Abstract
Mutations in the Notch pathway ligand Jagged1 (JAG1) cause Alagille syndrome (AGS), as well as cardiac defects in seemingly non-syndromic, individuals. To estimate the frequency of JAG1 mutations in cases with right-sided cardiac defects not otherwise diagnosed with AGS, we screened 94 cases with tetralogy of Fallot (TOF) and 50 with pulmonic stenosis/peripheral pulmonary stenosis (PS/PPS) or pulmonary valve atresia with intact ventricular septum (PA) for mutations. Sequence changes were identified in three TOF and three PS/PPS/PA patients,that were not present in 100 controls. We identified one frameshift and two missense mutations in the TOF cases, and one frameshift and two missense mutations in cases with PS/PPS/PA. The four missense mutations were assayed for their effect on protein localization, post-translational modification and ability to activate Notch signaling. The missense mutants displayed heterogeneous behavior in these assays, some with complete haploinsufficiency, suggesting that there are additional modifiers leading to organ specific features. We identified functionally significant mutations in 3% (2/94) of TOF patients and 4% (2/50) of PS/PPS/PA patients. Patients with right-sided cardiac defects should be carefully screened for features of AGS or a family history of cardiac defects that might suggest the presence of a JAG1 mutation.
Keywords: Alagille syndrome, AGS, heart, variable expressivity, tetralogy of Fallot, pulmonary stenosis, Jagged1, JAG1, Notch signaling
Introduction
Alagille Syndrome (AGS; MIM# 118450) is an autosomal dominant disorder that involves abnormalities of varying severity in multiple organ systems, including the heart (Alagille, et al., 1987; Krantz, et al., 1999). The diagnosis of AGS traditionally has been based on the finding of interlobular bile duct paucity, associated with at least three of five major features: chronic cholestasis, cardiac disease, skeletal abnormalities, ocular abnormalities, and a characteristic facial phenotype. Additional systems that are affected in a smaller percentage of cases include the kidneys and vasculature (Alagille, et al., 1987; Crosnier, et al., 2000; Emerick, et al., 1999; Kamath, et al., 2004). Congenital heart disease is seen in over 95% of cases with AGS and commonly involves the pulmonary valve and pulmonary artery branches, with the most common finding being peripheral pulmonic stenosis. Tetralogy of Fallot (TOF) is observed in 7–13% of cases and is the most common complex cardiac malformation seen (Alagille, et al., 1987; Deprettere, et al., 1987; Emerick, et al., 1999; McElhinney, et al., 2002).
Alagille syndrome is predominantly caused by mutations in Jagged1 (JAG1; MIM# 601920), a transmembrane protein that serves as a ligand for the Notch transmembrane receptors (Li, et al., 1997; Oda, et al., 1997). Less than 1% of AGS cases harbor mutations in NOTCH2 (MIM# 600275), a Notch signaling transmembrane receptor (McDaniell, et al., 2006). Notch signaling is a highly conserved pathway found in most vertebrates (Kopan and Ilagan, 2009). The pathway has been extensively studied in a variety of organisms including Drosophila, C. elegans, Xenopus and higher vertebrates. Notch signaling is instrumental in cell fate decisions in early development, and mutations in various proteins in the pathway have been implicated in numerous disorders (Artavanis-Tsakonas, et al., 1999; Ehebauer, et al., 2006; Gridley, 2003). The finding that a disease characterized by liver, heart, skeletal, and ocular findings is caused by mutations in a ligand for the Notch transmembrane receptor clearly implicates this fundamental signaling pathway in the normal development of these organs.
A significant proportion of congenital heart defects occurs in the context of genetic syndromes, which can be chromosomal (abnormalities of number or structure), Mendelian, or multifactorial in etiology. At least 8–10% of newborns with a heart defect have a gross chromosomal anomaly, 3–5% have a heart defect as a result of a single Mendelian gene mutation, with the remainder resulting from the interaction of genetic and environmental factors (Nora and Nora, 1988). Identification of disease related genes provides critical information for diagnosis and a fundamental understanding of the development of the heart. TOF is the most common cyanotic congenital heart disease (CHD), with a prevalence of 3.3 per 10,000 live births, and represents 6.8% of all cardiac malformations (Ferencz, 1993). The etiology of TOF is thought to be multifactorial, although some pedigrees suggest single gene inheritance (Cassidy and Allen, 1991; Der Kaloustian, et al., 1985; Krantz, et al., 1998; Miller and Smith, 1979; Pankau, et al., 1990; Wulfsberg, et al., 1991). TOF may occur in isolation or in combination with extra-cardiac features, as seen in several chromosomal anomalies and defined heritable syndromes. Peripheral pulmonary stenosis (PPS) can cause obstruction at the level of the main pulmonary artery, at its bifurcation, or at the bifurcation of more distal branches. PPS may occur at a single level, but multiple sites of obstruction or diffuse hypoplasia are more common in AGS (McElhinney, et al., 2002). PPS may be associated with other congenital heart anomalies such as valvar PS, atrial septal defect (ASD), ventricular septal defect (VSD), tetralogy of Fallot or patent ductus arteriosus.
This study was initiated by the observation that some individuals with known JAG1 mutations have apparently isolated cardiac disease, in combination with the known role of JAG1 in heart development. Alagille syndrome demonstrates highly variable expressivity. Family members of individuals with AGS who seem to have isolated cardiac disease [including TOF and pulmonic stenosis (PS)] have been found to carry the same familial JAG1 mutation as the proband with complete syndromic features (Kamath et al. 2003). The role of Notch signaling in coordinating proper cardiac development has been demonstrated (de la Pompa, 2009). With respect to JAG1, previous studies have demonstrated the significant expression of JAG1 in the developing mammalian heart and studies of mice with conditional knockout of JAG1 in endothelial cells have shown that JAG1 expression is crucial for proper heart formation (High, et al., 2008; Loomes, et al., 1999). These observations led us to hypothesize that mutations in JAG1 would be found in cases with cardiac disease of the type seen in Alagille syndrome who did not otherwise carry the diagnosis of AGS. A small number of cases with non-syndromic cardiac disease and JAG1 mutations have been reported in the literature (Eldadah, et al., 2001; Krantz, et al., 1999; Le Caignec, et al., 2002). Our lab performed functional analysis on one of these previously reported missense mutations (p.G274D) and determined that the mutant protein exhibited conformational sensitivity (Lu, et al., 2003). This mutant protein was partially expressed at the cell surface and partially retained in the secretory pathway at 37°C. We considered the p.G274D protein a “leaky” allele because some mutant protein was able to traffic correctly to the cell surface. Proper progression through the secretory pathway was restored when the protein was expressed at 32°C. However, the p.G274D protein was unable to activate a Notch-dependent luciferase reporter at either temperature. More recent in vitro oxidative folding studies of the second EGF repeat of JAG1, which includes residue 274, have indeed confirmed that the p.G274D substitution leads to a population of proteins with varying folding patterns (Guarnaccia, et al., 2009). We hypothesized at the time that this partial loss of function was enough to cause cardiac defects, but not the full spectrum of AGS findings, presumably because the developing heart was more sensitive to fluctuations in JAG1-dependent Notch signaling (Lu, et al., 2003).
While a few cases with non-syndromic cardiac disease and JAG1 mutations have been reported, the frequency of JAG1 mutations in these cardiac cohorts has not been explored, and functional analyses of some of the associated mutations (p.C234Y) have not been performed (Le Caignec, et al., 2002). In this study, we present our analysis of the frequency of JAG1 mutations in a cohort with right-sided cardiac defects. We identified multiple sequence variants, both protein truncating and missense, that in conjunction with a previously reported variant, were analyzed for localization and function.
Materials and Methods
Study Cohort
Cases were enrolled under a protocol of informed consent as part of the Specialized Center of Research in Cardiology (SCOR), which was approved by the IRB at The Children’s Hospital of Philadelphia. Cases with a recognized genetic syndrome were excluded from this study. In addition, cases with TOF were screened for deletions of 22q11.2 by fluorescent in situ hybridization and those with a deletion were excluded from the study. Since mutations in JAG1 cause other variable organ manifestations in AGS, available clinical and family information was reviewed on any case found to have a JAG1 mutation. If possible, cases were re-contacted for repeat proband and family blood samples. In addition, physical examinations by a dysmorphologist and ophthalmologist, and liver function testing were conducted on available mutation-positive cases to assess for sub-clinical features of AGS.
Mutation Screening
Cases with PS/PPS were screened for mutations in JAG1 (GenBank NM_000214.1) by conformation sensitive gel electrophoresis (CSGE) (Ganguly, 2002; Ganguly, et al., 1993). TOF cases were screened by single strand conformation analysis (SSCP) as previously described (Colliton, et al., 2001; Krantz, et al., 1998; Li, et al., 1997; Oda, et al., 1997). Eighteen TOF cases that were negative for a mutation by SSCP were further screened by direct sequencing and compared to JAG1 RefSeq NM_000214.2. Our previous work has demonstrated that we can capture 75% of sequence variants via CSGE and SSCP (Warthen, et al., 2006). We sequenced a small group of our patients to confirm that we were not missing a larger group of mutations in this cohort, and since we were not, we calculated that the extra sequencing was unlikely to reveal more than an additional 1–2 variants and elected not to continue. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG initiation codon in the reference sequence. Protein numbering reflects the initiation codon as codon 1. Primers for PCR analysis were designed to cover all exons as well as the intron/exon boundaries of JAG1 (Warthen, et al., 2006). Amplicons demonstrating band shifts on SSCP or CSGE were sequenced by the Nucleic Acid/Protein Core facility of The Children’s Hospital of Philadelphia.
cDNA sequencing
Lymphoblastoid cells from cases were grown in the presence of cycloheximide (1 mg/ml) to prevent the degradation of mutant transcripts by nonsense-mediated decay (Israel, et al., 1985). RNA was purified and total cDNA synthesized using Superscript II reverse transcriptase and random hexamers as per the manufacturers protocol (GibcoBRL). Primers were designed to amplify the region of the JAG1 cDNA predicted to be affected by the intronic sequence alterations. The amplicons included one exon flanking the potentially mutant exon in either direction to allow for identification of skipped exons. The following are the sequences of the forward and reverse primers used, preceded by the exon amplified. Exon 9: F – GTATTCAGGACCCAACT, R - TCTGACACTGGCCAAGGC3. Exon 11: F – GTGGACTGGGAAAACGTG, R – GGCAGGGATTAGGCTCAC. Exon 14: F – TCTCTGGAAACCTCTGT, R – GACACGTGCCCCCATTG. RT-PCR products were analyzed by gel electrophoresis to identify the presence of altered products and all RT-PCR products were sequenced in the event that the altered product was not visible by agarose gel analysis.
DNA constructs
The p.C234Y, p.P810L, p.C664S and p.R937Q mutant constructs were made by site-directed mutagenesis (Stratagene, QuikChange kit) using wild-type JAG1 as a template as described (Morrissette, et al., 2001). Two complementary oligonucleotides were synthesized containing the desired mutation, flanked by unmodified sequence for each mutation for the mutagenesis reaction. Several clones of each mutant were sequenced to assay for the presence of the mutation.
Cell Line Creation
Stably transfected cell lines containing wild-type JAG1 and JAG1 harboring missense mutations (p.C234Y, p.P810L, p.C664S or p.R937Q) were created by infection of NIH-3T3 cells using the retroviral expression vector pBABE-puro, into which JAG1 or JAG1-mutant was cloned as described (Morrissette, et al., 2001). All cell lines were maintained in Dulbecco’s modified Eagle’s media with 10% FBS and 2 mM glutamine in the presence of 1.5 µg/ml puromycin.
Trypsin analysis of cell surface JAG1
To determine if the mutant molecules were expressed on the cell surface, we treated JAG1 expressing cells with the protease trypsin, such that only JAG1 present on the cell surface would be degraded. NIH-3T3 cells expressing JAG1 or JAG1-mutants were exposed to trypsin at 37°C for 10 minutes. The trypsin was inactivated, the cells lysed in NP-40 lysis buffer and the whole cell lysates were analyzed by western blot.
EndoH analysis of mutant JAG1
To elucidate whether or not JAG1 variant proteins were being properly post-translationally modified, proteins were treated with the glycosidase EndoH to assay the maturation state of N-linked glycans. Lysates were made from stably transfected NIH3T3 cells as described above. Fifty micrograms of protein from whole cell extracts were treated with 3 µl of EndoH (New England Bio) at 37°C for 1 hour. Treated proteins were then visualized by western blot and compared to wild-type.
Notch Activation Assays
All cell lines were maintained in DMEM supplemented with 10% FBS, glutamine, and Pen/Strep. NIH3T3 cells grown in 24-well plates were transfected with 199 ng of the previously described 4xCBF-Luc reporter construct (Hsieh, et al., 1996) and 1 ng of an internal control SV40-Renilla construct (Promega). Twenty-four hours later, each well was washed twice with PBS and 5×104 stably transfected NIH3T3 cells expressing wild-type or mutant JAG1 were seeded onto the transfected cells. Luciferase activity was assayed 24-hours later (Dual-Luciferase System, Promega). Firefly luciferase readings were normalized to Renilla luciferase activity levels, and values were reported as fold change over vector (pBabe) alone. All luciferase assays were carried out in triplicate, and the experiments were repeated a minimum of three times.
Western Blot Analysis
NIH-3T3 cells expressing wild type JAG1, or the JAG1 mutants (p.C234Y, p.P810L, p.C664S or p.R937Q) were grown at 37°C and harvested as previously described (Morrissette, et al., 2001). To detect JAG1 protein, a polyclonal antibody to the C-terminal region was used (H-114, Santa Cruz Inc.); with an HRP-goat anti-rabbit secondary antibody (Amersham, Inc.) used for detection, and visualized using an ECL kit (Amersham, Inc.).
Immunofluorescence Studies
NIH 3T3 cells that had been stably transfected with JAG1 clones were plated on culture slides to 30% confluency. Following standard washing, fixation and blocking, they were treated with a JAG1 antibody (1/40) H114 (Santa Cruz) for 1 hour at 37°C. The direct-label secondary antibody (Alexa Fluor 488 or Alexa Fluor 594) was applied for 30 minutes at 37°C and cells were visualized using Hoechst. Confocal spectroscopy was performed in the pathology core of The Children’s Hospital of Philadelphia.
Results
Study Cohort
A total of 144 cases with right-sided cardiac defects were tested for JAG1 mutations, of which 70 (49%) were male and 101 (70%) were Caucasian. Ninety-four cases had TOF, and 50 cases had the primary diagnosis of valvar PS, PPS, PA/IVS or other pulmonary artery anomalies (Table 1). Although none of the cases had an identified genetic syndrome, 47 (32%) were noted to have highly variable non-cardiac anomalies at the time of consent that included: facial dysmorphia, developmental delay, as well as airway, skeletal, renal, genitourinary, or CNS anomalies.
Table 1.
Study Cohort
| Cardiac Defect | # Tested | JAG1 Variant |
|---|---|---|
| TOF/pulmonary valve stenosis | 73 | |
| TOF/pulmonary valve atresia | 18 | 3 |
| TOF/absent pulmonary valve | 3 | |
| PA/IVS | 16 | |
| Valvar PS | 21 | |
| PPS/branch PS/other PAA* | 13 | 3 |
| Total | 144 | 6 |
TOF, tetralogy of Fallot; PA, pulmonary valve atresia; IVS, intact ventricular septum; PS pulmonary stenosis; PPS, peripheral pulmonary stenosis.
Includes: branch PS (n=5), PPS (n=3), supravalvar PS (n=2), pulmonary artery anomaly (n=1), pulmonary stenosis, type unknown (n=2)
JAG1 mutation screening results
JAG1 sequence variants were identified in 3 of 94 (3%) cases with TOF and in 3 of 50 (6%) cases with PS/PPS. All of the variants, except one (p.H1104Q) were identified by CSGE or SSCP screening and confirmed by exonic sequencing. The missense mutation p.H1104Q was not seen by SSCP and was subsequently identified by direct sequencing. The variants in the TOF cases included one frameshift (c.414_415dupGT) and two missense changes (p.P810L, p.H1104Q). The three variants identified in cases with PS include one frameshift (c.1485_1486delCT) and two missense changes (p.C664S and p.R937Q) (Table 2). These mutations were not found in 100 ethnically matched controls (200 chromosomes), and were not reported in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). The JAG1 variants c.414_415dupGT, p.C664S, p.P810L, and p.H1104Q have not been previously reported in cases with AGS. The c.1485_1486delCT mutation has been observed in multiple cases with AGS (Spinner, et al., 2001). The missense variant p.R937Q has also been previously reported in a case with AGS (Ropke, et al., 2003). A number of common polymorphisms were also identified, including three intronic DNA changes in five cases (c.1349-11T>G, c.1721-21G>A, c.1121-25A>G). We hypothesized that these mutations could alter splicing and result in production of an abnormal RNA and protein product. However, analysis of cDNA from these cases revealed only normal mRNA, suggesting that these are benign polymorphisms (data not shown).
Table 2.
Missense variants in Jagged1 identified in patients with cardiac disease
| Patient # | Clinical Features |
Mutationa (exon) |
Inheritance | Protein Region |
|---|---|---|---|---|
| 1 | TOF with PA and branch PS |
c.414_415dupGT (3) |
NA | N-terminal Region |
| 2 | TOF with PS, PE |
c.2429C>T p.P810L (20) |
Maternal | 15th EGF-like repeat |
| 3 | PS | c.1485_1486delCT (8) |
NA (? Maternal based on clinical features) |
8th EGF-like repeat |
| 4 | PS | c.1991G>C p.C664S (15) |
Paternal | 11th EGF-like repeat |
| 5 | PS | c.2810G>A p.R937Q (23) |
NA | Cysteine Rich Region |
| 6 | TOF | c.3312C>G p.H1104Q (26) |
NA | Intracellular |
TOF – Tetrology of Fallot. PA – Pulmonary Atresia. PS – Pulmonic Stenosis. PE – Posterior Embryotoxon. NA – sample not available for analysis
The nomenclature is following journal guidelines (www.hgvs.org/mutnomen), JAG1 RefSeq accession number NM_000214.2. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG initiation codon in the reference sequence.
Clinical features of the six cardiac cases with JAG1 sequence variants are presented in Table 2. With the exception of one case with TOF who was found to have segmental posterior embryotoxon, no other case had identifiable extracardiac manifestations of AGS. Two of the variants were found to be familial, and one patient has a family history of several individuals affected with TOF or PPS. The missense change p.P810L was identified in the reportedly unaffected mother of a case with TOF, although no clinical evaluation of the mother was performed. The missense mutation p.C664S was identified in the apparently unaffected father of the proband, although the unaffected father refused additional testing. The protein truncating mutation c.1485_1486delCT was identified in a proband who had 3 of 4 siblings who also had cardiac disease (severe PPS in one sibling and TOF in two), and whose mother had PPS and mildly elevated liver function tests. One sibling with TOF also had butterfly vertebrae and normal liver function, and another sibling with TOF had a club foot deformity and normal liver function. We were unable to obtain samples to confirm the mutation in the other members of this family.
Analysis of missense mutations
We studied the effect of three of the missense changes we identified (p.C664S, p.P810L and p.R937Q), as well as the previously reported p.C234Y mutation (Le Caignec et al. 2002) on JAG1 localization and function. JAG1 is a cell surface protein that interacts with the human Notch receptors. This interaction results in cleavage and subsequent translocation of the Notch receptor intracellular domain (NICD) into the nucleus, where it interacts with a variety of transcription factors and repressors to affect the transcriptional repertoire of the cell (Kopan and Ilagan, 2009). We used several assays to assess the ability of mutant proteins to: 1) appear on the cell surface, 2) undergo appropriate post-translational modification, essential for proper trafficking, and 3) activate Notch signaling (Lu, et al., 2003; Morrissette, et al., 2001). For these experiments, the missense variants were created by site directed mutagenesis and stably over-expressed in NIH 3T3 cells, as described in Materials and Methods. We included the missense mutants p.L37S, a known null allele, and p.G274D, a previously reported “leaky allele,” as controls in our functional analysis (Lu, et al., 2003; Morrissette, et al., 2001). Results of the functional assays are summarized in Table 3.
TABLE 3.
Summary of functional studies
| Variant | EndoH | Trypsin | IF | Luciferase | Summary |
|---|---|---|---|---|---|
| WT JAG1 | Insenstitive | Degraded | Cell Membrane |
WT | WT |
| p.C234Y | Sensitive | Insensitive | Perinuclear | Null | Null |
| p.C664S | Leaky | Partially Degraded |
Perinuclear and Cell Membrane |
WT |
Partial Function |
| p.P810L | Sensitive | Insensitive | Perinuclear | Null | Null |
| p.R937Q | Insenstitive | Degraded | Cell Membrane |
WT | WT |
Glycosylation
We first analyzed the sensitivity of four variant JAG1 proteins to endoglycosidase H (EndoH). Previous work has demonstrated that wild-type JAG1 is insensitive to EndoH because it is modified with complex sugars. However, some missense mutants such as p.L37S, a known null allele of JAG1, are sensitive (Morrissette, et al., 2001). EndoH sensitivity suggests abnormal glycosylation, which is associated with improper trafficking and failure of the protein to localize to the cell surface (Morrissette, et al., 2001).
JAG1 variants p.C234Y and p.P810L are sensitive to EndoH, suggesting they are improperly modified as previously reported for JAG1 mutations associated with AGS (Morrissette, et al., 2001). p.R937Q was not sensitive to EndoH treatment, suggesting it is complexly modified in the secretory pathway, similar to wild-type JAG1. (Figure 1A). The analysis of p.C664S was more complex. The protein demonstrates some sensitivity to EndoH in contrast to wild-type JAG1, but it appears that there are two species of protein, one sensitive to the EndoH, and one that is insensitive (Figure 1A). This is the same “leaky” phenotype seen before in studies of the JAG1 missense mutation p.G274D, which was identified in a large family segregating with apparently isolated cardiac disease (Eldadah, et al., 2001; Lu, et al., 2003).
Figure 1.
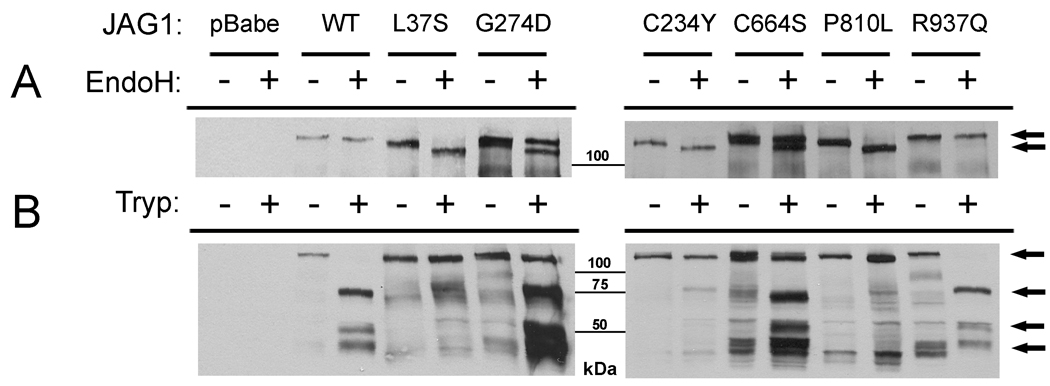
A: EndoH sensitivity of JAG1 missense mutations. The post-translational modification of the mutant JAG1 proteins was analyzed via treatment with the glycosidase EndoH and subsequent visualization by western blot with a JAG1 antibody (H114, Santa Cruz). Controls include wildtype JAG1 (not sensitive to EndoH), p.L37S (sensitive), and p.G274D (partially sensitive). p.C234Y and p.P810L appear to be EndoH-sensitive, whereas p.R937Q is insensitive, and p.C664S is partially sensitive. Arrows indicate the untreated or EndoH insensitive JAG1 proteins (top arrow) and the EndoH sensitive proteins (bottom arrows). B: Trypsin sensitivity as a measure of cell surface expression. Cells expressing variant proteins were treated with trypsin for 10 minutes, and JAG1 was visualized by western blot. Proteins present on the cell surface are degraded, with disappearance of the full length product and appearance of lower bands (wildtype, p.R37Q), whereas p.L37S, p.C234Y and p.P810L are insensitive. p.G274D and p.C664S are partially sensitive, consistent with two species of protein. Arrows indicate the full length, undigested JAG1 protein (top arrow) as well as the lower molecular weight degradation products (bottom 3 arrows).
Localization
Notch signaling initiates at the cell surface, and thus proper localization of ligands and receptors to the cell membrane is necessary for proper function. To determine if the mutant molecules were properly localized to the cell surface, we treated JAG1 expressing cells with the protease trypsin, such that only JAG1 present on the cell surface would be degraded. Trypsin was inactivated after 10-minutes and whole cell protein extracts were prepared and analyzed by western blot (Figure 1B). The western blots displayed multiple non-specific bands in some lanes independent of trypsin treatment or mutation type. As these bands did not interfere with the bands representative of JAG1 and its degradation products, they were not an impediment to analysis of our results. When cells expressing wild type JAG1 were treated, the amount of full length JAG1 present on subsequent western blots decreased due to proteolysis, coupled with the appearance of lower molecular weight products, as expected. However, cells expressing the p.L37S protein do not demonstrate any changes in band intensity, presumably because the mutant protein is not present on the cell surface, but rather retained intracellularly. The p.C234Y and p.P810L JAG1 proteins did not demonstrate any changes in size with increasing exposure, suggesting they are not accessible to trypsin degradation and therefore not present on the cell surface. The p.R937Q variant exhibited complete degradation by trypsin, similar to wild type. The p.C664S variant appears to undergo some trypsin degradation, consistent with it being localized on the cell surface, although there is less degradation than seen for wildtype.
To visualize the sub-cellular localization of the wild-type and mutant JAG1 proteins, immunofluorescence experiments were performed using an antibody directed to the intracellular domain of JAG1 (H-114, Santa Cruz, Inc.) (Figure 2). Wild-type JAG1 shows a characteristic staining at the cell membranes. There is also some light diffuse staining throughout the cell, which is representative of the normal pattern of JAG1 trafficking through the secretory pathway. The p.R937Q variant displayed cell surface staining comparable to wild-type JAG1. The p.C234Y variant displayed no cell surface staining, consistent with its being retained intracellularly, similar to previously reported AGS missense mutants (Morrissette, et al., 2001). The p.P810L variant showed very faint cell surface staining, with the majority of the protein retained intracellularly. The p.C664S variant appeared both on the cell surface and trapped intracellularly, with perinuclear staining not seen in wild-type JAG1 cells (Figure 2). These data supports the idea of two species of p.C664S protein being present. We conclude that the p.C234Y and p.P810L missense variants are predominantly localized intracellularly, while the p.C664S variant is partially expressed at the cell surface and partially retained in the cytoplasm. The p.R937Q variant continues to appear to function like wild-type protein.
Figure 2.
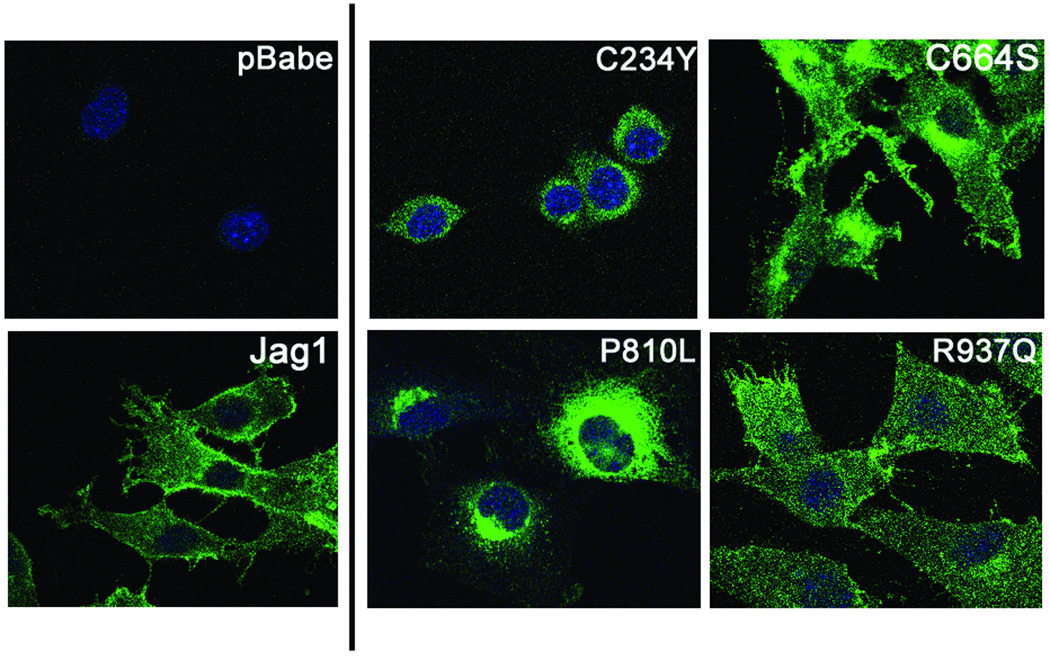
Sub-cellular localization of JAG1 wild-type and missense mutants in NIH3T3 cells by immunofluorescence. Wild-type JAG1 and p.R937Q show localization to the cell surface, while p.C234Y and p.P810L are localized to the perinuclear region. p.C664S shows both intense perinuclear staining as well as cell surface staining, corresponding to the two species of protein observed following EndoH treatment.
Signaling
To determine the ability of these variant proteins to successfully activate Notch signaling, we utilized a dual-luciferase reporter assay to measure signaling ability. Notch signaling is activated by ligand-receptor interaction at the cell surface, which is followed by multiple receptor cleavage events that lead to the release and translocation of the NICD to the nucleus, where it interacts with the CBF transcription factor to activate transcription of Notch downstream targets. We assayed the variant proteins for Notch signaling ability via a CBF-dependent luciferase assay. Results of these assays are shown in Figure 3. The p.R937Q variant protein was able to activate Notch signaling to wild-type levels. Conversely, p.C234Y and p.P810L were unable to activate Notch signaling, similar to the AGS missense mutation p.L37S. The p.C664S variant was surprisingly able to activate Notch signaling to wild-type levels, despite its “leaky” phenotype. The previously described “leaky” JAG1 allele p.G274D, found in cases with cardiac disease, was unable to initiate Notch signaling at 37° as previously reported (Lu, et al., 2003).
Figure 3.
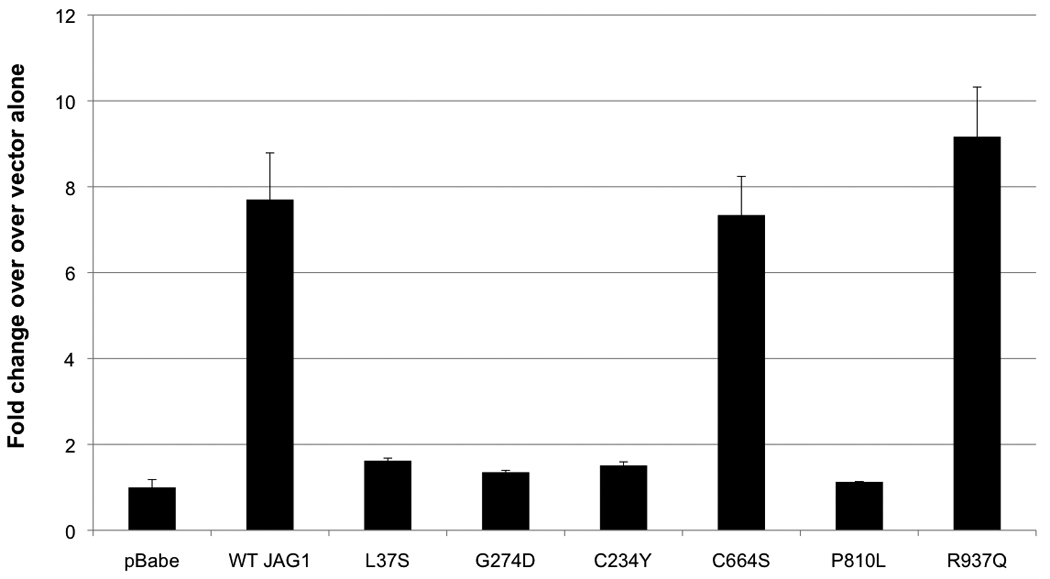
Notch signaling analysis. We used a firefly luciferase reporter construct with 4 Notch sensitive CBF binding sites upstream of the luciferase gene. RLU readings were normalized to an internal Renilla luciferase control. p.L37S and p.G274D were included as null and leaky controls, respectively. p.C664S and p.R937Q were able to activate Notch signaling to wild type levels. p.C234Y and p.P810L were unable to activate Notch signaling. Values reported are fold-change over the vector alone (pBabe) readings. Error bars represent standard error for technical triplicates.
Discussion
We screened a cohort with right-sided cardiac defects for mutations in JAG1, and identified functionally significant sequence variants in 3% (2/94) of TOF cases, and 4% (2/50) of PS/PPS/PA cases. These cases did not meet criteria for a diagnosis of AGS, nor did they have overt findings of any other known syndrome. These findings suggest that JAG1 mutations are present in a small percentage of seemingly non-syndromic cases of TOF or PS/PPS and add to the growing list of disease genes for specific types of cardiac defects.
The low incidence of JAG1 mutations the cohorts we studied, may be consistent with the high degree of heterogeneity for these phenotypes. A study of 114 patients with apparently isolated sporadic TOF using an array designed to detect deletions or duplications revealed that 1/114 individuals had a JAG1 deletion (Greenway, et al., 2009). On analysis of 230 patients with both syndromic and non syndromic TOF, genetic abnormalities were identified in 18% (42/230) (Rauch, et al., 2009). The most common genetic alteration was the 22q11.2 deletion associated with DiGeorge/VCFS syndrome, seen in 7.4%. Single gene mutations in JAG1, NKX2.5 (MIM# 600584), and TBX1 (MIM# 602054) were found in 1.3%, 0.9%, and 0.4% of the cohort, respectively, however, the JAG1 mutations were all in patients with Alagille Syndrome (Rauch, et al., 2009). Clearly, TOF is characterized by heterogeneity, with small contributions from various loci. Nonetheless, these studies further suggest that careful scrutiny for a family history of right-sided heart defects and careful examination for sub-clinical features of AGS is warranted in patients with right-sided heart defects, particularly those with isolated PPS. The clinical and reproductive implications of finding a JAG1 mutation carrier are significant, as identifying the patient with a JAG1 mutation may help identify the unsuspecting mutation-bearing parent with subclinical features of AGS and allow for more informed genetic counseling.
In this study six different JAG1 sequence variants were identified: two protein truncating and four missense changes (p.C664S, p.P810L, p.R937Q, p.H1104Q). We also investigated the functional significance of a previously reported missense variant, p.C234Y (Le Caignec, et al., 2002). The protein truncating mutations are predicted to lead to the creation of premature termination codon, and the transcripts of these alleles should undergo nonsense-mediated decay leading to 50% normal JAG1 expression. These mutations are very similar to nonsense mutations seen in AGS, which is caused by haploinsufficiency of JAG1. We therefore predict that these mutations would lead to JAG1 haploinsufficiency.
Three of the missense variants occurred in the epidermal growth factor-like (EGF) repeats of the JAG1 protein. These repeats are known to be important in ligand-receptor interaction (Kopan and Ilagan, 2009). Residue 234 (p.C234Y) is located in the first EGF repeat, while 664 (p.C664S) is in the eleventh, and 810 (p.P810L) is in the fifteenth (Figure 4A). Cysteine residues are required for proper folding of EGF repeats due to their role in the formation of disulfide bridges. A partial crystal structure of JAGGED1 was recently produced which encompassed the DSL domain and first 3 EGF repeats (Cordle, et al., 2008). This region of the protein was shown to be crucial for receptor-ligand interactions. Residue 234 lies in the first EGF repeat within this critical region, suggesting that loss of this cysteine residue would have deleterious effects on Notch signaling. Unfortunately, no available crystal structures of JAGGED1 include the other residues found altered in this study. Both affected cysteine residues show conservation not only through all vertebrates, but also in both the Delta and Serrate ligands in Drosophila (Figure 4B). The proline at position 810 is conserved in vertebrates, but not in the Drosophila ligands. The p.R937 residue lies within a region of the JAG1 protein whose significance is unknown, and this residue showed far less conservation (Figure 4A,B). While conserved in mammals such as chimp, mouse, and dog, it was not present in lower vertebrates like zebrafish. The p.H1104 residue resides in the intracellular portion of JAG1, and exhibits the least conservation of the five missense variants (Figure 4A,B). No functionally significant variants in the intracellular domain of JAG1 have been reported in AGS patients to date. Residue p.H1104 also does not lie in any of the intracellular regions known to be required for proper function of JAG1 (Glittenberg, et al., 2006) A missense variant in the intracellular region of JAG1 was reported in a patient with Biliary Atresia, but the functional significance of this variant was never investigated (Kohsaka, et al., 2002). The low conservation of this residue combined with the lack of precedent for functionally significant intracellular missense variants suggests that p.H1104Q is likely a benign rare variant.
Figure 4.
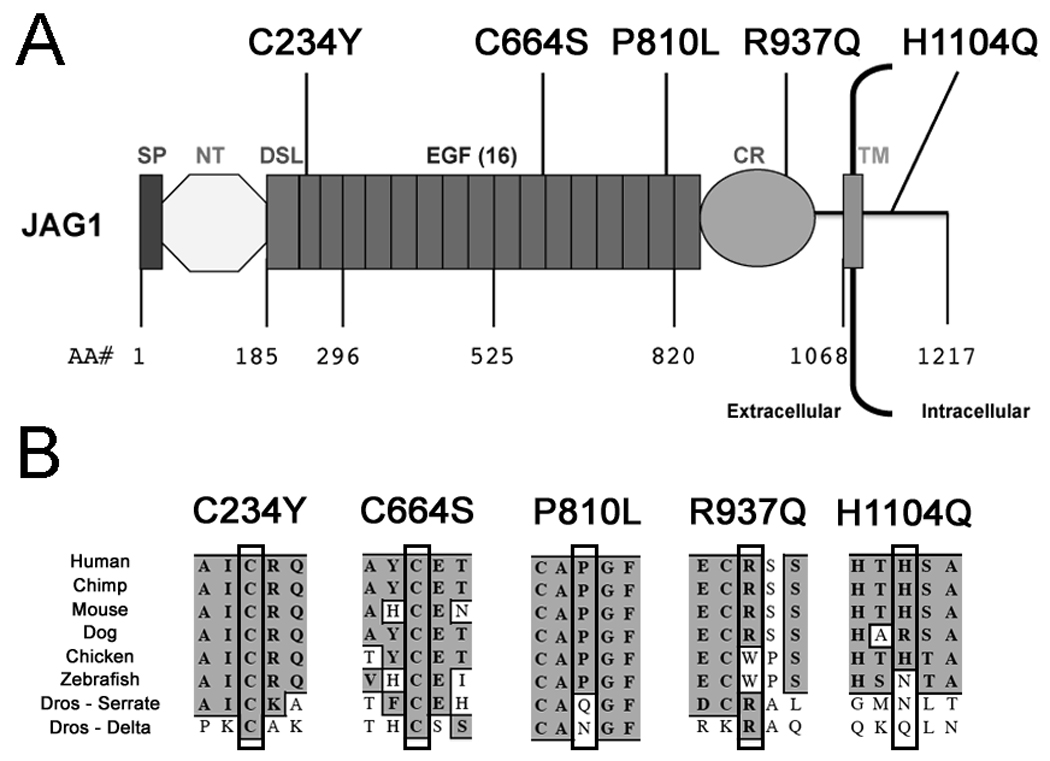
A: The locations of the five sequence variants indentified in the JAG1 protein are shown, as is the domain structure of JAG1. Domains are labeled as such; SP, Signal Peptide; NT, N-Terminal Region; DSL, Delta-Serrate-Lag2 conserved region; EGF, Epidermal Growth Factor-like Repeat; CR, Cysteine Rich region; TM, Transmembrane region. B: We demonstrate the conservation of the 5 altered residues of JAG1 investigated in this study amongst a variety of species. In Drosophila, we compared human JAG1 to both the Serrate and Delta ligands.
We performed functional analysis on four of these missense variants to determine the extent to which these residue substitutions affect normal cell-surface localization, post-translational modification, and signaling ability. As mentioned, the p.R937Q variant protein appears to function like wild-type, in that it is expressed at the cell surface, properly post-translationally modified, and is able to activate Notch signaling. Furthermore, the residue is not as highly conserved as the other residues in question, suggesting it is of less importance for correct ligand function. This variant has been previously reported in the literature as an AGS mutation, although parental samples were not screened (Ropke, et al., 2003). The results of our functional analysis of p.R937Q raise some questions as to its pathogenicity. Although the p.R937Q variant clearly has no affect on the ligand’s function in canonical Notch signaling, this does not rule out the potential for a secondary effect of the substitution on a non-canonical JAG1 function that we did not test.
The p.C234Y and p.P810L mutations were not present at the cell surface, were not properly post-translationally modified, and could not initiate Notch signaling. We would therefore predict that these mutations lead to JAG1 haploinsufficiency, with only the wild-type allele in carriers of this dominant mutation appearing on the cell surface. The p.P810L and p.C234Y missense mutations and the two nonsense mutations are therefore all similar to those that have been studied in cases with Alagille syndrome. It is worth noting that the p.P810L missense mutation is in fact a maternally inherited mutation, although the mother did not have any cardiac defects and she was not available for evaluation of other sub-clinical features of AGS. The p.C234Y variant is a familial mutation segregating with cardiac defects, deafness, and posterior embryotoxon, yet all carriers have normal liver function (Le Caignec, et al., 2002). Familial JAG1 mutations commonly exhibit variable expressivity in AGS, with very mildly affected parents harboring a disease causing mutation (Kamath, et al., 2003). It is not completely surprising to us to see the same scenario in the case of these two CHD-related mutations, although the lack of any liver dysfunction in the nine family members studied by Le Caignec et al. is unusual.
In contrast, the missense mutation p.C664S appears to have a “leaky” phenotype, with some of the protein appearing on the cell surface and some being improperly trafficked. This mutation is similar in that regard to the previously reported p.G274D mutation seen in a large family segregating isolated cardiac disease in the absence of other features of Alagille syndrome (Eldadah, et al., 2001; Lu, et al., 2003). We have previously shown via reporter assays that p.G274D cannot initiate Notch signaling (Lu, et al., 2003). However, p.C664S appears to be able to initiate Notch signaling, suggesting that the mutation may only be affecting ligand trafficking, not receptor binding and activation. This finding implies that p.C664S is not a null allele, and that complete haploinsufficiency is not required for manifestation of a cardiac phenotype. Since we have only examined two “leaky” mutations, further studies will be required to understand the extent and impact of this “leaky” signaling.
TOF and PS/PPS are two relatively common structural cardiac defects that are heterogeneous in etiology. Both of these disorders can be seen as isolated findings, or in the context of multiple different congenital anomalies or genetic syndromes. For example, TOF is commonly seen in patients with 22q11.2 deletions and/or AGS, and PS is commonly seen in patients with Noonan syndrome (MIM# 163950), LEOPARD syndrome (MIM# 151100), and AGS, while PPS is rather uniquely found in patients with AGS and Williams syndrome (MIM# 194050). Our functional analysis of JAG1 missense mutations clearly confirms that the activity of most mutation-bearing proteins differs from wild-type JAG1 in signaling ability, subcellular localization, and post-translational modification suggesting that these mutations are indeed responsible for the cardiac manifestations seen in these cases. What remains unknown is why these cases present with only a cardiac phenotype and not the full clinical features of AGS, particularly hepatic disease. The first mutation we examined, p.G274D, was identified in a family with apparently isolated cardiac disease (Eldadah, et al., 2001; Lu, et al., 2003). The p.G274D mutant clearly displayed the “leaky” phenotype, similar to p.C664S. We hypothesized at the time that the peculiar nature of these mutations could result in hypomorphic activity and not true haploinsufficiency, and that this increased level of functionality was sufficient to prevent the typical liver manifestations seen in AGS, but the developing heart was too sensitive to slightly decreased Notch signaling and was still affected. However, the p.C234Y and p.P810L mutations appear to be completely haploinsufficient, yet the patients with these mutations do not have the full spectrum of AGS, consistent with the presence of additional modifying factors. In an era of increasing whole genome analysis, future studies may elucidate what these modifying factors are and allow for better predictive diagnosis in humans with JAG1 mutations.
Acknowledgments
We thank the participating families for their cooperation and the members of the Cardiac Center for facilitating ascertainment. This research was supported by grants from the NIH/NHLBI (P50 HL62177-05 to EG and NBS, P50 HL74731 to EG) and by NIDDK grant DK53104 (NBS). This project was also supported by Grant Number M01-RR-000240 and UL1-RR-024134 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
References
- Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr. 1987;110(2):195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Cassidy SC, Allen HD. Tetralogy of Fallot in triplet siblings. Am J Cardiol. 1991;67(16):1442–1444. doi: 10.1016/0002-9149(91)90481-y. [DOI] [PubMed] [Google Scholar]
- Colliton RP, Bason L, Lu FM, Piccoli DA, Krantz ID, Spinner NB. Mutation analysis of Jagged1 (JAG1) in Alagille syndrome patients. Hum Mutat. 2001;17(2):151–152. doi: 10.1002/1098-1004(200102)17:2<151::AID-HUMU8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, others A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15(8):849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Lykavieris P, Meunier-Rotival M, Hadchouel M. Alagille syndrome. The widening spectrum of arteriohepatic dysplasia. Clin Liver Dis. 2000;4(4):765–778. doi: 10.1016/s1089-3261(05)70140-9. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL. Notch signaling in cardiac development and disease. Pediatr Cardiol. 2009;30(5):643–650. doi: 10.1007/s00246-008-9368-z. [DOI] [PubMed] [Google Scholar]
- Deprettere A, Portmann B, Mowat AP. Syndromic paucity of the intrahepatic bile ducts: diagnostic difficulty; severe morbidity throughout early childhood. J Pediatr Gastroenterol Nutr. 1987;6(6):865–871. doi: 10.1097/00005176-198711000-00008. [DOI] [PubMed] [Google Scholar]
- Der Kaloustian VM, Ratl H, Malouf J, Hatem J, Slim M, Tomeh A, Khouri J, Kutayli F. Tetralogy of Fallot with pulmonary atresia in siblings. Am J Med Genet. 1985;21(1):119–122. doi: 10.1002/ajmg.1320210117. [DOI] [PubMed] [Google Scholar]
- Ehebauer M, Hayward P, Arias AM. Notch, a universal arbiter of cell fate decisions. Science. 2006;314(5804):1414–1415. doi: 10.1126/science.1134042. [DOI] [PubMed] [Google Scholar]
- Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet. 2001;10(2):163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology. 1999;29(3):822–829. doi: 10.1002/hep.510290331. [DOI] [PubMed] [Google Scholar]
- Ferencz C. Epidemiology of congenital heart disease : the Baltimore-Washington infant study 1981–1989. Mount Kisco, N.Y.: Futura Publishing; 1993. [Google Scholar]
- Ganguly A. An update on conformation sensitive gel electrophoresis. Hum Mutat. 2002;19(4):334–342. doi: 10.1002/humu.10059. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci U S A. 1993;90(21):10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 2006;25(20):4697–4706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, others De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41(8):931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12:R9–R13. doi: 10.1093/hmg/ddg052. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Guarnaccia C, Dhir S, Pintar A, Pongor S. The tetralogy of Fallot-associated G274D mutation impairs folding of the second epidermal growth factor repeat in Jagged-1. FEBS J. 2009;276(21):6247–6257. doi: 10.1111/j.1742-4658.2009.07333.x. [DOI] [PubMed] [Google Scholar]
- High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2008;105(6):1955–1959. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16(3):952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DI, Estolano MG, Galeazzi DR, Whitlock JP., Jr Superinduction of cytochrome P1-450 gene transcription by inhibition of protein synthesis in wild type and variant mouse hepatoma cells. J Biol Chem. 1985;260(9):5648–5653. [PubMed] [Google Scholar]
- Kamath BM, Bason L, Piccoli DA, Krantz ID, Spinner NB. Consequences of JAG1 mutations. J Med Genet. 2003;40(12):891–895. doi: 10.1136/jmg.40.12.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Spinner NB, Emerick KM, Chudley AE, Booth C, Piccoli DA, Krantz ID. Vascular anomalies in Alagille syndrome: a significant cause of morbidity and mortality. Circulation. 2004;109(11):1354–1358. doi: 10.1161/01.CIR.0000121361.01862.A4. [DOI] [PubMed] [Google Scholar]
- Kohsaka T, Yuan ZR, Guo SX, Tagawa M, Nakamura A, Nakano M, Kawasasaki H, Inomata Y, Tanaka K, Miyauchi J. The significance of human jagged 1 mutations detected in severe cases of extrahepatic biliary atresia. Hepatology. 2002;36(4 Pt 1):904–912. doi: 10.1053/jhep.2002.35820. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB. Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet. 1998;62(6):1361–1369. doi: 10.1086/301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Smith R, Colliton RP, Tinkel H, Zackai EH, Piccoli DA, Goldmuntz E, Spinner NB. Jagged1 mutations in patients ascertained with isolated congenital heart defects. Am J Med Genet. 1999;84(1):56–60. [PubMed] [Google Scholar]
- Le Caignec C, Lefevre M, Schott JJ, Chaventre A, Gayet M, Calais C, Moisan JP. Familial deafness, congenital heart defects, and posterior embryotoxon caused by cysteine substitution in the first epidermal-growth-factor-like domain of jagged 1. Am J Hum Genet. 2002;71(1):180–186. doi: 10.1086/341327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, others Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16(3):243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Loomes KM, Underkoffler LA, Morabito J, Gottlieb S, Piccoli DA, Spinner NB, Baldwin HS, Oakey RJ. The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum Mol Genet. 1999;8(13):2443–2449. doi: 10.1093/hmg/8.13.2443. [DOI] [PubMed] [Google Scholar]
- Lu F, Morrissette JJ, Spinner NB. Conditional JAG1 mutation shows the developing heart is more sensitive than developing liver to JAG1 dosage. Am J Hum Genet. 2003;72(4):1065–1070. doi: 10.1086/374386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79(1):169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney DB, Krantz ID, Bason L, Piccoli DA, Emerick KM, Spinner NB, Goldmuntz E. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106(20):2567–2574. doi: 10.1161/01.cir.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- Miller ME, Smith DW. Conotruncal malformation complex: examples of possible monogenic inheritance. Pediatrics. 1979;63(6):890–893. [PubMed] [Google Scholar]
- Morrissette JD, Colliton RP, Spinner NB. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet. 2001;10(4):405–413. doi: 10.1093/hmg/10.4.405. [DOI] [PubMed] [Google Scholar]
- Nora JJ, Nora AH. Familial risk of congenital heart defect. Am J Med Genet. 1988;29(1):231. doi: 10.1002/ajmg.1320290134. 233. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, others Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16(3):235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Pankau R, Siekmeyer W, Stoffregen R. Tetralogy of Fallot in three sibs. Am J Med Genet. 1990;37(4):532–533. doi: 10.1002/ajmg.1320370421. [DOI] [PubMed] [Google Scholar]
- Rauch R, Hofbeck M, Zweier C, Koch A, Zink S, Hoyer J, Kaulitz R, Singer H, Rauch A. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet. 2009 doi: 10.1136/jmg.2009.070391. [DOI] [PubMed] [Google Scholar]
- Ropke A, Kujat A, Graber M, Giannakudis J, Hansmann I. Identification of 36 novel Jagged1 (JAG1) mutations in patients with Alagille syndrome. Hum Mutat. 2003;21(1):100. doi: 10.1002/humu.9102. [DOI] [PubMed] [Google Scholar]
- Spinner NB, Colliton RP, Crosnier C, Krantz ID, Hadchouel M, Meunier-Rotival M. Jagged1 mutations in alagille syndrome. Hum Mutat. 2001;17(1):18–33. doi: 10.1002/1098-1004(2001)17:1<18::AID-HUMU3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Warthen DM, Moore EC, Kamath BM, Morrissette JJ, Sanchez P, Piccoli DA, Krantz ID, Spinner NB. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27(5):436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- Wulfsberg EA, Zintz EJ, Moore JW. The inheritance of conotruncal malformations: a review and report of two siblings with tetralogy of Fallot with pulmonary atresia. Clin Genet. 1991;40(1):12–16. doi: 10.1111/j.1399-0004.1991.tb03063.x. [DOI] [PubMed] [Google Scholar]


