To the Editor
Cyclin-dependent kinases (CDKs) are protein kinases involved in critical cellular processes, such as cell cycle or transcription, whose activity requires association with specific cyclin subunits. Based on sequence similarity, the human genome contains 21 genes encoding CDKs and five additional genes encoding a more distant group of proteins known as CDK like (CDKL) kinases. The current nomenclature for CDK proteins includes 11 classical CDKs (CDK1-11), two newly proposed family members (CDK12-13) and additional proteins whose names are based on the presence of a cyclin-binding element (PFTAIRE and PCTAIRE proteins) or simply based on sequence relationship with the original CDKs, such as CDC2-like kinases (CDC2L) or Cell cycle-related kinases (CCRK)1. We propose here the use of ‘CDK’ for all CDK family members (CDK1-20) based on similarities in sequence and function as described below. This nomenclature will facilitate the rational and comparative study of the poorly understood family members and the analysis of their relevance in human disease.
During the Cold Spring Harbor Symposium on the Cell Cycle in 1991, a group of interested scientists proposed that members of this kinase family would be called cyclin dependent kinases (CDKs). The consensus was that no kinase should be called a ‘CDK’ until it was proven rigorously that its activity depended on association with some cyclin-like regulatory subunit. Known family members were then were renamed CDK1-6 and further members (CDK7-CDK10) were cloned and characterized in the following years1. CDK11 has been used to refer to the protein encoded by three different human loci (CDC2L1, CDC2L2 and CDC2L6) in different publications. CDC2L1 and CDC2L2 are two highly similar human genes originated by duplication in human chromosome 1 and they encode almost identical protein kinases. Each of these loci encode at least two major peptides: a protein kinase of 110 kDa, and a smaller 58 kDa isoform that is expressed following alternative initiation of translation. To make the situation more complex, some publications refer to CDK11 as the protein encoded by CDC2L6, including the original description of the human kinome2 (Table S1). The single Cdc2l1 gene in the mouse genome encodes a protein more similar to human CDC2L2 (91% identity) than CDC2L1 (87%). We therefore propose the use of Cdk11 for the mouse gene and CDK11A and CDK11B for the human CDC2L2 and CDC2L1 loci, respectively (Figure 1 and Table S1). The corresponding human proteins should be referred to as CDK11Ap58 or CDK11Ap110 for the CDC2L2-encoded proteins, and CDK11Bp58 or CDK11Bp110 for the CDC2L1-encoded proteins. Two additional kinases, formerly known as Crk7 (CrkRS) and Ched (Cdc2L5), were recently renamed CDK12 and CDK13, as they were reported to interact with cyclin L1 and cyclin L23,4.
Figure 1. Human CDK and CDKL proteins.
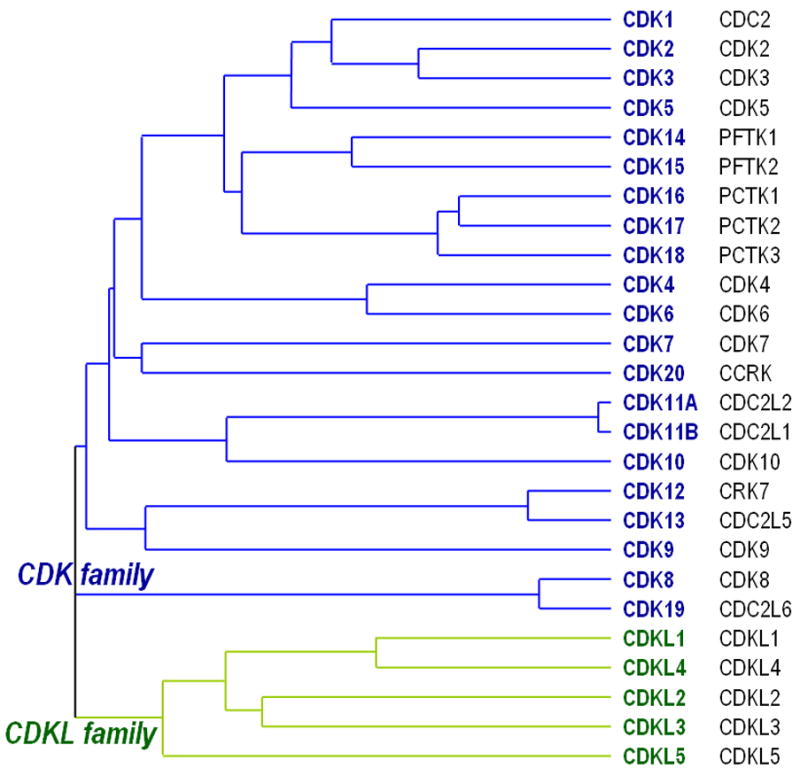
The proposed nomenclature for CDK (blue) proteins is shown in boldface whereas current symbols are indicated in the right column (black font). CDK-like proteins (green) cluster together in a separate branch close to MAP kinases and GSK3 kinases2 and the absence of evidence for cyclin association preclude us from considering these proteins as CDKs. The sequences of the kinase domains were obtained from UniProt (http://www.uniprot.org/) and analyzed using Clustal2 (http://www.ebi.ac.uk/Tools/clustalw2/) using the neighbour-joining method.
Additional members of the family, such as PCTAIRE or PFTAIRE proteins, CCRK or CDC2L6, although being very similar to other CDKs in their primary structure (Figure 1), and their predicted cyclin-binding domain (Figure S1) still maintain their original names (Table S1). Ironically, some these proteins have not received CDK names because their association with cyclins has not been demonstrated, and yet they are known by their predicted cyclin association element sequence (PFTAIRE or PCTAIRE; Figure S1). Although the strict ‘cyclin dependent’ rule has been useful during the gradual characterization of the CDK family, we feel that there is now enough evidence to group all these CDK-related proteins into a single protein family with sufficient sequence and functional similarities to justify the use of the ‘CDK’ term for all family members. PFTAIRE (PFTK) and PCTAIRE (PCTK) proteins are encoded by 5 genes more related to CDK1 than other classical family members such as CDK4 or CDK6 (Figure 1). Recent studies indicate that PFTK1 is activated by cyclin D3 and cyclin Y, and phosphorylates the retinoblastoma protein (pRb) (Refs 5,6 and Table S1). PFTK1 should therefore be renamed CDK14. PFTK2 displays 60% identity with CDK14/PFTK1 but no studies of this protein have been published since its identification in 2001. Based on conservation of the PFTAIRE motif and surrounding sequences, as well as overall similarity, APFTK2 seems very likely to have a cyclin partner and should therefore be renamed as CDK15. The related PCTAIRE kinases (PCTK1-3) are also poorly studied, but retain a cyclin binding motif and have some evidence for cyclin interaction. PCTK3 was found to interact with cyclin K in a large-scale interactions study7. The two other PCTK kinases also interact with p35, the major activator of CDK5 (Table S1). Given the interaction with cyclin K, p35 and the high similarity between PCTAIRE proteins and CDK5, we suggest that these proteins should be renamed as CDK16, CDK17 and CDK18. The same rationale can be applied to the remaining members of the family, CDC2L6 and CCRK. CDC2L6 is highly similar to CDK8, contains an identical SMSACRE motif and it is also a component of the Mediator complex8, suggesting not only sequence homology but also functional similarities. CDC2L6 form complexes with cyclin C although they interaction has not been described in detail (Table S1). CDK19 should therefore be used instead of CDC2L6. Finally, CCRK (also known as CAKp42) is selectively similar to CDK7, the catalytic subunit of the CDK-activating kinase (CAK) complex (Figure 1), and it has also been suggested to function similarly by phosphorylating and activating CDK2 (Refs. 9,10). We therefore propose to use the CDK20 symbol for this protein.
Although a few of these kinases do not have an identified cyclin partner, we believe that the CDK term should be applied to the whole family. First, it is quite likely that all these proteins are activated by cyclins or cyclin-related proteins, many of which are still poorly studied. Second, even if some family members are not activated by cyclins, this does not invalidate the use of the ‘CDK’ root to denominate all family members. There are multiple examples in gene nomenclature where some family members do not share the major features that originally defined that family. For example, 45 human protein kinases lack at least one of the conserved catalytic residues2 and may not have kinase activity. Kinase activity has not been detected for several CDK-related proteins but they are still known as PFTK2, PCTK2 or CCRK, where “K” stands for “kinase”. Finally, most ‘cyclins’ do not cycle.
The current nomenclature has probably contributed to the lack of consideration of the therapeutic value of the lesser-known CDKs, and also a failure to examine these protein kinases for specificity when developing CDK inhibitors. Protein names like PFTAIRE-1 do not encourage the consideration of the therapeutic value of these proteins, which are usually not tested for therapeutic purposes. In summary, we believe that sequence and functional relationships suggest that all of the 21 human CDKs should be given the ‘CDK’ prefix. This expanded CDK family nomenclature has also been agreed upon by the HGNC and the Mouse Genomic Nomenclature Committee, and will be listed in all the major databases for the human and mouse genes encoding the CDK family proteins. The analysis of sequence and functional trends in the CDK family may facilitate new research (e.g. compensatory functions in loss-off function studies) directed toward their relevance in the cell cycle, transcription or other cellular processes. In addition, this simplified nomenclature may increase awareness of their potential importance as therapeutic targets in human disease. Researchers and drug discoverers should keep the whole family picture in mind.
Supplementary Material
References
- 1.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–41. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Chen HH, Wang YC, Fann MJ. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol Cell Biol. 2006;26:2736–45. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HH, Wong YH, Geneviere AM, Fann MJ. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem Biophys Res Commun. 2007;354:735–40. doi: 10.1016/j.bbrc.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 5.Shu F, et al. Functional characterization of human PFTK1 as a cyclin-dependent kinase. Proc Natl Acad Sci U S A. 2007;104:9248–53. doi: 10.1073/pnas.0703327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanyon CA, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 8.Sato S, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–91. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Wu C, Galaktionov K. p42, a novel cyclin-dependent kinase-activating kinase in mammalian cells. J Biol Chem. 2004;279:4507–14. doi: 10.1074/jbc.M309995200. [DOI] [PubMed] [Google Scholar]
- 10.Ng SS, et al. Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma. J Natl Cancer Inst. 2007;99:936–48. doi: 10.1093/jnci/djm011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


