Abstract
Eukaryotic DNA replication is highly stratified, with different genomic regions shown to replicate at characteristic times during S phase. Here we observe that mutation rate, as reflected in recent evolutionary divergence and human nucleotide diversity, is markedly increased in later-replicating regions of the human genome. All classes of substitutions are affected, suggesting a generalized mechanism involving replication time-dependent DNA damage. This correlation between mutation rate and regionally stratified replication timing may have substantial evolutionary implications.
Evolutionary divergence and inferred mutation rates are known to vary across the human genome1–3, and it has long been speculated that this is a consequence of covariance with an epigenetic feature1,2. In human cells, the time of DNA replication exhibits marked regional variability during an S-phase lasting approximately 10-hours4,5. To parallel the conventional division of S-phase into four sequential temporal states (S1-S4), we used a hidden Markov model6 to perform unbiased four-state partitioning of continuous, high-resolution replication timing measurements across 1% of the human genome7. We then determined human-chimpanzee nucleotide divergence rates and the density of SNPs8 at putatively neutrally evolving sites within each temporal state, excluding any bases within annotated exons, repetitive elements, CpG islands, 2kb-regions upstream and downstream of genes, intronic splice sites, and conserved non-coding sequences9 (Supplementary Table S1).
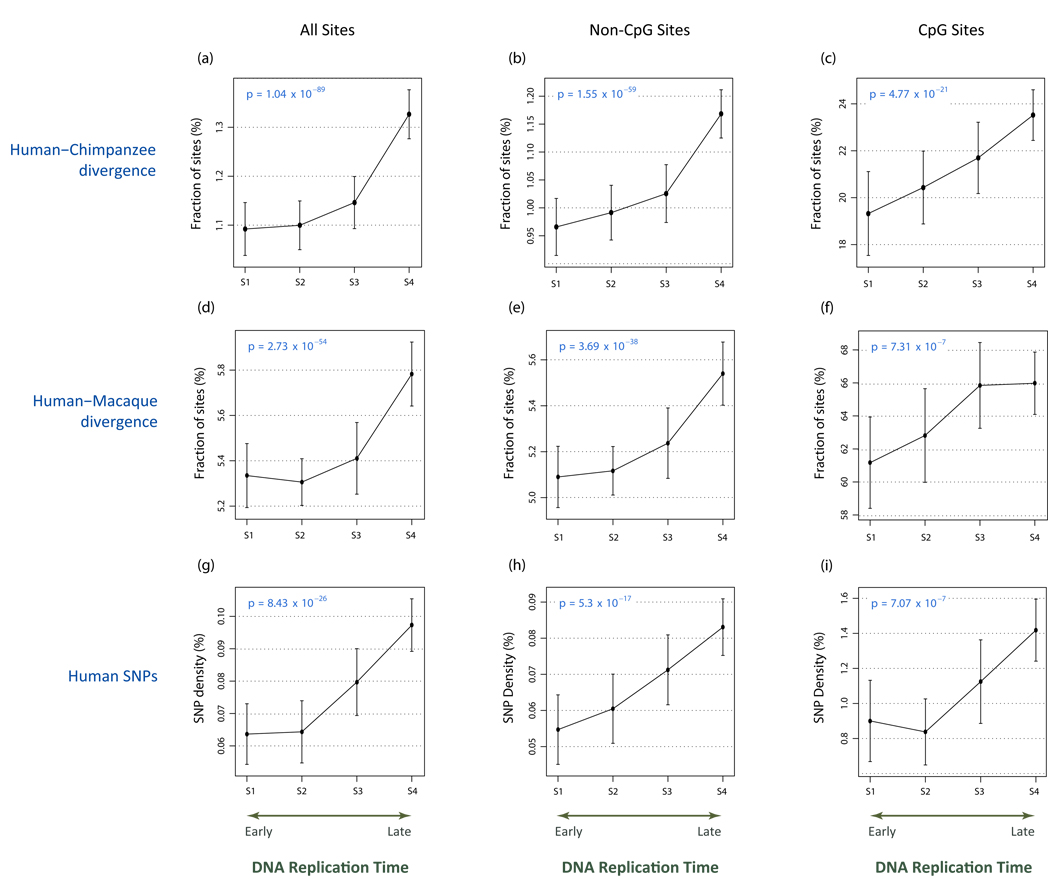
We observed a striking trend relating the rate of evolutionary divergence and the density of human SNPs to the progress of DNA replication (Fig. 1). Human-chimpanzee substitutions and human SNP density increase 22% and 53%, respectively, during the temporal course of replication, both of which are highly statistically significant (p < 8.43 × 10−26, Cochran-Armitage; Fig. 1a–c, g–i). To rule out potential confounding by the overall low genome-wide rate of human-chimpanzee divergence, we also analyzed human-macaque divergence, with similar results (p < 2.7 × 10−54; Fig. 1d–f). We confirmed the absence of bias due to a sampling or stratification effect across different genomic regions by testing (Cochran-Mantel-Haenszel) for three-way interactions, treating region assignment as controlling variable (p < 7.2 × 10−12, p < 0.00026 for human-chimpanzee divergence and human SNPs, respectively). Additionally, we repeated all analyses with an independent set of randomly ascertained SNPs10, with nearly identical effect (p < 9.69 × 10−22).
FIGURE 1. Replication time-dependence of evolutionary divergence and human SNP density.
Shown are replication time-dependence of (a–c) human-chimpanzee divergence (fraction sites changed); (d–f) human-macaque divergence; and (g–i) human polymorphism rate (SNP density), computed across 44 regions (ENCODE) comprising 1% of the human genome. Analyses are presented for all putatively neutral sites (a,d,f – left column), for neutral sites with all CpG-prone dinucleotides removed (b,e,h – middle column), and for CpG dinucleotides only (c,f,i – right column). CpG-prone dinucleotides were defined as all sites immediately preceded by C of followed by G in either species, CpG dinucleotides were defined as sites for which C was immediately followed by G at least in one of the species. Plots show mutation rates averaged over all 50kb non-overlapping windows within each of four temporal replication states (S1–S4), together with 95% confidence intervals for the mean. Significance of each trend is shown within the corresponding panel as a p-value (Cochran-Armitage trend test for proportions). For divergence analysis, human-chimpanzee hg17vsPanTro1 and human-macaque hg17vsRheMac2 axtNet alignments9 were processed to exclude all regions with more than a single substitution per sliding 5-nucleotide window and axtNet fragments which were either shorter than 500 bases or demonstrated average substitution rate above 3% (12% in case of macaque) were also discarded. For polymorphism analysis, we used Version 1 bulk SNPs data set published as part of the Personal Genome Sequence project8 (Watson).
Next we examined whether the observed correlation between mutation rate and replication time could be explained by variation in another genomic feature for which replication timing might be acting as a surrogate. Regional variation in G+C content2,3 and, independently, recombination rate2,3 have been invoked as potential causes of human mutation rate variation. We therefore obtained the distribution of G+C content, CpGs, recombination hotspots9, and gene, exon, and conserved non-coding sequence9 densities in sliding non-overlapping 50kb windows (approximating the size of chromosomal domains linked to replicons) across each temporal replication state (Supplementary Fig. S1). We binned each distribution into three classes (low, medium and high content), with an equal number of windows at each level and performed separate tests for three-way interactions using each factor as a controlling variable (total 12 tests). All were highly significant with p-values not exceeding 3.0 × 10−12 (Table 1), as were repeated tests with the additional permutation re-sampling of temporal states (p < 5.0 × 10−6 for divergence; p < 2.2 × 10−4 for SNPs; Table 1).
TABLE 1. Significance of replication time-dependence of evolutionary divergence and human.
Human-chimpanzee and human-macaque divergence and human SNP density were stratified by one of six controlling variables including G+C content; the number of CpGs; exons (RefSeq); gene units (RefSeq); conserved non-coding sequences (CNS); and recombination hotspots (see Supplemental Fig. S1). p-Values were computed by both stratification (generalized Cochran-Mantel-Haenszel) and permutation re-sampling (Monte-Carlo) approaches. The distribution of each compositional feature was produced by sliding a non-overlapping 50kb window across each of the four temporal replication states (S1–S4) and binning composition values obtained into three classes (low, medium and high content), with an equal number of pooled windows under each level. Cochran-Mantel-Haenszel tests for three-way interactions were then performed using each compositional factor as a controlling variable. Monte-Carlo tests were carried out by running 200,000 random permutations of replication timing assignments for a set of generated 50kb windows for each feature, followed by calculation of Cochran-Mantel-Haenszel statistic as described above.
| p-Value | G+C | CpG | Exons | Genes | CNS | Recombination hotspots | |
|---|---|---|---|---|---|---|---|
| Human-Chimpanzee divergence | Stratification | 2.8·10−47 | 1.1·10−81 | 3.3·10−43 | 1.5·10−49 | 7.1·10−44 | 1.2·10−43 |
| Permutation | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 | |
| Human SNP density | Stratification | 2.0·10−13 | 1.2·10−22 | 2.9·10−13 | 8.1·10−14 | 3.0·10−12 | 1.8·10−13 |
| Permutation | 1.5·10−4 | 1.0·10−5 | 1.8·10−4 | 1.1·10−4 | 2.2·10−4 | 1.7·10−4 | |
| Human-Macaque divergence | Stratification | 2.9·10−30 | 3.7·10−43 | 8.9·10−29 | 1.0·10−30 | 1.4·10−28 | 1.5·10−28 |
| Permutation | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 | < 5·10−6 |
To address potential interplay between more than one variable, we developed multiple regression models of both divergence and diversity, confirming the independent effect of replication timing (Supplementary Table S2 and Supplementary Fig. S2). These models suggest that replication time alone may explain 40–70% of the variability explained by the full model, and ∼8% of overall variability in diversity and divergence. The observed correlation between rates of nucleotide change and replication timing is therefore highly unlikely to be caused by variation in G+C content or by a mutagenic effect of recombination. To rule out any hidden dependence on window size, we repeated all analyses conditioned on smaller (30kb) and larger (100kb) windows, with equivalent results (Supplementary Fig. S3).
The effects of replication timing on evolutionary divergence and SNP density are highly similar when all other genomic features are controlled. These findings are compatible with a process that impacts mutation rate, which should affect both diversity and divergence in a stable fashion over evolutionary time. Furthermore, the findings persist across the spectrum of selected sites, from ancestral repeats and 4-fold degenerate sites to conserved non-coding sequences and non-degenerate coding sites (Supplementary Fig. S4), and across the human and chimpanzee lineages following the split from macaque (Supplementary Fig. S5).
We next considered whether the relationship with mutation rate might be due to a consequence of transcription such as transcription-coupled repair11. To rule this out, we examined introns and intergenic regions separately, and found no significant difference in any parameter (data not shown).
Finally, we examined the possibility that the mutational effect might be restricted to the subset of the genome we analyzed. To test this, we examined a lower-resolution genome-wide data set comprising early- and late-replicating regions mapped in lymphoblastoid cells5. These data also evince a mutational effect analogous with that reported above (Supplementary Fig. S6), confirming the generality of our observations.
What molecular mechanism might underlie a monotonic increase in mutation rate during S-phase? One possibility is that late stages of DNA replication are associated with the slowing or stalling of replication forks due to exhaustion of the dNTP pool or difficulty in negotiating heterochromatinized templates, with consequent accumulation of single-stranded DNA (ssDNA) regions12. ssDNA is more susceptible to endogenous and environmental damage, and can potentiate mutagenesis directly31 or via triggering of intra-S-phase checkpoints that set in motion low-fidelity polymerases. Another possibility is that the mismatch repair system might erode during S-phase, or that lesions in late replicating regions simply lack adequate time to undergo effective repair.
To differentiate these scenarios, we examined mutations at CpG dinucleotides, which arise overwhelmingly from spontaneous deamination of methylcytosine into thymine, a process which escapes DNA mismatch repair. Surprisingly, we found that both evolutionary divergence and human nucleotide diversity at CpG sites (Fig. 1c,f,i) correlate with replication timing, closely paralleling other types of sites (Fig. 1a,b,d,e,g,h). The parallelism between CpG and non-CpG sites cannot be explained by alterations in the dNTP pool, nor by reduced polymerase fidelity, nor by defective mismatch repair. In addition, we found all classes of evolutionary transitions and transversions to display strong replication timing-dependence with a characteristically similar trend (Supplementary Fig. S7). This indicates that the effect is not due to biases in the genesis of specific mutational events nor to their handling by the repair machinery.
Our results therefore suggest that a simple consequence of the process of DNA replication – accumulation of single-stranded DNA within later replicating regions – may provide the most parsimonious explanation. Because ssDNA is highly susceptible to endogenous DNA damage, including alkylation, oxidation and deamination13, accumulation of ssDNA in late-replicating regions would be expected to increase mutation rate across all classes of substitutions, consistent with our observations.
In conclusion, we find a clear and striking relationship between the time at which human genomic DNA sequences replicate and their corresponding mutation rates. Our results affirm longstanding speculation concerning the existence of such a relationship, and they explain limited prior observations of increased SNP density near later replicating genes14. In order for mutations to be propagated, they must arise in the germ line. Our results were obtained using replication timing measurements from somatic cells, suggesting that the somatic replication program largely parallels the temporal landscape of replication in germ cells, which have evaded study owing to their scarcity. Because the replication timing of tissue-specific genes is expected to vary between cell types, it is reasonable to expect that there will be discrepancies between our calculations and those that might be made from germ cells were data available. The correlation reported herein should therefore be regarded as a lower limit estimate of actual dependence of mutation rate on replication timing.
Interestingly, exons preferentially reside in early replicating regions (Supplementary Fig. S1) and, consequently, in regions with reduced mutation rate. This observation may have either a mechanistic or a selection-based explanation. We found that replication timing is the dominant factor responsible for the reduced nucleotide diversity around exons. It is further observable that a significant number of human genes controlling developmental fate, differentiation, and cell proliferation are exceptions and undergo replication late in S-phase in most adult cell types15, and that late replication timing is associated with repression of cell fate-modifying genes15. This suggests that increased mutation rate affecting late replicating regions of the human genome may reflect a significant evolutionary cost for sequestering specific gene subsets within a repressed nuclear compartment15.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alexey Kondrashov, Molly Przeworski and Josep Cameron for helpful discussions. This work was supported by NIH grants U54HG003042 and R01GM071852 to J.A.S., R01GM078598, R01MH084676, U54LM008748 to S.R.S., and R01GM60987 (NIGMS) to S.M.M..
REFERENCES
- 1.Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- 2.Hellmann I, et al. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyekucheva S, et al. Human-macaque comparisons illuminate variation in neutral substitution rates. Genome Biol. 2008;9:R76. doi: 10.1186/gb-2008-9-4-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeon Y, et al. Temporal profile of replication of human chromosomes. Proc Natl Acad Sci U S A. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodfine K, et al. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 6.Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–1426. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- 7.Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 9.Karolchik D, et al. The UCSC Genome Browser Database: 2008 Update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randomly ascertained SNPs were identified by comparing Celera individual A with the public genome build (NCBI Build 35)
- 11.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 12.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe Y, et al. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum Mol Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 15.Chuang JH, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:E29. doi: 10.1371/journal.pbio.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.