasmic microtubules in budding yeast and uncovering the reasons for these differences will likely lead to a better understanding of kinesin-8 function. In a recent study, Gupta and colleagues made detailed measurements of microtubule dynamic parameters in yeast cells lacking kip3 (kip3Δ cells) [11]. They found that cortical microtubules in kip3Δ cells spent less time in an attenuated state and more time growing or shrinking, indicating that microtubules are more dynamic in the absence of Kip3p (Figure 1B). In addition, the rate of microtubule depolymerization was significantly increased in the absence of Kip3p, suggesting that the motor might be needed to govern the velocity of depolymerization. When considering the results of these two studies, it appears that the yeast kinesin-8 suppresses microtubule dynamics and reduces shortening velocity while the human motor promotes dynamics and increases shortening velocity. The similar biochemical activities measured for Kif18A and Kip3p suggest that these differences are not likely due to intrinsic differences between the two motors. Are the differences an indication of motor regulation or indirect effects on other regulatory factors? Obviously, these questions warrant further investigation.
Mayr et al.’s interesting study functionally dissects a novel component of the machinery for chromosome congression [2]. The complexity of the problem illuminated by their work and recent studies of Kip3p function [11, 12, 15] suggest that unraveling the molecular mechanisms that kinesin-8 family motors utilize to regulate dynamic microtubules will remain quite a fascinating problem for a number of years to come.
References
- 1.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007 doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 3.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- 9.West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- 10.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional Analysis of Human Microtubule-based Motor Proteins, the Kinesins and Dyneins, in Mitosis/Cytokinesis Using RNA Interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 12.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 13.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 14.Pearson CG, Gardner MK, Paliulis LV, Salmon ED, Odde DJ, Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol Biol Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]