Abstract
Recent structural studies of receptor tyrosine kinases (RTKs) have revealed unexpected diversity in the mechanisms of their activation by growth factor ligands. Strategies for inducing dimerization by ligand binding are surprisingly diverse, as are mechanisms that couple this event to activation of the intracellular tyrosine kinase domains. As our understanding of these details becomes increasingly sophisticated, it provides an important context for therapeutically countering the effects of pathogenic RTK mutations in cancer and other diseases. Much remains to be learned, however, about the complex signaling networks downstream from RTKs and how alterations in these networks are translated into cellular responses.
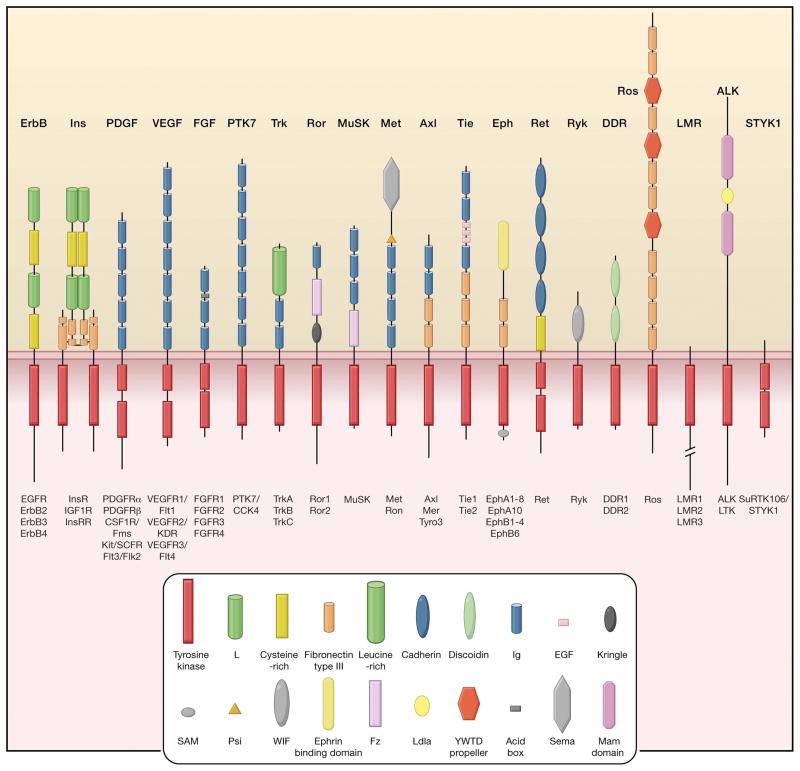
Since the discovery of the first receptor tyrosine kinase (RTK) more than a quarter of a century ago, many members of this family of cell surface receptors have emerged as key regulators of critical cellular processes, such as proliferation and differentiation, cell survival and metabolism, cell migration and cell cycle control (Blume-Jensen and Hunter, 2001; Ullrich and Schlessinger, 1990). Humans have 58 known RTKs, which fall into twenty subfamilies (Figure 1). All RTKs have a similar molecular architecture, with a ligand-binding region in the extracellular domain, a single transmembrane helix, and a cytoplasmic region that contains the protein tyrosine kinase (TK) domain plus additional carboxy (C-) terminal and juxtamembrane regulatory regions. The overall topology of RTKs, their mechanism of activation, and key components of the intracellular signaling pathways that they trigger are highly conserved in evolution from the nematode Caenorhabditis elegans to humans, which is consistent with the key regulatory roles that they play. Furthermore, numerous diseases result from genetic changes or abnormalities that alter the activity, abundance, cellular distribution, or regulation of RTKs. Mutations in RTKs and aberrant activation of their intracellular signaling pathways have been causally linked to cancers, diabetes, inflammation, severe bone disorders, arteriosclerosis and angiogenesis. These connections have driven the development of a new generation of drugs that block or attenuate RTK activity.
Figure 1. Receptor tyrosine kinase families.
Human receptor tyrosine kinases (RTKs) contain 20 subfamilies, shown here schematically with the family members listed beneath each receptor. Structural domains in the extracellular regions, identified by structure determination or sequence analysis, are marked according to the key presented in Supplementary Figure 1, where all 58 RTKs in the human proteome are listed. The intracellular domains are shown as red rectangles.
In this Review, we discuss insights into the mechanism of RTK regulation that have emerged from recent structural and functional studies. We examine prevailing concepts that underlie the activation of intracellular signaling pathways following growth factor binding to RTKs. We also consider recent systems biology approaches for understanding the complicated circuits and networks that result from the interplay among the multiple signaling pathways activated by RTKs. Finally, we describe the impact of these advances on the discovery and application of new therapies for cancers and other diseases driven by activated RTKs.
Mechanisms of Receptor Activation
In general, growth factor binding activates RTKs by inducing receptor dimerization (Ullrich and Schlessinger, 1990). However, before discussing this aspect of RTK regulation, it is important to note that a subset of RTKs forms oligomers even in the absence of activating ligand. For example, the insulin receptor and IGF1-receptor are expressed on the cell surface as disulfide-linked (αβ)2 dimers (Ward et al., 2007). Binding of insulin or IGF1 induces structural changes within these dimeric receptors that stimulate tyrosine kinase activity and cell signaling. Some studies have suggested that epidermal growth factor (EGF) binds to and activates pre-existing oligomers of its receptor (Clayton et al., 2005; Gadella and Jovin, 1995), but the precise nature and size of these oligomers is not known. Moreover, there is evidence that activation of certain RTKs, such as Tie2 (an angiopoietin receptor) and Eph receptors, may require the formation of larger oligomers (Barton et al., 2006; Himanen and Nikolov, 2003).
Whether the ‘inactive’ state is monomeric or oligomeric, activation of the receptor still requires the bound ligand to stabilize a specific relationship between individual receptor molecules in an ‘active’ dimer or oligomer. Structural studies of the extracellular regions of RTKs have provided clear views of how ligand binding can drive dimerization. In addition, the single membrane-spanning α-helix may contribute to dimerization in some cases, although the precise role is not yet clear. In the ligand-bound receptor, self-association of the extracellular region is thought to guide the intracellular domains into a dimeric conformation that activates their tyrosine kinase domains through the mechanisms discussed below. One receptor in the dimer/oligomer then phosphorylates one or more tyrosines in a neighboring RTK, and the phosphorylated receptor then serves as a site for assembly (and activation) of intracellular signaling proteins (Ullrich and Schlessinger, 1990).
Ligand-induced dimerization of RTK extracellular regions
Early studies of RTKs and cytokine receptors suggested a conceptually straightforward mechanism for ligand-induced dimerization: a bivalent ligand interacts simultaneously with two receptor molecules and effectively cross-links them into a dimeric complex. This ‘ligand-mediated’ mode of receptor dimerization was further supported by crystal structures of several fragments of the ligand-binding domains from RTKs bound to their relevant ligands. Examples include the stem cell factor receptor KIT (Liu et al., 2007), the Flt1 vascular endothelial growth factor (VEGF) receptor (Leppänen et al., 2010; Wiesmann et al., 1997), the nerve growth factor (NGF)/neurotrophin receptor TrkA (Wiesmann et al., 1999), Axl (Sasaki et al., 2006), Tie2 (Barton et al., 2006), and Eph receptors (Himanen and Nikolov, 2003). In each of these cases, the ligand is itself a dimer and simply cross-links the ligand-binding fragments of two receptor molecules. Recent structures of more complete extracellular regions of RTKs have provided important additional insight into the range of mechanisms used for ligand-induced dimerization. Figure 2 illustrates two mechanistic extremes and two intermediate cases. At one extreme, receptor dimerization is entirely “ligand-mediated” and the two receptors make no direct contact (Figure 2A). At the other extreme, dimerization is instead entirely “receptor-mediated” (Figure 2D, and the ligand makes no direct contribution to the dimer interface. Alternatively, dimerization can involve both ligand-mediated and receptor-mediated components (Figure 2B and 2C. Dimerization of most RTKs is likely to resemble one of these four modes. However, additional paradigms are likely to emerge from more comprehensive studies of other RTK families.
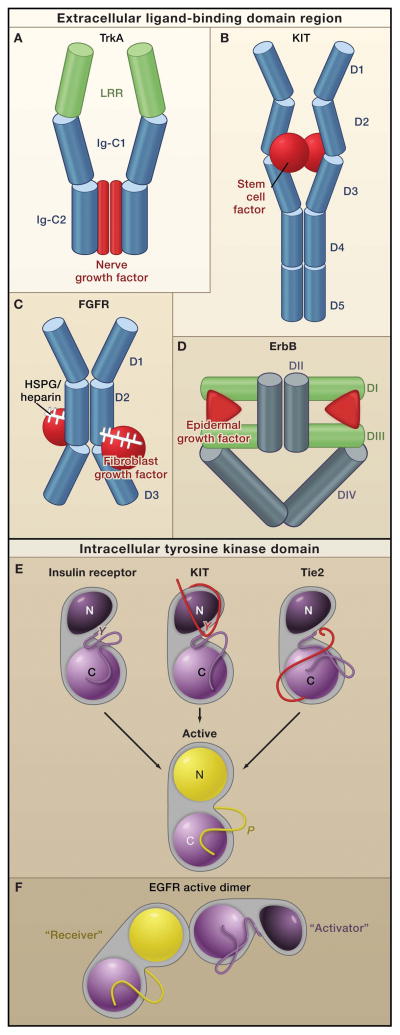
Figure 2. Receptor Tyrosine Kinase Dimerization and Kinase Activation.
Top: In general, receptor tyrosine kinases (RTKs) associate into dimers when ligand (red) binds to their extracellular regions. The bound ligand, which can form all, a portion, or none of the dimer interface, activates the receptors by stabilizing a specific relationship between two individual receptor molecules.
A. A nerve growth factor dimer (red) cross-links two TrkA molecules without any direct contact between the two receptors (Wehrman et al. 2007). B. A stem cell factor dimer (red) also cross links two KIT molecules. In addition, two Ig-like domains (D4 and D5), which reorient upon receptor activation, interact across the dimer interface (Yuzawa et al., 2007). Thus, KIT combines ligand-mediated and receptor mediated dimerization modes. C. Two fibroblast growth factor receptor (FGFR) molecules contact one another through the Ig-like domain D2, and the accessory molecule heparin or heparin sulfate proteoglycans (white sticks) also contacts this domain (Schlessinger et al., 2000). In addition, each fibroblast growth factor molecule (red) contacts Ig-like domains D2 and D3 of both FGFR molecules. D. Dimerization of ErbB receptors is mediated entirely by the receptor. Binding simultaneously to two sites (DI and DIII) within the same receptor molecule, the ligand drives conformational changes in epidermal growth factor receptor (EGFR) that expose a previously-occluded dimerization site in Domain II
Bottom: Dimerization of the extracellular regions of RTKs activates the intracellular tyrosine kinase domains (TKDs), which contain a C-lobe (light purple or yellow), N-lobe (dark purple or yellow in the inactive and active states), and an activation loop (dark purple or yellow in the inactive and active states, respectively). Although the crystal structures of the activated TKDs are very similar (Huse and Kuriyan, 2002), structures of inactive TKDs differ substantially among the receptors (top row), reflecting the diversity in their regulatory mechanisms. However, many receptors are inhibited by a set of intramolecular (or cis) interactions:
E. Insulin receptor-like (activation loop inhibition). In FGFR, insulin receptor, and IGF1-receptor the activation loop interacts directly with the active site of the kinase and blocks access to protein substrates (in FGFR) or to both ATP and protein substrates (in insulin and IGF1 receptors). Phosphorylation of key tyrosines (‘Y’) disrupts these autoinhibitory interactions and allows the kinase to ‘relax’ to the Active state.
KIT-like (juxtamembrane inhibition). In KIT, PDFGR, and Eph receptors the juxtamembrane region (red) interacts with elements within the active site of the kinase (including the αC helix and the activation loop) to stabilize an inactive conformation. Phosphorylation of key tyrosines in the juxtamembrane region destabilizes these autoinhibitory interactions and allows the TKD to assume an active conformation.
Tie1-like (C-terminal tail inhibition). In Tie1 and Tie2(and possibly Met and Ron), the C-terminal tail (red) interacts with the active site of the TKD to stabilize an inactive conformation(Shewchuk et al., 2000).
F. The EGFR TKD is allosterically activated by direct contacts between the C-lobe of one TKD, the ‘Activator,’ and the N-lobe of another TKD, ‘Receiver’ (Zhang et al., 2006). The Activator TKD destabilizes autoinhibitory interactions that involve the activation loop of the Receiver TKD. No activation loop phosphorylation is required in this mechanism (Jura et al., 2009; Red Brewer et al., 2009).
TrkA: ligand-mediated dimerization (Figure 2A)
The TrkA (NGF receptor) extracellular region contains a solenoid-like leucine-rich repeat (LRR) region followed by two immunoglobulin-like domains (Ig-C1 and Ig-C2). In the NGF-induced dimer, the extracellular regions of the two TrkA molecules do not contact one another. Only the Ig-C2 domain of each receptor molecule contacts the dimeric NGF ligand, with each Ig-C2 contacting both chains of the NGF dimer (Wehrman et al., 2007; Wiesmann et al., 1999). Thus, the bound NGF appears to provide the entire dimer interface of TrkA, with the caveat that a 30 amino acid juxtamembrane region is missing from the most complete TrkA structure (Wehrman et al., 2007).
KIT: a ligand-mediated dimer with receptor contacts (Figure 2B
The KIT ligand, stem cell factor, or SCF) is a homodimer of two four-helix bundles. Each SCF molecule binds to one molecule of KIT through contacts with the first three (of five) Ig-like domains in the KIT extracellular region (Liu et al., 2007; Yuzawa et al., 2007). This D1–D3 region is structurally unaltered upon ligand binding, and association with the SCF dimer simply ‘cross-links’ the two receptors. However, the two Ig-like domains closest to the plasma membrane (D4 and D5) undergo a significant reorientation upon KIT dimerization (Yuzawa et al., 2007). D4 and D5 make important homotypic interactions across the dimer interface (Figure 2B), which properly orient the two KIT molecules for activation. Oncogenic gain-of-function mutations are found in the D5 domain of KIT and are thought to stabilize these activating interactions.
Recent studies of the CSF-1 (colony stimulating factor-1) receptor suggest that it has an activated structure similar to that of KIT (Figure 2B). However the study also indicated that receptor-receptor interactions are required for dimerization in this case (Chen et al., 2008). Additional variations on this theme are likely for other RTKs. For example, direct interactions between membrane proximal portions of Eph receptors appear to be important for their oligomerization and activation (Seiradake et al., 2010).
FGFR: multiple contacts with FGF and heparin molecules (Figure 2C)
Dimerization of fibroblast growth factor (FGF) receptors uses a combination of bivalent ligand binding, direct receptor-receptor contacts, and the involvement of an accessory molecule (Schlessinger et al., 2000; Stauber et al., 2000). The extracellular regions of FGFRs contain three Ig-like domains (D1–D3). Domains D2 and D3, plus an intervening linker, are crucial for binding to FGF ligands (Plotnikov et al., 1999), which are monomeric (unlike NGF and SCF). An X-ray crystal structure of a dimeric complex containing the FGFR1c extracellular region, FGF2, and heparin in a 2:2:2 ratio (Schlessinger et al., 2000) showed that each receptor molecule simultaneously contacts both FGF and heparin (the accessory molecule). Each FGF molecule also contacts both receptor molecules in the dimer, forming a major interaction with one and a more minor interaction with the other. Heparin simultaneously contacts both ligands in the dimer and both receptor molecules (through domain D2). Moreover, the two receptors interact directly with each other through their D2 domains. Thus, receptor-ligand, receptor-heparin, ligand-heparin and receptor-receptor interactions all cooperate to stabilize the FGFR dimer. Although several structures of FGF/FGFR display this type of arrangement (Ibrahimi et al., 2005; Plotnikov et al., 1999; Schlessinger et al., 2000; Stauber et al., 2000), an alternative configuration has also been observed in X-ray crystal structures, in which heparin bridges two FGF/FGFR complexes. This creates an asymmetric dimer without significant contribution of protein-protein interactions to the dimer interface (Pellegrini et al., 2000). Detailed analyses of both engineered and disease-related mutations in FGFR support the physiological relevance of the symmetric arrangement shown in Figure 2C rather than the alternative heparin-bridged dimer (Ibrahimi et al., 2005).
FGFR has an additional intramolecular control mechanism that involves ‘autoinhibition’ of ligand binding. The affinity of FGFRs for FGFs or HSPGs (heparan sulfate proteoglycans) is increased when either D1 or an 8-residue ‘acid box’ in the D1–D2 linker (Figure 1) is removed. An intramolecular interaction between D1 and the ligand-binding site formed by D2 and D3 competes with ligand binding to FGFR. At the same time, the acid-box binds to a positively charged ‘canyon’ within the same receptor that would otherwise accommodate HSPGs/heparin (Plotnikov et al., 1999; Schlessinger et al., 2000; Stauber et al., 2000). FGFR monomers adopt a “closed” or autoinhibited configuration when D1 and the acid-box occupy their intramolecular binding sites. This autoinhibited state is thought to be in equilibrium with an “open” configuration in which the two binding sites are empty, and are poised to interact with FGF and HSPGs, allowing FGFR activation.
The EGFR/ErbB family: the ‘receptor-mediated’ extreme (Figure 2D)
Receptors in the epidermal growth factor receptor (EGFR or ErbB) family represent the other extreme of activation mechanisms, where activating ligands make no direct contribution to the dimerization interface. These receptors also display a dramatic form of intramolecular autoinhibition (Burgess et al., 2003). The extracellular regions of ErbB receptors contain four domains (I-IV). Domains I and III are each ~160 amino acids in length, comprise β-helix LRR-like ‘solenoid’ domains, and both bind to activating ligands. Domains II and IV are cysteine-rich domains consisting of ~150 amino acids each. Structures of the EGFR extracellular region revealed that dimerization is entirely ‘receptor-mediated,’ as depicted in Figure 2D (Garrett et al., 2002; Ogiso et al., 2002). Although the ligand is bivalent like those discussed above, in this case it contacts two distinct sites within a single receptor molecule (on Domains I and III) rather than crosslinking two separate receptor molecules as seen for NGF, SCF or FGF. This bivalent ligand binding promotes substantial conformational changes in the extracellular region of EGFR, which unmask a dimerization arm in Domain II (Burgess et al., 2003). Before ligand binds, this arm is completely buried by intramolecular interactions with Domain IV that stabilize a ‘tethered’ conformation in which both ligand binding and dimerization are autoinhibited (Bouyain et al., 2005; Burgess et al., 2003; Cho and Leahy, 2002; Ferguson et al., 2003). Ligand binding breaks the tether, allowing the dimerization arm of Domain II to interact with a second ligand-bound receptor molecule (Figure 2D). As with KIT (Figure 2B), the membrane-proximal domain of EGFR (Domain IV) also appears to make contacts across the dimer interface (Burgess et al., 2003), which may orient the dimers in the configuration required for maximal activation.
New lessons from other RTKs
Although many of the 58 RTKs in humans are likely to use one of the four mechanisms outlined above for activation, further studies will certainly identify new variations. For example, the two human Discoidin Domain Receptors (DDR1 and DDR2) are activated by collagen fibers rather than soluble growth factors (Shrivastava et al., 1997; Vogel et al., 1997). Little is known about how collagen binding promotes receptor dimerization and activation. The kinetics of DDR1/2 activation is unusually slow, suggesting that these receptors may reveal a new twist on the theme of receptor crosslinking by multivalent ligands.
The Ryk (related to receptor tyrosine kinase), Ror (RTK-like orphan receptor), and MuSK (muscle-specific kinase) families of RTKs, which all have unexpected links to Wnt signaling (van Amerongen et al., 2008), probably also use a unique activation mechanism. Ryk contains a Wnt-inhibitory factor-1 (WIF-1) domain in its extracellular region and is reported to function as a receptor (or co-receptor) for Wnts. However, the mechanistic details remain unclear, and it is not clear whether Ryk contains an active tyrosine kinase domain. The two Ror RTKs also appear to bind Wnts through a domain in their extracellular regions that is closely related to the cysteine-rich Wnt-binding domain found in the Frizzled receptors. Moreover, Ror2 appears to mediate certain responses to Wnt5a (van Amerongen et al., 2008). The MuSK RTK also has a cysteine rich domain related to Frizzled receptors (Figure 1), suggesting a Wnt connection. MuSK is regulated by the HSPG agrin, but efforts to detect direct interactions between agrin and the MuSK extracellular region have been unsuccessful (Stiegler et al., 2006). Recent studies showed that Lrp4 (LDL receptor-related protein-4) functions as an accessory molecule for MuSK; it binds to agrin and mediates its effects on MuSK activity (Kim et al., 2008). Although not yet visualized structurally, such indirect effects of activating ligands on RTKs are likely to represent another paradigm in RTK activation mechanisms.
The Ret (rearranged during transfection) receptor is also activated indirectly and may use a mechanism related to that of MuSK. Ret responds to homodimeric ligands in the glial-derived neurotrophic factor (GDNF) family, but these ligands must first bind to a GDNF-family receptor-α (GFRα) chain (Runeberg-Roos and Saarma, 2007) that is glycosyl phosphatidylinositol (GPI) anchored. The GFRα/GDNF complex appears to promote dimerization of Ret, and the steps in this process have been identified by quantitative studies in a cellular context (Schlee et al., 2006). Ret illustrates another variation on the ligand-mediated dimerization theme and may also provide a model for understanding how agrin/Lrp4 complexes promote MuSK dimerization.
Activation of intracellular kinase domains
The crucial question of how ligand-induced dimerization of the extracellular regions of RTKs leads to activation of the intracellular tyrosine kinase domain (TKD) has been addressed in detail for several RTKs, including KIT, FGFR, the insulin receptor, and EGFR. The activation mechanisms are surprisingly different. All TKDs have an N-lobe and a C-lobe (Figure 2E), and crystal structures of the activated forms of TKDs of RTKs (and indeed of activated TKDs in general) are all very similar (Huse and Kuriyan, 2002). Key regulatory elements including the ‘activation loop’ and the αC helix in the kinase N-lobe adopt a specific configuration in all activated TKDs that is required for catalysis of phosphotransfer (Nolen et al., 2004). By contrast, the structures of inactive TKDs differ substantially from receptor to receptor, and this variation reflects the diversity in their regulatory mechanisms. Each TKD is uniquely cis-autoinhibited by a set of intramolecular interactions specific for its receptor. Release of cis-autoinhibition, following ligand-induced receptor dimerization, is the key event that triggers RTK activation.
TKD autoinhibition by the activation loop: insulin and FGF receptors
A structure of the insulin receptor TKD was the first to illustrate RTK autoinhibition (Hubbard, 2004). A key tyrosine (Y1162) in the activation loop of the insulin receptor TKD projects into the active site as if poised to be autophosphorylated by its own kinase domain (i.e., in cis) (‘Insulin receptor’ in Figure 2E). This interaction stabilizes an activation loop configuration that occludes the active site, blocking access of both ATP and protein substrates. Thus, the insulin receptor TKD is autoinhibited in cis by its own activation loop. When insulin activates the receptor, Y1162 in one TKD within the dimer becomes phosphorylated by its partner (together with two additional tyrosines), and this trans-phosphorylation disrupts the cis-autoinhibitory interactions. The phosphorylated activation loop of the insulin receptor TKD is then free to adopt the ‘active’ configuration seen in all other activated TKDs (Huse and Kuriyan, 2002; Nolen et al., 2004). The αC helix in the N-lobe also reorients, enabling it to contribute to stabilization of the ATP binding site. Thus, ‘release’ of cis-autoinhibition via autophosphorylation allows the TKD of the insulin receptor to ‘relax’ into an active state (Figure 2E).
FGFR1 uses a conceptually similar mechanism, although its autoinhibition involves a different set of activation loop interactions than does the insulin receptor (Mohammadi et al., 1996). Tyrosines in the activation loop of FGFR1 do not block the substrate-binding site directly. Instead, they participate in a unique set of intramolecular contacts that stabilize the inactive conformation of the kinase, in turn cis-autoinhibiting the FGFR1 TKD by occluding the protein-substrate binding site but not the ATP binding site. When FGF induces dimerization of its receptor, trans-phosphorylation of tyrosines in the activation loop disrupts the cis-autoinhibitory configuration so that both the activation loop and the αC helix can adopt the characteristic active configuration (Bae et al., 2009; Chen et al., 2007).
Juxtamembrane autoinhibition
Phosphorylation of the activation loop plays a crucial regulatory role in most kinases. It is required both to stabilize the activated configuration (Nolen et al., 2004) and to destabilize cis-autoinhibitory interactions. In addition to this mechanism of regulation, many RTKs are cis-autoinhibited by elements outside the TKD itself. The best known example is ‘juxtamembrane autoinhibition,’ exemplified by MuSK (Till et al., 2002), Flt3 (Griffith et al., 2004), KIT (Mol et al., 2004) and Eph family RTKs (Wybenga-Groot et al., 2001). In each case, sequences in the juxtamembrane region make extensive contacts with several parts of the TKD, including the activation loop, and stabilize an autoinhibited conformation (‘KIT’ in Figure 2E). These autoinhibitory interactions differ in detail among the TKDs, but in all cases they involve key tyrosines in the juxtamembrane region. Receptor dimerization promotes trans-phosphorylation of these tyrosines, which disrupts the cis-autoinhibitory interactions and promotes receptor activation (Hubbard, 2004). Mutations that disrupt the autoinhibitory juxtamembrane interactions in the KIT/PDGFR family constitutively activate these RTKs and are frequently found in cancers (Dibb et al., 2004).
Autoinhibition by C-terminal sequences
Another variation on this theme is seen with Tie2. Although the activation loop of its TKD exists in an active-like conformation even without phosphorylation (Shewchuk et al., 2000), the nucleotide-binding loop of Tie2 adopts an inactive configuration. Moreover, a region in the C-terminal tail that contains tyrosine autophosphorylation sites blocks substrate access to the active site, representing a third form of reversible cis-autoinhibition (Niu et al., 2002) generalized in Figure 2E (‘Tie1’). Autophosphorylation of the Tie2 C-terminal tail may disrupt these autoinhibitory interactions (and thus activate Tie2) in a manner similar to reversal of juxtamembrane autoinhibition. A similar situation may also exist for PDGFR and Ron although structural details have yet to be described.
Allosteric activation of TKDs
For all of the TKDs discussed above, activation requires trans-phosphorylation of tyrosines in the activation loop, the juxtamembrane segment, and/or the C-terminal region. However, even when autoinhibited, the TKD is thought to have sufficient kinase activity to trans-phosphorylate its partner in an RTK dimer stabilized by ligand-binding. The cis-autoinhibitory interactions outlined above are thought to ‘breathe,’ such that each TKD is, for a portion of the time, both competent to phosphorylate its neighbor and susceptible to phosphorylation at key regulatory sites. Bringing two TKDs together in a specific stable dimer increases their respective local concentrations and may also promote allosteric effects. These influences dramatically increase the probability that a transiently active TKD will encounter another TKD that can be phosphorylated in a way that promotes activation. Trans-autophosphorylation ensues, and the receptors become activated.
The EGFR/ErbB family and Ret stand out as clear exceptions to this rule because they do not require trans-phosphorylation of their activation loops (or elsewhere) for activation (Knowles et al., 2006; Zhang et al., 2006). How, then, are these RTKs regulated? Ret activation may involve disruption of trans-autoinhibitory interactions that were seen in crystallographic dimers of its TKD (Knowles et al., 2006). For EGFR, crystallographic and mutational studies have identified an allosteric mechanism that resembles activation of cyclin-dependent kinases (CDKs) by cyclins (Zhang et al., 2006). The EGFR TKD forms an asymmetric dimer (Figure 2F) in which the C-lobe of one TKD, called the ‘Activator,’ makes intimate contacts with the N-lobe of the second TKD, called the ‘Receiver.’ These contacts induce conformational changes in the N-lobe of the Receiver kinase that disrupt cis-autoinhibitory interactions seen in the monomer. As a result, the Receiver kinase can adopt the characteristic active configuration without activation loop phosphorylation. EGFR can also be activated without ligand binding when the monomer’s cis-autoinhibitory interactions are disrupted by oncogenic mutations found in a subset of patients with non-small cell lung cancer (Sharma et al., 2007). ErbB4 is regulated through a similar mechanism as seen for EGFR (Qiu et al., 2008), and mutations in the asymmetric TKD dimer interface of EGFR or ErbB4 impair normal activation of the intact receptors (Qiu et al., 2008; Zhang et al., 2006).
Recent studies show that the intracellular juxtamembrane region of EGFR also plays a key part in promoting the allosteric mechanism of its activation (Jura et al., 2009; Red Brewer et al., 2009), instead of serving the autoinhibitory role described above for juxtamembrane regions of several other RTKs. Part of the juxtamembrane region of the Receiver kinase ‘cradles’ the C-lobe of the Activator kinase in the dimer represented in Figure 2F. This interaction promotes dimerization and thus, allosteric activation of the Receiver. The remainder of the Receiver’s juxtamembrane region may interact with its counterpart in the Activator to further stabilize the asymmetric dimer (Jura et al., 2009). Certain mutations in lung cancer appear to promote EGFR activation by stabilizing these juxtamembrane interactions (Red Brewer et al., 2009).
Interestingly, Jura et al. (2009) also identified a structure of a potential inactive dimer for the EGFR TKD, in which C-terminal sequences occlude the site on the Activator onto which the Receiver juxtamembrane region must dock for receptor activation. This suggests an autoinhibitory role for the EGFR C-terminus, as suggested previously (Walton et al., 1990). Studies of intact EGFR also suggest that the juxtamembrane region is involved in allosteric control of ligand binding by the receptor (Macdonald-Obermann and Pike, 2009). Clearly, many important lessons still remain to be learned about the details of EGFR activation.
Linking RTK activation to cell signaling
The first and primary substrates that RTKs phosphorylate are the receptors themselves. Autophosphorylation sites in the kinase domain itself play an important regulatory role in most RTKs, with EGFR and Ret as exceptions. Autophosphorylation of the activation loop in the insulin receptor TKD increases its catalytic efficiency by 50–200 fold (Cobb et al., 1989). Additional tyrosines are then autophosphorylated in other parts of the cytoplasmic region of most RTKs (IRS proteins fulfill this function for the insulin receptor). The resulting phosphotyrosines function as specific sites for the assembly of downstream signaling molecules that are recruited to the receptor and activated in response to growth factor stimulation.
Phases of RTK autophosphorylation
Studies of several RTKs in vitro demonstrated that autophosphorylation occurs in trans (Favelyukis et al., 2001; Furdui et al., 2006; Honegger et al., 1989; Till et al., 2002) and that autophosphorylation sites are phosphorylated in a precise order. For example, in the closely related insulin receptor and IGF-1 receptor tyrosine kinases, the three sites in the activation loop are phosphorylated in the same order: Y1162, Y1158, and then Y1163 (using insulin receptor numbering) (Favelyukis et al., 2001). Each successive event has a significant effect on catalytic properties by destabilizing the cis-autoinhibitory interactions outlined above. The first phosphorylation event causes the largest increase in Vmax; the second event causes the largest drop in KM for substrates (Favelyukis et al., 2001); and the third event has a modest effect on both. In the case of MuSK, phosphorylation of Y553 in the autoinhibitory juxtamembrane region and Y754 in the activation loop precede two additional phosphorylation events in the activation loop (Till et al., 2002). These are ‘first-phase’ autophosphorylation events that primarily serve to enhance the catalytic activity of the kinase once the receptor binds its activating ligand.
Autophosphorylation events in a ‘second-phase’ require prior (first-phase) activation of the kinase and create the phosphotyrosine-based binding sites that recruit cytoplasmic signaling molecules containing Src homology-2 (SH2) and phosphotyrosine-binding (PTB) domains. Recent studies indicate that a third phase also exists for some RTKs, involving trans-autophosphorylation events that maximize the ability of the kinase to phosphorylate its downstream targets. Studies with FGFR1 showed that autophosphorylation of Y653 in the activation loop is the key autophosphorylation event during the first-phase, which increases kinase activity by ~10–50 fold (Furdui et al., 2006). Second-phase autophosphorylation events then occur in an unexpectedly precise order. Y583, which lies in an extra loop within the TKD called the kinase insert, is trans-autophosphorylated first. Y463 in the juxtamembrane region is phosphorylated next and is followed by Y585 in the kinase insert. These three second-phase sites are likely to be SH2/PTB domain docking sites, and their phosphorylation promotes recruitment of downstream signaling molecules rather than increasing kinase activity. After this second-phase, a further autophosphorylation event occurs in the FGFR1 activation loop (at Y654) that elevates kinase activity an additional 10 fold (Furdui et al., 2006) to reach 100–500 times basal levels. This modification of Y654 represents a ‘third-phase’ of autophosphorylation that maximally stimulates the FGFR1 kinase domain for phosphorylation of downstream targets such as phospholipase C-γ (PLCγ) and the FGF receptor substrate-2 (FRS2). Phosphorylation of Y654 is not needed for receptor autophosphorylation events in the second phase that direct the assembly of signaling molecules on the activated receptor.
Receptor activation nucleates formation of signaling complexes
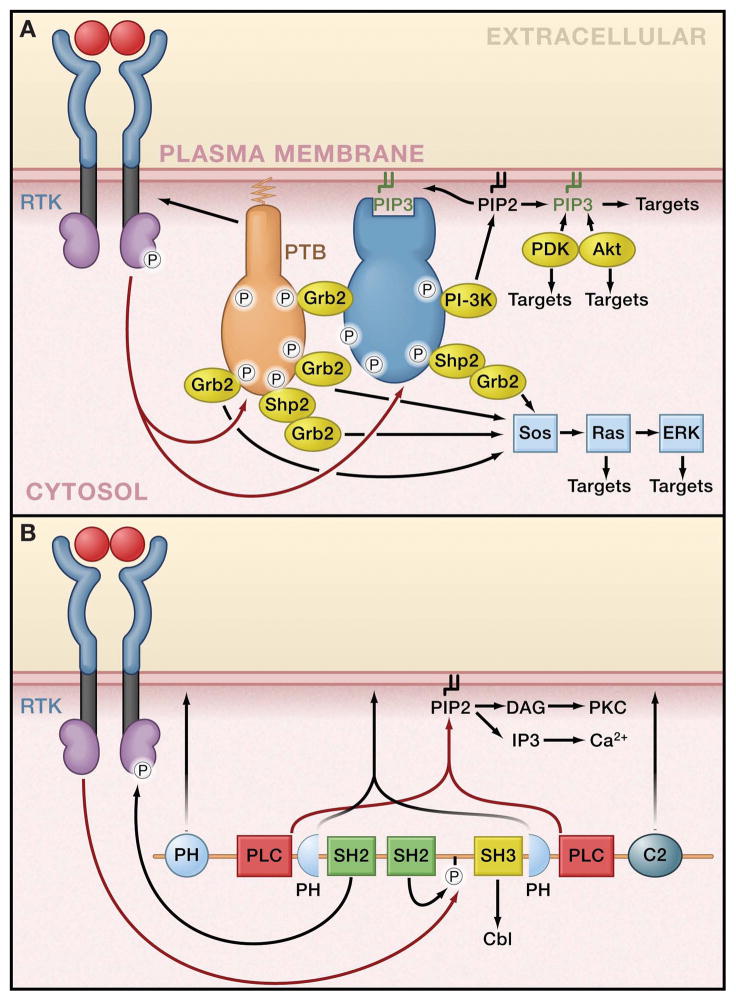
The first response to autophosphorylation of RTKs is the recruitment and activation of a host of downstream signaling molecules. These molecules contain SH2 or PTB domains that specifically bind to phosphotyrosine (Pawson, 2004; Schlessinger and Lemmon, 2003). They may be directly recruited to phosphotyrosines in the receptor, or they may be recruited indirectly by binding to docking proteins that are phosphorylated by RTKs with which they associate (Schlessinger, 2000). These docking proteins include FRS2, IRS1 (insulin receptor substrate-1), and Gab1 (the Grb2-associated binder). Phosphorylation of docking proteins is functionally equivalent to second-phase RTK autophosphorylation. Docking proteins typically contain a membrane targeting site at their amino (N-) terminus, followed by an array of tyrosine phosphorylation sites that serve as binding sites for a distinct repertoire of downstream signaling proteins (Figure 3A). Although a number of docking proteins (such as Gab1) are recruited by multiple RTKs, others are restricted to particular subsets of receptors. For example, the two members of the FRS2 family (FRS2α and FRS2β) mediate signaling primarily by FGF and NGF receptors (Schlessinger, 2000). The four members of the IRS family (IRS1–4) play crucial roles in mediating signaling by the insulin and IGF1-receptors, which rely entirely on these docking proteins for recruitment of downstream signaling molecules. With multiple phosphotyrosines in most receptors and the involvement of numerous docking proteins, activated RTKs clearly can recruit and influence a large number of different signaling molecules. Therefore, an activated RTK can be thought of as a node in a complex signaling network that transmits information from the exterior to the interior of the cell.
Figure 3. Coincidence detection and network branching in RTK Signaling.
A. Coordinated assembly of multiprotein complexes in receptor tyrosine kinase (RTK) signaling provides branching points in a signaling network. The docking protein FGF receptor substrate-2 (FRS2α forms a complex with activated fibroblast growth factor (FGF) or nerve growth factor (NGF) receptors via its phosphotyrosine-binding domain (PTB). The activated RTK phosphorylates FRS2α on multiple tyrosines, and the resulting phosphotyrosines recruit multiple Grb2 and Shp2 molecules, which bring a second docking protein, Gab1, into the complex. Gab1 is tyrosine phosphorylated and recruits additional signaling proteins, including phosphoinositide 3-kinase (PI3K). PI3K initiates a positive feedback loop in which PtdIns(3,4,5)P3 (PIP3), generated by PI3K, recruits more Gab1, leading to further PI3K activation. B. The multiple domains of phospholipase C-γ (PLCγ cooperate to integrate multiple signals at the plasma membrane. The N-terminal SH2 domain is responsible for complex formation with activated receptor tyrosine kinases (RTKs). The C2 and PH domains cooperate with the SH2 domain to target PLCγ to the plasma membrane. One or both of the PH domains may also specifically recognize products of RTK-activated PI3K. RTK-mediated tyrosine phosphorylation of PLCγ leads to intramolecular binding of the C-terminal SH2 domain to phosphotyrosine 783. This stimulates enzymatic activity of PLCγ, leading to hydrolysis of PtdIns(4,5)P2 (PIP2) and consequently leads to the formation of Ins(1,4,5)P3 (IP3) and diacylglycerol (DG).
Multi-domain interactions specify signaling complex formation
It is now well appreciated that a wide array of interaction modules is responsible for communication among signaling molecules in the networks influenced by RTKs (Seet et al., 2006). These signaling molecules frequently contain multiple modules, as illustrated in Figure 3B for phospholipase C-γ (PLCγ). Table 2 in the Supplemental Material online depicts various protein modules known to mediate intermolecular interactions in RTK signaling networks. Some modules bind RTKs directly in ‘receptor-proximal’ interactions (e.g., SH2, PTB), whereas others are involved in interactions that are spatially and temporally more distal. The receptor-proximal interactions require modification of the receptor. SH2 and PTB domains bind only to the tyrosine-phosphorylated receptor (with a few exceptions), and subsequently link RTK autophosphorylation to the initiation of other events in the signaling network (Pawson, 2004; Schlessinger and Lemmon, 2003). Several ubiquitin-binding modules (Hurley et al., 2006) also bind directly to RTKs that have been ubiquitylated. Ubiquitylation, which frequently depends on receptor activation, directs termination of the RTK’s influence on the signaling network by promoting receptor degradation (Kirkin and Dikic, 2007), creating an important negative feedback mechanism (Hunter, 2007). The remaining modules listed and defined in Table 2 (Supplemental Material online) fall into two categories. SH3, WW and PDZ domains are all examples of protein-protein interaction modules, whereas PH (Pleckstrin Homology), PX, C1, C2, and FYVE domains are all best known as phospholipid-binding modules. Domains in these classes are frequently located alongside SH2 domains in the typical multi-domain proteins that signal downstream of RTKs (Figure 3B).
Table 2. Protein modules that mediate intracellular signaling pathways downstream of RTKs and the targets they recognize.
| Receptor Tyrosine Kinase Module | Targeta |
|---|---|
| SH2 | pYXXX |
| PTB | NPXpY peptides |
| FHA | pTXXD |
| 14-3-3 | SXpSXP |
| SH3 | PXXP |
| WW | PXPX |
| CH | F-actin |
| LIM | Protein-protein |
| SAM | Homotypic oligomerization |
| PDZ | C-terminal valine |
| FERM | Membrane and cytoskeleton |
| PX | Phosphoinositides |
| C1 | Membrane |
| C2 | Membrane and calcium |
| PH | Phosphoinositides |
| FYVE | PtdIns3P |
| UIM | Ubiquitin |
| UBA | Ubiquitin |
Protein sequence motifs are shown in bold.
Although these domains certainly drive and specify the formation of key signaling complexes (reviewed by Seet et al., 2006), few of these modules show sufficient binding selectivity or affinity on their own to explain the precise specificity of signaling-complex formation (Ladbury and Arold, 2000). Multivalency appears to be a key solution, with several domains in a single signaling protein cooperating with one another to drive formation of the signaling complex (or network node) in response to several cues (Pawson, 2004; Seet et al., 2006). The example of PLCγ illustrates this point nicely (Figure 3B). Two SH2 domains, two PH domains (one split into two parts), one C2 domain, and one SH3 domain all participate in multivalent signal-dependent targeting of PLCγ to its site of action at the membrane. The SH2 domains bind phosphotyrosines in the receptor or docking protein; the PH domains bind phosphoinositides at the plasma membrane, including the PI 3-kinase product PtdIns(3,4,5)P3; the C2 domain also binds membrane components; and the SH3 domain binds Cbl (Casitas B-lineage lymphoma) that has been recruited into the signaling complex (Tvorogov and Carpenter, 2002). PLCγ thus integrates multiple signal inputs through a combination of recognition modules, which permits ‘coincidence detection’ (Pawson, 2007). Another example involves tandem SH2 domains that engage their target receptor only if two tyrosines in the target are phosphorylated such that both SH2 domains can bind simultaneously (Eck et al., 1996). Indeed, the phosphotyrosines in RTKs that bind to downstream signaling molecules with tandem SH2 domains (e.g., PLCγ, ZAP-70, PI3K, and Shp phosphatases) have been the easiest sites to map, implying a stricter specificity in these multivalent cases.
New perspectives on well-known signaling domains
It has long been thought that the determinants of specificity in SH2 domain interactions with their target proteins are limited largely to the primary sequence surrounding the phosphotyrosine (Songyang and Cantley, 2004). Structural details of SH2 domains recognizing short phosphopeptides, which are well described elsewhere (Waksman and Kuriyan, 2004), reinforced this hypothesis by explaining the results of in vitro studies of peptide-binding specificity. The assumption that all cellular SH2 domain-mediated interactions can be recapitulated using short phosphopeptides prompted their use in protein microarray studies to generate quantitative potential interaction networks for certain RTKs. Although the resulting network for EGFR (Jones et al., 2006), for example, contained many of the SH2 domain-mediated interactions already known for this receptor, it also predicted a larger number of interactions not identified in prior studies with intact EGFR. It is tempting to assume that many such interactions were simply missed in the earlier more targeted, lower-throughput studies of EGFR. However, it is equally possible that these additional hypothetical interactions are not physiologically relevant and arise because comparative microarray-based binding studies were undertaken with short peptides rather than the intact proteins.
Recent structural studies with SH2 domains provide important insights into this issue and suggest that studies with short peptides may indeed miss crucial specificity determinants. Bae et al. (2009) reported the first structure of an SH2 domain from an RTK substrate bound to a phosphorylated RTK, rather than to a phosphopeptide. A fragment of PLCγ1 containing both SH2 domains was crystallized in complex with the phosphorylated FGFR1 intracellular domain. Although the two PLCγ1 SH2 domains bind with similar affinities to phosphopeptides corresponding to FGFR1 pY766, the N-terminal SH2 domain binds >15 times more strongly than the C-terminal SH2 to phosphorylated FGFR1 intracellular domain (Bae et al., 2009). The KD for PLCγ1 N-terminal SH2 domain binding to phosphorylated FGFR1 intracellular domain is 33nM, compared with the more ‘normal’ KD value for its binding to phosphopeptides of more than one micromolar. Crystallography and mutagenesis studies showed that the enhanced affinity results from a secondary binding site on the N-terminal PLCγ1 SH2 domain that contacts key regions in the C-lobe of the FGFR1 kinase domain. In addition to the canonical ‘two-pronged plug’ binding mode seen in studies of SH2 domain/phosphopeptide complexes (Waksman and Kuriyan, 2004), the N-terminal PLCγ1 SH2 domain uses a separate specificity-determining site that buries almost the same surface area and is crucial for FGFR1-mediated PLCγ1 activation in cells. It will be very important to determine whether or not the case of the PLCγ N-terminal SH2 domain is typical for cellular SH2 domain interactions. This will require extensive future studies of SH2 domain binding to intact protein partners rather than to peptide mimetics.
It is important to note that a similar situation was described for SH3 domains, which are best known for binding, albeit weakly, to peptides that contain PxxP motifs (Li, 2005). The SH3 domains of p47phox and Src family kinases bind a great deal more strongly to their respective intact protein partners than to PxxP-containing peptides. In doing so, they ‘augment’ the canonical PxxP-binding modes with additional contacts that increase affinity and specificity for the intact protein (Li, 2005). Although the key experiments have yet to be performed for many of the other domains, these views of SH2 and SH3 binding to their intact protein targets argue that short synthetic peptides do not represent good mimetics of the physiological interactions that define signaling pathways. Understanding the full spectrum of specificity determinants will be crucial for future systems-level analysis of signaling networks.
RTKs as nodes in complex signaling networks
As the first SH2 domain-containing proteins, Ras exchange factors, MAP kinases, and other proteins involved in RTK signaling were being identified, the prevailing view was that these components comprise linear signaling pathways. A key example that appeared to support this hypothesis was the RTK-Grb2-Sos-Ras/MAP kinase pathway. Genetic experiments further supported this linear-pathway view (Noselli and Perrimon, 2000). However, several key emerging facts clearly suggested that a more complicated and intertwined signaling network must exist for RTKs. First, RTKs such as EGFR have multiple (5–12) autophosphorylation sites that can each recruit different SH2 and PTB domain-containing proteins; second, a given adaptor or scaffold protein can interact with multiple signaling molecules (Pawson, 1995; Schlessinger, 2000). Moreover, as biochemical analyses advanced, it became clear that all pathways previously thought to be linear are in fact highly interconnected into a complex and dynamic signaling network (Figure 4A), with RTKs functioning as key regulatory nodes. An important conundrum arose as different RTKs that elicit quite distinct cellular responses (e.g., proliferation versus differentiation) were shown nonetheless to engage overlapping and similar complements of receptor-proximal components in the cellular signaling network. If both the EGF and NGF receptors activate similar sets of RTK-proximal downstream signaling molecules, how can activating the EGFR lead to cell proliferation whereas activating the NGF receptor promotes neurite outgrowth and differentiation in the same cell (Marshall, 1995)? In other words, what defines the specificity of signaling?
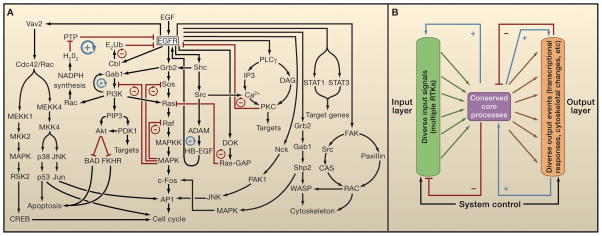
Figure 4. Intracellular signaling networks activated by EGFR.
A. A subset of intracellular signaling components influenced by epidermal growth factor receptor (EGFR) activation are intertwined in a complex network. Through a combination of stimulatory (black arrows) or inhibitory (red lines) signals, several key positive feedback loops (blue circular arrows) and negative feedback loops (red circular arrows) emerge in the network and exert significant influence on its behavior. For example, inhibition of Ras by Ras-GAP or EGFR by protein kinase C (PKC) serve a negative feedback function. On the other hand, H2O2 inhibits protein tyrosine phosphatases (PTPs) and thus, prolongs or increases activity of EGFR by a positive-feedback mechanism. B. A conceptual representation of a ‘bow tie’ or ‘hourglass’ network, as described by Kitano (2004). A wide ‘input layer’ (green) includes multiple RTKs that all influence a relatively small number of ‘core processes’ (magenta), including phosphoinositide 3-kinase (PI3K) signaling, MAPK signaling, and Ca2+ signaling. Feedback processes within the core define specific emergent properties of the system. The behavior of the core processes is ‘read out’ by a wide ‘output layer’ (orange) that consists of diverse transcriptional responses and cytoskeletal changes. Extensive negative and positive feedback loops exists between the core processes and the input layer. Similar feedback exists between the output layer and the core processes, in addition to ‘feed forward’ regulation by core processes (e.g., MAPK signaling) of immediate early gene products described by Murphy and Blenis (2006). An additional layer of ‘system control’ also occurs between the input and output layers.
Simply knowing the components of a signaling pathway or network is not sufficient to predict qualitative outcomes of its activation. Neither is it sufficient to know the components of the RTK signaling node. Instead, a quantitative understanding of how the network behaves as a whole is crucial. Experimentally, this fact is underscored by very early experiments showing that activation of a given RTK by its ligand has dramatically different effects depending on the RTK’s expression level and the kinetics of its activation (Marshall, 1995). Indeed, the dynamics and extent of network activation are critical for determining outcome (Kholodenko, 2006; Murphy and Blenis, 2006), but they are extremely difficult to intuit in a qualitative sense. For example, simply overexpressing EGFR or the insulin receptor in PC12 cells switches the outcome of stimulation with EGF or insulin from a proliferative response with transient MAPK activation (as seen in wild-type PC12 cells) to cell differentiation and neurite outgrowth, with more sustained MAPK activation (Marshall, 1995). This dramatic shift in cellular response is partly due to the differential engagement of positive and negative feedback mechanisms (Santos et al., 2007).
Current systems biology efforts to understand RTK signaling are focused on quantitative analysis of signaling networks. However, this is a very daunting challenge. For example, a map of the signaling network influenced by the EGFR node (Oda et al., 2005) contains 211 reactions involving 322 components (a small subset of which are shown in Figure 4A). Quantitative information is scant for the vast majority of these reactions. Thus, full deterministic modeling is currently impossible, even when spatial and stochastic aspects are ignored, and this is likely to be true for the foreseeable future. Readers are directed to recent reviews (Huang and White, 2008; Lazzara and Lauffenburger, 2009) for detailed discussions of the current challenges and opportunities in combining quantitative proteomics and computational modeling approaches to describe ErbB receptor signaling.
Several key organizational features have emerged from systems views of EGFR/ErbB receptor signaling that provide useful insights into the structure and behavior of the underlying networks. One key concept is the ‘bow-tie’ or ‘hourglass’ structure of the network (Citri and Yarden, 2006; Oda et al., 2005), where diverse inputs and outputs are linked through a conserved ‘processing’ core (Figure 4B). The four ErbB receptors (EGFR, ErbB2, ErbB3 and ErbB4) are regulated by multiple ligands to yield a broad array of signaling inputs into the network. These inputs (and those from other RTKs) converge on a relatively limited set of highly conserved ‘core processes’ (Figure 4B), mirroring the initial impression that a surprisingly small group of downstream signaling intermediates propagate signals from all RTKs. The bow tie or hourglass then widens again as the core processes are linked to the control of transcriptional, cytoskeletal, and other ‘output’ events that define the cellular response (e.g., proliferation, differentiation, apoptosis). Several of the core processes are shown in Figure 4A, including small GTPase cycles, kinase cascades, phosphoinositide signaling, non-receptor tyrosine kinase activities, and ubiquitylation/deubiquitylation cycles. There is significant redundancy and substantial cross talk between the subnetworks or modules that constitute the conserved core processes, and this has been noted as a characteristic feature of robust and evolvable systems (Kitano, 2004).
Unfortunately, even details of the input level of the bow tie network remain poorly understood, making deterministic modeling of the system impossible at present. It is still not clear exactly how EGFR itself is activated. Neither is it clear which ErbB receptors are activated by each of the several ErbB ligands (and to what extent), how ErbB receptors heterodimerize (and with what stoichiometry), or how rapidly different activated ErbB receptor complexes are internalized and disabled. However, conceptualization of network organization has aided and simplified efforts to model ligand/receptor dynamics and has provided substantial insights (Lazzara and Lauffenburger, 2009). Moreover, it has become clear that positive and negative feedback loops, both within and between different levels, play key roles in regulating the ‘information flow’ through the bow tie network, and can explain many emergent properties of the system (Citri and Yarden, 2006; Kholodenko, 2006; Kitano, 2004; Oda et al., 2005).
Positive and negative feedback in RTK signaling networks
Figure 4A depicts selected key elements of the EGFR signaling network, illustrating the substantial redundancy in the core processes as well as the numerous positive and negative feedback loops. Positive feedback increases the sensitivity of the system to signaling inputs by amplifying the stimulus. It can also lead to bistability (Kholodenko, 2006), or ‘switch-like’ ligand-induced transitions between two distinct steady states, leading to differentiation versus proliferation for example. Negative feedback in the network serves in part to dampen noise, thus preventing stochastic minor fluctuations in RTK activation from promoting signaling. Negative feedback in a system can define the steady-state level of a response, keeping it constant over a wide range of signaling inputs and thus imparting substantial robustness to the system (Kitano, 2004; Stelling et al., 2004). Moreover, characteristic oscillations in the system can result from positive feedback loops (with substrate depletion), negative feedback loops, or a combination of the two types of feedback (Kholodenko, 2006). The nature of these oscillations is a characteristic property of the network, and their full understanding requires quantitative modeling of the system.
Positive feedback mechanisms
Phosphorylation/dephosphorylation cycles represent a universal motif in cell signaling (Kholodenko, 2006), and stochastic RTK autophosphorylation is reversed by a variety of protein tyrosine phosphatases (PTPs). In principle, RTK activation can be promoted by ligand-stimulated kinase activity, ligand-inhibited PTP activity, or both. Indeed, it is well known that inhibiting PTPs with pharmacological agents promotes general activation of RTKs in cells (Ostman and Böhmer, 2001). A similar reaction can occur in RTK signaling as a mechanism of positive feedback. Sustained activation of EGFR is accompanied by ligand-induced production of H2O2 and other reactive oxygen species (ROS), mediated by PI-3 kinase and Rac dependent activation of NADPH-oxidase (Bae et al., 1997). These ROS transiently inhibit PTP activity by oxidizing a crucial cysteine in the phosphatase active site (Tonks, 2006). This temporary inactivation of negative regulatory proteins (the PTPs), represents a positive feedback mechanism (Figure 4A) and constitutes a bistable switch. It has been proposed that EGF-induced H2O2 production drives global activation of EGFR molecules in a cell; diffusion of H2O2 through the cells inhibits distant PTPs that activate unoccupied EGFR molecules, resulting in lateral signaling propagation (Reynolds et al., 2003). Significant questions remain about the range of this phenomenon (Schlessinger, 2002). Indeed, ROS have a very limited lifetime and are, therefore, likely to function as ‘messengers’ over only very short distances within the cell. Moreover, it is not clear how such a mechanism would selectively activate EGFR in preference to other phosphorylation-dependent signals.
Another positive feedback loop occurs at the level of Gab1 (Figures 3A and 4A). Gab1 is recruited to activated EGFR (or FRS2 in FGFR signaling) via the Grb2 adaptor. Gab1 becomes tyrosine phosphorylated at sites that recruit the SH2 domains of the PI3K p85 subunit, providing an indirect mechanism for EGFR (or FGFR) to activate PI3K (Figures 3A and 4A). Gab1 also contains an N-terminal PH domain with clear specificity for PI3K products (Gu and Neel, 2003). The PH domain, therefore, promotes PtdIns(3,4,5)P3-dependent translocation of Gab1 to the membrane in response to PI3K activation, creating a positive feedback loop that stimulates Akt-dependent anti-apoptotic signaling.
EGFR-dependent autocrine activation represents a third type of positive feedback loop. This loop is shown in Figure 4A as EGFR-driven activation of Shc and Src, which promotes the ability of ADAM family proteases to cleave membrane-bound heparin-binding EGF-like growth factor (HB-EGF) from the cell surface. The shed HB-EGF drives autocrine EGFR signaling, which is an important factor in several cancers (Hynes and Schlange, 2006). Similarly, in both mammalian and Drosophila systems, activation of the Ras-MAPK pathway induces production of EGFR/ErbB receptor ligands, providing another key positive feedback mechanism (Schulze et al., 2004; Shilo, 2005).
Negative feedback mechanisms
Negative feedback mechanisms operate at many steps in the signaling networks controlled by RTKs. Direct activation of PTPs (as opposed to the transient ROS-dependent inhibition of phosphatases) represents one obvious negative feedback mechanism. The SH2 domain-containing phosphatases Shp1 (PTPN6) and Shp2 (PTPN11) are recruited to activated EGFR and promote its dephosphorylation in a negative feedback loop (not shown in Figure 4A). RTK activity is also attenuated by negative feedback loops that result from receptor-dependent stimulation of heterologous protein kinases. For example, EGFR stimulation promotes activation of protein-kinase-C (PKC) via PLCγ. In turn, PKC can phosphorylate T654 in the juxtamembrane domain of EGFR (Figure 4A). This eliminates high affinity binding of EGF to cell-surface EGFR (Ullrich and Schlessinger, 1990) and thus inhibits EGFR activation in a negative feedback loop.
Several negative feedback mechanisms also control RTK-mediated MAPK-responses, as expected given their central role in the network’s core processes. As shown in Figure 4A, two critical upstream regulators of MAPK (Sos and Raf) are also direct MAPK substrates. Direct phosphorylation of Sos by MAPK impairs Sos/Grb2 interactions and thus reduces Sos recruitment to the membrane, in turn dampening Ras activation (Buday et al., 1995). In addition, MAPK phosphorylates its upstream regulator Raf (Ueki et al., 1994), leading to a reduction in Raf kinase activity and consequently diminished phosphorylation of MAPKK and MAPK. MAPK also phosphorylates docking proteins in another mode of negative feedback that links core processes to the input layer of the bow tie (Figure 4B). MAPK activation leads to Gab1 phosphorylation, which diminishes the ability of Gab1 to recruit and activate PI3K (Gual et al., 2001). The IRS1 and FRS2 docking proteins are also phosphorylated by MAPK in similar negative feedback loops in insulin and FGF receptor signaling, respectively (Lax et al., 2002; Mothe and Van Obberghen, 1996). Moreover, MAP kinase inactivates the EGFR itself, by phosphorylating T669 in the juxtamembrane region (Li et al., 2008). Naturally, another key element of negative feedback in RTK signaling involves downregulation of the receptors following their activation. As discussed below, there remains significant debate about the molecular mechanisms that control RTK internalization and degradation (Sorkin and Goh, 2009; Zwang and Yarden, 2009).
A slower negative feedback mechanism involves signal-dependent transcription of negative regulators of the network. Amit et al. (2007) identified several signal attenuators among the group of ‘delayed early gene’ products induced by EGFR signaling with a delay of more than 40 minutes. These included several known transcriptional repressors, including ID2, NAB2, FOSL1, and JUNB, which attenuate EGF-driven transcription (including that driven by immediate early gene products). Several other delayed early gene products, including Krüppel-like factors (KLF) −2 and −6, were also shown to repress EGF-dependent transcription events when expressed. Still other attenuators encoded by delayed early genes include the RNA-binding protein ZFP36, which may promote degradation of short-lived inducible mRNAs, and several dual-specificity phosphatases (the identity of which was cell- and stimulus-specific), which feed back to attenuate MAP kinase activity in the network (Amit et al., 2007).
There are several other inhibitors of EGFR family signaling, including Sprouty, LRIG-1, Mig6, as well as Argos and Kekkon-1 in Drosophila (Shilo, 2005). LRIG-1, Kekkon-1 and Mig6 appear to inhibit receptor function itself. In contrast, Argos sequesters activating ligand, and Sprouty has diverse effects on downstream signaling (Citri and Yarden, 2006; Shilo, 2005). EGFR signaling induces the expression of each of these inhibitors, but apparently with a longer delay than the delayed early genes (Amit et al., 2007). This is consistent with their importance in coordinating EGFR signaling at the organismal level during development (Shilo, 2005).
Feedback/system control differences explain distinct responses of PC12 cells to EGF and NGF
We began this section on networks with the conundrum that NGF and EGF induce diametrically opposed cellular responses, with very different dynamics, despite engaging a broadly similar set of signaling intermediates (Marshall, 1995). Given the discussion above, it seems reasonable to expect that the distinct cellular responses to NGF and EGF reflect differences in the input layer of the network or its feedback mechanisms. Both appear to be true, and several mechanisms appear to be at play.
Using a combined computational and experimental approach, Bastiaens and colleagues have provided evidence for a difference in the topology of the MAPK network in PC12 cells when activated by NGF and EGF, respectively (Santos et al., 2007). The key difference is additional positive feedback from MAPK to Raf when the system is activated by NGF, in contrast with only negative feedback loops (Figure 4A) in EGF-treated cells. As a result, NGF-induced activation of Raf and MAPK has switch-like, or bistable, properties that are required for the sustained response. Although the mechanism of this NGF-specific positive feedback is not yet clear, it appears to involve PKCδ because combined treatment of cells with EGF and a PKC Activator could mimic NGF-induced sustained MAPK activation (and differentiation).
In addition to this change in ‘wiring’ of the core processes, another key factor appears to be the ability of NGF (but not EGF) stimulation to promote sustained activation of Rap1, a Ras family member (York et al., 1998), leading to sustained B-Raf activation. Studies of TrkA indicate that NGF-induced activation of this receptor promotes the formation of a long-lived signaling complex containing FRS2, the adaptor protein Crk, and the Rap-specific GEF C3G. This allows Rap1 activation by NGF to be prolonged (Kao et al., 2001) whereas Rap1 is only transiently activated by EGFR. Both EGFR and TrkA activate Ras itself only transiently because of negative feedback by Ras-GAP activation (Figure 4A). The sustained MAPK activation seen specifically in response to NGF may arise from the ability of TrkA to promote prolonged Raf-MAPK activation via Rap1.
Through these mechanisms and likely others, the dynamics of activation of the core processes in the bow tie signaling network are dramatically different for NGF and EGF. Activation of the MAPK pathway becomes sustained (rather than transient) when PC12 cells are treated with NGF rather than EGF or when overexpressed EGFR or insulin receptor are stimulated. How, then, is the sustained MAPK pathway activation interpreted by the ‘output layer’ to trigger differentiation rather than proliferation? A key here appears to be feed-forward mechanisms (Figure 4B) between the ‘core’ MAP kinase pathway and the ‘output layer’ products of the immediate early genes (Murphy and Blenis, 2006). Several immediate early gene products, such as c-Fos are inherently unstable, and are rapidly degraded. MAP kinase-dependent phosphorylation of c-Fos, directed by a DEF domain in c-Fos, stabilizes the protein and prolongs its existence. This appears also true for several immediate early gene products (Murphy and Blenis, 2006). If MAP kinase activation is transient and has waned by the time that c-fos is transcribed and translated, newly synthesized c-Fos will not be phosphorylated and will be rapidly degraded. In contrast, if MAP kinase activation is sufficiently sustained that significant kinase activity remains when new c-Fos appears, then the nascent c-Fos will be stabilized by phosphorylation, allowing it induce expression of late-response genes. Immediate early gene products thus appear to serve as ‘sensors’ or ‘interpreters’ for MAP kinase signaling dynamics (Murphy and Blenis, 2006).
Receptor endocytosis in down-regulation and signaling
An early and general response in the activation of all cell surface receptors, including RTKs, is receptor “down-regulation”. This involves ligand-stimulated endocytosis of occupied receptors and subsequent intracellular degradation of both ligand and receptor molecules (Sorkin and Goh, 2009; von Zastrow and Sorkin, 2007; Zwang and Yarden, 2009). Activated EGFR and other RTKs are internalized primarily by clathrin-mediated endocytosis following growth factor-induced receptor clustering in clathrin coated-pits. In addition, clathrin-independent endocytosis can play an important role under particular conditions, such as cell stimulation with high ligand concentrations (Sadowski et al., 2009; Sorkin and Goh, 2009).
EGFR kinase activity is required for trafficking through multi-vesicular bodies (MVBs) into lysosomes where both EGF and EGFR are degraded (Sorkin and Goh, 2009). It has typically been assumed that RTK activation and cell signaling take place primarily at the cell surface, and that the primary role of endocytosis is to terminate RTK activation. However, several studies show that activated RTKs continue to recruit and activate intracellular signaling pathways from within intracellular vesicles following their internalization (Di Guglielmo et al., 1994; Miaczynska et al., 2004; von Zastrow and Sorkin, 2007). After internalization, activated RTKs are dephosphorylated, ubiquitylated, and the activating ligand becomes dissociated in the lower pH environment of the endosomal lumen. The stage in the endocytic pathway at which these events occur varies with both ligand and receptor. In addition, depending on the specific receptor and ligand pair, RTKs may be recycled from endosomes to the plasma membrane or sorted for degradation, which effects receptor number and thus signaling. As seen for the TrkA and TrkB neurotrophin receptors (Chen et al., 2005), these types of differences can profoundly alter biological response.
Intriguingly, it has become clear that the signaling molecules associated with an activated receptor can be significantly different at the plasma membrane compared with that seen at endosomal (and other) compartments. This adds a key spatial dimension, not captured in Figure 4A, to what is required to understand signaling networks. For example, one recent study found that clathrin-mediated endocytosis is required for Akt and MAPK activation by EGFR (presumably in endosomes) but not for Shc phosphorylation. The authors also suggested that clathrin-mediated endocytosis prolongs the duration of EGFR signaling by directing the receptor toward a recycling fate, rather than a degradative one (Sigismund et al., 2008). In signaling by the RTK Met, trafficking to a perinuclear endosomal compartment appears to be required for tyrosine phosphorylation of STAT3 and its nuclear translocation but not for MAP kinase activation (Kermorgant and Parker, 2008). Experiments with the PDGF receptor that allow it to be activated in endosomes, but not at the plasma membrane, also support the hypothesis of signaling following internalization (Wang et al., 2004). Moreover, anchoring constitutively active forms of Flt3 in the endoplasmic reticulum diminishes activation of Akt and MAP kinase but elevates activation of STAT5 (Choudhary et al., 2009).
These studies all demonstrate that RTKs can transmit intracellular signals from endosomes and other intracellular compartments (Miaczynska et al., 2004). They also illustrate that the precise subcellular location can define signaling specificity in a manner that is not yet understood. Certain RTKs, notably Eph receptors, require internalization for their signaling function (Sadowski et al., 2009). Understanding the spatial aspects of RTK signaling in different membrane compartments will, therefore, be crucial for quantitative understanding and modeling of the complete network (Kholodenko, 2006).
Ubiquitylation of RTKs
It is now well established that protein phosphorylation regulates protein ubiquitylation and conversely, that ubiquitylation regulates the function of a variety of protein kinases and phosphatases. Moreover, crosstalk between these two post-translational modifications plays a critical role in cell signaling by regulating protein degradation, processing and cellular trafficking (Hunter, 2007). One of the RTK-proximal proteins that associates with phosphorylated EGFR and other RTKs is Cbl. This E3 ubiquitin ligase is a modular oncogenic protein that catalyzes ubiquitylation of the cytoplasmic domains of a variety of RTKs.
Several studies have claimed that ubiquitylation serves as a sorting signal for targeting activated EGFR to coated-pits for endocytosis and intracellular degradation. These studies also suggested that mono- (rather than poly-) ubiquitylation is the principal signal for EGFR endocytosis and degradation (Mosesson et al., 2003). More recent quantitative analyses with tandem mass spectrometry have shown that the EGFR cytoplasmic domain is polyubiquitylated for the most part (Huang et al., 2006). Moreover, a mutated EGFR that cannot be ubiquitylated was endocytosed normally following EGF binding at physiological concentrations of growth factor, demonstrating that ubiquitylation is not necessary for EGFR internalization (Huang et al., 2007). This mutated form of EGFR did show impaired intracellular degradation, despite being endocytosed efficiently (Huang et al., 2007). Similarly, deletion mutants of EGFR that are unable to recruit Cbl or B-Cbl (and thus fail to become ubiquitylated) exhibit impaired intracellular degradation and down regulation but are internalized normally (Pennock and Wang, 2008). Thus, it now appears that the main role of ubiquitylation is to target activated EGFR for degradation rather than directing internalization itself. Ubiquitylation thus controls a crucial negative feedback element. There is much debate in the literature about the molecular mechanisms that control RTK internalization and degradation, and this topic has been well reviewed recently from a few different perspectives (Citri and Yarden, 2006; Sorkin and Goh, 2009; Zwang and Yarden, 2009).
RTK mutations in disease
It was appreciated in the 1960s that virally-transformed cells rely less on exogenous growth factors for cell proliferation than their normal cell counterparts (Temin, 1966). This observation suggested that aberrant growth factor signaling might play a key role in cell transformation. Nearly two decades later, studies demonstrated that the v-sis oncogene from simian sarcoma virus originated by viral transduction of the PDGF gene (Doolittle et al., 1983; Waterfield et al., 1983) and that its protein product (p28sis) promotes cell transformation by activating the PDGFR in an autocrine loop. Subsequently, the product of the v-erbB oncogene from avian erythroblastosis virus was found to correspond to a truncated and constitutively activated form of EGFR (Downward et al., 1984). The gene encoding EGFR was later shown to be amplified and mutated in primary human brain tumors (Libermann et al., 1985), leading to overexpression and constitutive activation of EGFR tyrosine kinase activity in tumor tissues.
Since this time, a wealth of information has accumulated implicating deregulated and dysfunctional RTKs in a variety of human diseases. Aberrant RTK activation in human cancers is mediated by four principal mechanisms: autocrine activation, chromosomal translocations, RTK overexpression, or gain-of-function mutations. Recent sequencing efforts in a wide variety of tumors have identified mutations in numerous RTKs, collected in the COSMIC (Catalogue of Somatic Mutations in Cancer) database (Forbes et al., 2010). Examples in the KIT/PDGFR, ErbB, FGF receptor families illustrate key mechanisms of tumorigenic RTK mutations.
Mutations in KIT and PDGFR families
Gain-of-function mutations in KIT have been found in a variety of human cancers, including gastrointestinal-stromal tumors (GIST), acute myeloid leukemia (AML), mast cell leukemia (MCL) and in melanoma. These activating mutations cluster in key locations within the KIT protein (Supplemental Figure 1), namely the TKD (exon 17), the intracellular juxtamembrane segment (exon 11), and in exons 8 and 9 that encode D5 of the extracellular ligand-binding region (Corless and Heinrich, 2008). There is good evidence that mutations in the juxtamembrane domain and TKD found in cancers constitutively activate KIT by relieving the normal cis-autoinhibitory constraints in the TKD. In the KIT extracellular region, gain-of-function mutations in D5 map to the interface formed between two neighboring receptor molecules in a dimer induced by SCF binding (Figure 2B) (Yuzawa et al., 2007). These mutations are thought to stabilize intermolecular interactions between two KIT D5 domains, promoting constitutive receptor-mediated dimerization by sufficiently strengthening receptor-receptor contacts to overcome the need for ligand.
Oncogenic mutations and alterations in the EGFR/ErbB family
EGFR family receptors are overexpressed or mutated in several human cancers. ErbB2, an orphan receptor that fails to form the tethered structure seen in the extracellular regions of other ErbB receptors (Burgess et al., 2003), is highly overexpressed as a result of gene amplification in ~30% of breast cancer patients. In addition, ErbB2 overexpression correlates with poor prognoses (Slamon et al., 1989). Although EGFR is overexpressed in several cancers, amplification of the EGFR gene appears to be restricted to glioblastomas, where it occurs in ~35% of cases and leads to overexpression of both wild-type and mutated forms of the receptor (Libermann et al., 1985). Several single-residue mutations in the EGFR extracellular region (Supplemental Figure 1) were reported in glioblastoma patients (Lee et al., 2006). A subset of these mutations may promote activation of the receptor by weakening the autoinhibitory tether. The most well known EGFR mutant in glioblastoma is variant III (vIII) or Δ2–7 EGFR (Supplemental Figure 1), which lacks residues 6-273 (encoded by exons 2–7) in the extracellular region of the wild type receptor. This deletion encompasses all of Domain I and most of Domain II (Figures 1 and 2), including the key dimerization arm (Burgess et al., 2003). Biochemical studies of vIII EGFR indicate that it has constitutive (but low level) tyrosine kinase activity. Reports of its ability to bind EGF and to dimerize vary (Pedersen et al., 2001), but the absence of the dimerization arm means that it cannot dimerize through the same mechanism as the wild-type receptor. One possibility is that misfolding, due to the large extracellular deletion, causes vIII EGFR to aggregate in the endoplasmic reticulum resulting in constitutive active of the receptor.
Recent studies in non-small cell lung cancer (NSCLC) provided important insights into regulation of the EGFR TKD (Sharma et al., 2007; Zhang et al., 2006). In clinical trials, only ~10% of NSCLC patients responded to EGFR-targeted TKD inhibitors, and these patients showed dramatic initial responses (Lynch et al., 2004). Sequencing of EGFR showed that all responding patients have somatic mutations in key regulatory elements of the EGFR TKD that disrupt normal cis-autoinhibitory interactions (Sharma et al., 2007; Zhang et al., 2006). Each of these EGFR mutations causes constitutive activation of the TKD by mimicking the conformational changes ordinarily promoted by the ‘Activator’ kinase in the asymmetric dimer (Figure 2F). Intracellular juxtamembrane mutations have also been described in EGFR that activate the receptor, apparently by promoting ligand-independent dimerization of the TKD (Red Brewer et al., 2009).
FGFR mutations in cancer and other pathologies
FGFR family members are also mutated in a variety of cancers. Chromosomal translocations, which result in the expression of dimeric (activated) fusion proteins containing the FGFR1 or FGFR3 TKDs, have been identified in lymphoblastic lymphoma, multiple myeloma, peripheral T-cell lymphoma and chronic myelogenous leukemia (CML). Germ line activating FGFR3 mutations (Figure 5) have been identified in 30% of bladder cancer and 25% of cervical carcinoma patients (Eswarakumar et al., 2005). Gain-of-function mutations in FGFR1, FGFR2 and FGFR3 also cause a variety of severe skeletal dysplasias (Eswarakumar et al., 2005).
Whereas activating germ line mutations in FGFRs can cause craniosynostosis, skeletal dysplasia, and many cancers, mutations that impair FGFR functions have a distinct set of important pathological consequences. One example is seen in Kallmann Syndrome; this developmental disease is caused by inactivating FGFR1 mutations and is characterized by hearing loss, cleft palate, and tooth agenesis. Another developmental disease, LADD (lacrimo-ariculo-dento-digital) syndrome is caused by mutations that impair the tyrosine kinase activity of the ‘b’ isoform of FGFR2 or the activity of its ligand FGF10 (Rohmann et al., 2006).
RTKs are important drug targets
Several drugs have been developed and approved by the US Food and Drug Administration (FDA) for treating cancers and other diseases caused by activated RTKs (Table 1). These drugs fall into two categories: small molecule inhibitors that target the ATP-binding site of the intracellular TKD (Shawver et al., 2002) and monoclonal antibodies that both interfere with RTK activation and target RTK-expressing cells for destruction by the immune system (Reichert and Valge-Archer, 2007).
Table 1.
FDA approved small molecule inhibitors and monoclonal antibodies against RTKs for cancer therapy.
| Small Molecule | Mab | Target | Disease | Year of approval |
|---|---|---|---|---|
| Imatinib (Gleevec) | PDGFR, KIT, Abl, Arg | CML, GIST | 2001 | |
| Gefitinib (Iressa) | EGFR | Esophageal cancer, Glioma | 2003 | |
| Erlotinib (Tarceva) | EGFR | Esophageal cancer, Glioma | 2004 | |
| Sorafenib (Nexavar) | Raf, VEGFR, PDGFR, Flt3, KIT | Renal cell carcinoma | 2005 | |
| Sunitinib (Sutent) | KIT, VEGFR, PDGFR, Flt3 | Renal cell carcinoma, GIST, Endocrine pancreatic cancer | 2006 | |
| Desatinib (Sprycel) | Abl, Arg, KIT, PDGFR, Src | Gleevec-resistant CML | 2007 | |
| Nilotinib (Tasigna) | Abl, Arg, KIT, PDGFR | Gleevec-resistant CML | 2007 | |
| Lapatinib (Tykerb) | EGFR, ErbB2 | Mammary carcinoma | 2007 | |
| Trastuzumab (Herceptin) | ErbB2 | Mammary carcinoma | 1998 | |
| Cetuximab (Erbitux) | EGFR | Colorectal cancer, Head and neck cancer | 2004 | |
| Bevacizumab (Avastin) | VEGF | Lung cancer, Colorectal cancer | 2004 | |
| Panitumumab (Vectibix) | EGFR | Colorectal cancer | 2006 | |
As might be expected for agents that target the ATP-binding site of protein kinases, many small-molecule RTK inhibitors affect multiple tyrosine kinases in addition to their initially intended target. For example, imatinib (Gleevec) was initially identified in a program to develop PDGFR inhibitors, but it also potently inhibits KIT and the non-receptor tyrosine kinase Abl. Imatinib has shown clinical activity in CML, which arises from constitutive tyrosine kinase activity of the aberrant Bcr-Abl fusion protein (Shawver et al., 2002). Imatinib has also been successfully applied to the treatment of gastrointestinal stromal tumors (GISTs), cancers that are primarily driven by constitutively activated KIT. Sunitinib (Sutent) also blocks the tyrosine kinase activities of several RTKs, including KIT, VEGFR2, PDGFR, Flt3 and Ret, and it has been successfully applied in the treatment of GIST and renal cell carcinoma (Chow and Eckhardt, 2007). On the other hand, the EGFR inhibitors erlotinib (Tarceva) and gefitinib (Iressa) show much greater specificity (Shawver et al., 2002) and are capable of selectively inhibiting EGFR under conditions where the closely-related ErbB2 TKD is unaffected. Although several tyrosine kinase inhibitors have been successfully applied to treat different cancers, key difficulties encountered during treatment include side effects that arise from a lack of selectivity towards an individual target and the acquisition of drug resistance.
Monoclonal antibodies that bind to the extracellular domain of ErbB2 (trastuzumab/Herceptin) or EGFR (cetuximab/Erbitux and panitumumab/Vectibix) have been used to treat mammary carcinoma, colorectal cancer, and head and neck cancers, respectively (Reichert and Valge-Archer, 2007). A monoclonal antibody directed against the VEGF ligand (bevacizumab/Avastin) has also been used as an inhibitor of tumor angiogenesis for the treatment of colorectal, lung, and other cancers or diseases in which angiogenesis is important. Notably, the efficacy of the currently approved monoclonal antibodies for cancer therapy as single agents is very limited unless applied in combination with conventional chemotherapeutic agents. Moreover, because of their bivalent characteristics, several therapeutic antibodies that bind to receptors have been shown to function as weak RTK agonists that do not inhibit the activity of oncogenic RTKs with activating mutations in their TKDs.
It is now clear that drug resistance almost invariably develops in cancer patients treated with tyrosine kinase inhibitors or with monoclonal anti-receptor antibodies. Selective pressure leads to the emergence of drug-resistant variants of the targets or to compensations in the signaling networks that overcome the need for the inhibited RTK for continued growth (Engelman and Settleman, 2008; Sergina and Moasser, 2007). Mutations that abrogate the inhibitory activity of tyrosine kinase inhibitors are frequently seen in the TKD of the targeted RTK of tumor cells. For example, resistance of EGFR to inhibition by gefitinib is caused by T766M or T830A mutations (mature EGFR numbering) in the EGFR kinase domains of patients with lung adenocarcinomas (Bean et al., 2008; Sharma et al., 2007). Drug resistance to tumors treated with tyrosine kinase inhibitors or monoclonal antibodies against an RTK target has also emerged from compensatory activation or overexpression of a different RTK or other cell signaling proteins. For example, upregulation of ErbB3 signaling via amplification of the Met gene (Engelman et al., 2007) or through other mechanisms (Sergina and Moasser, 2007) overcomes growth inhibition by EGFR inhibitors. Upregulation of Met expression in breast cancer cells also promotes resistance to the ErbB2-targeted Trastuzumab antibody (Shattuck et al., 2008). These findings and numerous other studies suggest that combining treatments that inhibit ErbB receptors and Met is likely to be a useful strategy in the clinic, either to overcome Met-induced resistance to ErbB-targeted therapy (Engelman and Settleman, 2008) or, conversely, ErbB receptor-induced resistance to Met targeted therapy. However, upregulation of still other RTKs can promote escape from targeted therapy. For example, VEGFR-1 upregulation was found in cell lines with acquired resistance to EGFR-targeted inhibitors (Bianco et al., 2008).
It is anticipated that resistance will eventually develop towards every inhibitor that is targeted against RTKs. A key challenge for the future is to ascertain the range of possible escape mechanisms. One obvious approach to resolving the problem of resistance in the clinic is to apply combinations or series of different agents targeted against RTKs. However, as knowledge of the network properties of RTKs becomes more sophisticated, therapeutic approaches that target the core processes engaged by all RTKs, such as PI3 kinase signaling (Engelman, 2009), may prove to be more robust than RTK-targeted agents alone.
Conclusions and perspectives
Advances in our understanding of RTK signaling over the past quarter century have been dramatic. Basic investigations into the genetics, cellular biology, biochemistry, and structural biology of these receptors have yielded quite a sophisticated view of how this family of proteins functions. These findings have led to the development of numerous important therapeutics and serve as excellent examples of laboratory-driven translational research. Equally important, clinical analyses of RTKs in disease have provided substantial mechanistic insight (i.e., reverse-translational research), which in turn has allowed further refinement of therapeutic strategies. The molecular mechanisms underlying activation of RTKs share common themes among the families but differ markedly in their details. Only approximately half of the RTK families (Figure 1) are well understood, and completing this picture is a major challenge for the future; one that will surely provide many new lessons about RTK activation and signaling. At the level of the receptor molecules themselves, the importance of direct receptor cross talk or heterodimerization for signaling specificity remains unclear, as does the exact role played by intracellular trafficking. A quantitative understanding of these issues is another key challenge for the future.
Finally, as we appreciate the spatial, temporal and chemical complexity of the signaling networks controlled by RTKs, it becomes increasingly clear that a quantitative biochemical understanding of the behavior of the systems is crucial for predicting qualitative outcomes. Although genetic and biochemical approaches have defined many signaling components in RTK networks, the next major challenge is to put these components into mechanistic and quantitative contexts. This information can then be used to develop new effective treatments for cancers and other diseases driven by activated RTKs.
Supplementary Material
Supplemental Figure 1. RTK mutations in diseases.
Locations of gain-of-function (green arrows) and loss of function (red arrows) mutations in epidermal growth factor receptor (EGFR) (left), KIT (middle), and fibroblast growth factor receptor (FGFR) (right). Receptors are depicted as in Figure 1.
EGFR Mutations in the EGFR tyrosine kinase domain (TKD) relieve autoinhibitory interactions and constitutively elevate kinase activity. These mutations were identified in patients with non-small cell lung carcinoma (Sharma et al., 2007). Additional activating mutations are found in the juxtamembrane region (Red Brewer et al., 2009). Point mutations in Domains I, II and III of the extracellular region were also identified in glioblastoma patients (Libermann et al., 1985; Pedersen et al., 2001). In addition, variant III (vIII) or Δ2–7 EGFR, which lacks Domain I and the majority of Domain II was identified in glioblastoma patients.
KIT Gain-of-function mutations in KIT have been identified in gastrointestinal stromal tumors (GIST), acute myeloid leukemia (AML) and in melanoma. Activating mutations in the juxtamembrane region release the autoinhibitory constraints and constitutively elevate tyrosine kinase activity. The oncogenic mutations in the KIT D5 extracellular region enhance contacts between neighboring KIT molecules, resulting in enhanced tyrosine kinase activity. Loss-of-function mutations in KIT give rise to the disorder Piebaldism. Examples include point mutations in the catalytic domain that impair tyrosine kinase activity and point mutations in D2 of the extracellular region that compromise stem cell factor binding.
FGFRs Gain-of-function FGFR1, FGFR2 and FGFR3 have been identified in severe skeletal dysplasias such as Crouzon, Jackson-Weiss, Apert and Pfeiffer Syndromes. Similar activating mutations were identified in bladder cancer, cervical carcinoma and in multiple myeloma among other cancers. Activating mutations in the extracellular region are primarily seen in D3 of FGFR2; these result in aberrantly disulfide-linked and activated FGFR2 dimers. Other point mutations in the FGFR2 extracellular region enhance fibroblast growth factor (FGF) binding or alter FGF selectivity. Activating mutations in the cytoplasmic domain enhance tyrosine kinase activity of FGFR2 and FGFR3. Loss-of-function FGFR2 mutations in the TKD were identified in Lacrimo-Auriculo-Dento-Digital Syndrome. A loss-of-function point mutation in D3 of FGFR1C that impairs its ability to bind FGF8 was also shown to be responsible for the autosomal dominant Kallmann Syndrome.
Supplementary Figure 1. Receptor tyrosine kinase families.
The scheme depicts all 58 human RTKs. Key domain types are immunoglobulin-like and fibronectin-III domains (blue and orange rectangles respectively), Frizzled-related (Fz) domains (mauve rectangles), solenoid domains (green rectangles containing coils), and cysteine-rich domains (yellow rectangles). Ryk also contains a WIF domain (gray oval). The extracellular region of each receptor is connected by a single transmembrane helix (black) to the cytoplasmic region (red) that contains the tyrosine kinase domain together with regulatory regions. Splits in the intracellular kinase domains represent the kinase-insert domain. The four members of the ErbB receptor family are EGFR, ErbB2, ErbB3, and ErbB4. The insulin receptor (InsR) family has three members: InsR, IGF1R and the InsR-related receptor (InsRR). The PDGF receptor family has 5 members: PDGFRα, PDGFRβ, CSF1R/Fms, KIT/SCFR, and Flt3/Flk2. The three VEGF receptors are VEGFR1/Flt1, VEGFR2/KDR and VEGFR3/Flt4. The four FGF receptors are FGFR1–4, the three Trks are TrkA, TrkB, and TrkC, and the two Ror receptors (for orphan receptor tyrosine kinase) are Ror1 and Ror2. The Met family has two members (Met and Ron), and Axl, Mer and Tyro3 make up the three Axl family RTKs. Tie1 and Tie2 make up the Tie family, and the Eph receptors have fourteen members, named EphA1–8, EphA10, EphB1–4 and Eph6. The DDR (for discoidin domain receptor) family has two members – DDR1 and DDR2, and the LMR family (for lemur) has three members (LMR1–3). Finally, ALK and LTK make up the two members of the ALK (Anaplastic lymphoma kinase) family.
Acknowledgments
This work was supported by NIH grants R01-AR051448, R01-AR051886 and P50-AR054086 to J.S. and R01-CA079992, R01-CA096768 and R01-CA125432 to M.A.L. We thank members of the Lemmon and Schlessinger labs, as well as Kathryn Ferguson, for helpful comments on the manuscript, and Mary Leonard for expert assistance with figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Bae JH, Lew ED, Yuzawa S, Tomé F, Lax I, Schlessinger J. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell. 2009;138:514–524. doi: 10.1016/j.cell.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, Rhee SG. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:217–221. [PubMed] [Google Scholar]
- Barton WA, Tzvetkova-Robev D, Miranda EP, Kolev MV, Rajashankar KR, Himanen JP, Nikolov DB. Crystal structures of the Tie2 receptor ectodomain and the angiopoietin-2-Tie2 complex. Nat Struct Mol Biol. 2006;13:524–532. doi: 10.1038/nsmb1101. [DOI] [PubMed] [Google Scholar]
- Bean J, Reiley GJ, Balak M, Marks JL, Ladanyi M, Miller VA, Pao W. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco R, Rosa R, Damiano V, Daniele G, Gelardi T, Garofalo S, Tarallo V, DeFalco S, Melisi D, Albini A, et al. Vascular endothelial growth factor receptor-1 contributes to resistance to anti-epidermal growth factor receptor drugs in human cancer cells. Clin Cancer Res. 2008;14:5069–5080. doi: 10.1158/1078-0432.CCR-07-4905. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci U S A. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buday L, Warne PH, Downward J. Downregulation of the Ras activation pathway by MAP kinase phosphorylation of Sos. Oncogene. 1995;11:1327–1331. [PubMed] [Google Scholar]
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TPJ, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Ieraci A, Tanowitz M, Lee FS. A novel endocytic recycling signal distinguishes biological responses of Trk neurotrophin receptors. Mol Biol Cell. 2005;16:5761–5772. doi: 10.1091/mbc.E05-07-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Böhmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Müller-Tidow C, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell. 2009;36:326–339. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Chow LQ, Eckhardt SG. Sunitinib: from rational design to clinical efficacy. J Clin Oncol. 2007;25:884–896. doi: 10.1200/JCO.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- Cobb MH, Sang BC, Gonzalez R, Goldsmith E, Ellis L. Autophosphorylation activates the soluble cytoplasmic domain of the insulin receptor in an intermolecular reaction. J Biol Chem. 1989;264:18701–18706. [PubMed] [Google Scholar]
- Corless CL, Heinrich MC. Molecular pathobiology of gastrointestinal stromal sarcomas. Annu Rev Pathol. 2008;3:557–586. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ, Dilworth SM, Mol CD. Switching on kinases: oncogenic activation of BRAF and the PDGFR family. Nat Rev Cancer. 2004;4:718–727. doi: 10.1038/nrc1434. [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Hunkapiller MW, Hood LE, Devare SG, Robbins KC, Aaronson SA, Antoniades HN. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983;221:275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- Eck MJ, Pluskey S, Trub T, Harrison SC, Shoelson SE. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature. 1996;379:277–280. doi: 10.1038/379277a0. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Mol Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucl Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Gadella TW, Jr, Jovin TM. Oligomerization of epidermal growth factor recptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol. 1995;129:1543–1558. doi: 10.1083/jcb.129.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TPJ, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- Gu H, Neel BG. The “Gab” in signal transduction. Trends Cell Biol. 2003;13:122–130. doi: 10.1016/s0962-8924(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Gual P, Giordano S, Anguissola S, Parker PJ, Comoglio PM. Gab1 phosphorylation: a novel mechanism for negative regulation of HGF receptor signaling. Oncogene. 2001;20:156–166. doi: 10.1038/sj.onc.1204047. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- Honegger AM, Kris RM, Ullrich A, Schlessinger J. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proc Natl Acad Sci U S A. 1989;86:925–929. doi: 10.1073/pnas.86.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci U S A. 2007;104:16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Huang PH, White FM. Phosphoproteomics: unraveling the signaling web. Mol Cell. 2008;31:777–781. doi: 10.1016/j.molcel.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Schlange T. Targeting ADAMS and ERBBs in lung cancer. Cancer Cell. 2006;10:7–11. doi: 10.1016/j.ccr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Yeh BK, Eliseenkova AV, Zhang F, Olsen SK, Igarashi M, Aaronson SA, Linhardt RJ, Mohammadi M. Analysis of mutations in fibroblast growth factor (FGF) and a pathogenic mutation in FGF receptor (FGFR) provides direct evidence for the symmetric two-end model for FGFR dimerization. Mol Cell Biol. 2005;25:671–684. doi: 10.1128/MCB.25.2.671-684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Jaiswal RK, Kolch W, Landreth GE. Identification of the mechanisms regulating the differential activation of the mapk cascade by epidermal growth factor and nerve growth factor in PC12 cells. J Biol Chem. 2001;276:18169–18177. doi: 10.1074/jbc.M008870200. [DOI] [PubMed] [Google Scholar]
- Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a stratedy employed by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–863. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. Lrp4 Is a Receptor for Agrin and Forms a Complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Knowles PP, Murray-Rust J, Kjaer S, Scott RP, Hanrahan S, Santoro M, Ibáñez CF, McDonald NQ. Structure and chemical inhibition of the RET tyrosine kinase domain. J Biol Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- Ladbury JE, Arold S. Searching for specificity in SH domains. Chem Biol. 2000;7:R3–8. doi: 10.1016/s1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, Schlessinger J. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol Cell. 2002;10:709–719. doi: 10.1016/s1097-2765(02)00689-5. [DOI] [PubMed] [Google Scholar]
- Lazzara MJ, Lauffenburger DA. Quantitative modeling perspectives on the ErbB system of cell regulatory processes. Exp Cell Res. 2009;315:717–725. doi: 10.1016/j.yexcr.2008.10.033. [DOI] [PubMed] [Google Scholar]
- Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppänen VM, Prota AE, Jeltsch M, Anisimov A, Kalkkinen N, Strandin T, Lankinen H, Goldman A, Ballmer-Hofer K, Alitalo K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci U S A. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang Y, Jiang J, Frank SJ. ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cell Signal. 2008;20:2145–2155. doi: 10.1016/j.cellsig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Nusbaum HR, Raxon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangment of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Macdonald-Obermann JL, Pike LJ. The intracellular juxtamembrane domain of the epidermal growth factor (EGF) receptor is responsible for the allosteric regulation of EGF binding. J Biol Chem. 2009;284:13570–13576. doi: 10.1074/jbc.M109.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Pelkmans L, Zerial M. Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang BC, Wilson KP. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Mothe I, Van Obberghen E. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731, modulates insulin action. J Biol Chem. 1996;271:11222–11227. doi: 10.1074/jbc.271.19.11222. [DOI] [PubMed] [Google Scholar]
- Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Niu XL, Peters KG, Kontos CD. Deletion of the carboxyl terminus of Tie2 enhances kinase activity, signaling, and function. Evidence for an autoinhibitory mechanism. J Biol Chem. 2002;277:31768–31773. doi: 10.1074/jbc.M203995200. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Noselli S, Perrimon N. Signal transduction. Are there close encounters between signaling pathways? Science. 2000;290:68–69. doi: 10.1126/science.290.5489.68. [DOI] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Ostman A, Böhmer FD. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001;11:258–266. doi: 10.1016/s0962-8924(01)01990-0. [DOI] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr Opin Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28:3020–3037. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal S, Bornmann WG, Lemmon MA, Cole PA, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Disc. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- Reynolds AR, Tischer C, Verveer PJ, Rocks O, Bastiaens PI. EGFR activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat Cell Biol. 2003;5:447–453. doi: 10.1038/ncb981. [DOI] [PubMed] [Google Scholar]
- Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nürnberg G, Lew ED, Dobbie A, Eswarakumar VP, Uzumcu A, Ulubil-Emeroglu M, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat Genetics. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos P, Saarma M. Neurotrophic factor receptor RET: structure, cell biology, and inherited diseases. Ann Med. 2007;39:572–580. doi: 10.1080/07853890701646256. [DOI] [PubMed] [Google Scholar]
- Sadowski L, Pilecka I, Miaczynska M. Signaling from endosomes: location makes a difference. Exp Cell Res. 2009;315:1601–1609. doi: 10.1016/j.yexcr.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Knyazev PG, Clout NJ, Cheburkin Y, Göhring W, Ullrich A, Timpl R, Hohenester E. Structural basis for Gas6-Axl signalling. EMBO J. 2006;25:80–87. doi: 10.1038/sj.emboj.7600912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee S, Carmillo P, Whitty A. Quantitative analysis of the activation mechanism of the multicomponent growth-factor receptor Ret. Nat Chem Biol. 2006;2:636–644. doi: 10.1038/nchembio823. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. All signaling is local? Mol Cell. 2002;10:218–219. doi: 10.1016/s1097-2765(02)00607-x. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;191:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Schulze A, Nicke B, Warne PH, Tomlinson S, Downward J. The transcriptional response to Raf activation is almost completely dependent on Mitogen-activated Protein Kinase Kinase activity and shows a major autocrine component. Mol Biol Cell. 2004;15:3450–3463. doi: 10.1091/mbc.E03-11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17:398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergina NV, Moasser MM. The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends Mol Med. 2007;13:527–534. doi: 10.1016/j.molmed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Carraway KL, 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- Shawver LK, Slamon D, Ullrich A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell. 2002;1:117–123. doi: 10.1016/s1535-6108(02)00039-9. [DOI] [PubMed] [Google Scholar]
- Shewchuk LM, Hassell AM, Ellis B, Holmes WD, Davis R, Horne EL, Kadwell SH, McKee DD, Moore JT. Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure. 2000;8:1105–1113. doi: 10.1016/s0969-2126(00)00516-5. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. Regulating the dynamics of EGF receptor signaling in space and time. Development. 2005;132:4017–4027. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, Davis S, Goldfarb MP, Glass DJ, Lemke G, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell. 1997;1:25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Cantley LC. ZIP codes for delivering SH2 domains. Cell. 2004;116(2 Suppl):S41–S43. doi: 10.1016/s0092-8674(04)00041-8. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci U S A. 2000;97:49–45. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Stiegler AL, Burden SJ, Hubbard SR. Crystal structure of the agrin-responsive immunoglobulin-like domains 1 and 2 of the receptor tyrosine kinase MuSK. J Mol Biol. 2006;364:424–433. doi: 10.1016/j.jmb.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin HM. Studies on carcinogenesis by avian sarcoma viruses. 3 The differential effect of serum and polyanions on multiplication of uninfected and converted cells. J Natl Cancer Inst. 1966;37:167–175. [PubMed] [Google Scholar]
- Till JH, Becerra M, Watty A, Lu Y, Ma Y, Neubert TA, Burden SJ, Hubbard SR. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Tvorogov D, Carpenter G. EGF-dependent association of phospholipase C-gamma1 with c-Cbl. Exp Cell Res. 2002;277:86–94. doi: 10.1006/excr.2002.5545. [DOI] [PubMed] [Google Scholar]
- Ueki K, Matsuda S, Tobe K, Gotoh Y, Tamemoto H, Yachi M, Akanuma Y, Yazaki Y, Nishida E, Kadowaki T. Feedback regulation of mitogen-activated protein kinase kinase kinase activity of c-Raf-1 by insulin and phorbol ester stimulation. J Biol Chem. 1994;269:15756–15761. [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci Signal. 2008 Sep 2;1:re9. doi: 10.1126/scisignal.135re9. [DOI] [PubMed] [Google Scholar]
- Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Kuriyan J. Structure and specificity of the SH2 domain. Cell. 2004;116(2 Suppl):S45–S48. doi: 10.1016/s0092-8674(04)00043-1. [DOI] [PubMed] [Google Scholar]
- Walton GM, Chen WS, Rosenfeld MG, Gill GN. Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. J Biol Chem. 1990;265:1750–1754. [PubMed] [Google Scholar]
- Wang Y, Pennock SD, Chen X, Kazlauskas A, Wang Z. Platelet-derived growth factor receptor-mediated signal transduction from endosomes. J Biol Chem. 2004;279:8038–8046. doi: 10.1074/jbc.M311494200. [DOI] [PubMed] [Google Scholar]
- Ward CW, Lawrence MC, Streltsov VA, Adams TE, McKern NM. The insulin and EGF receptor structures: new insights into ligand-induced receptor activation. Trends Biochem Sci. 2007;32:129–137. doi: 10.1016/j.tibs.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Waterfield MD, Scrace GT, Whittle N, Stroobant P, Johnsson A, Wasteson A, Westermark B, CHH, Huang JS, Deuel TF. Platelet-derived growth factor is structurally related to the putative transforming protein p28sis of simian saccoma virus. Nature. 1983;304:35–39. doi: 10.1038/304035a0. [DOI] [PubMed] [Google Scholar]
- Wehrman T, He X, Raab B, Dukipatti A, Blau H, Garcia KC. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Ultsch MH, Bass SH, de Vos AM. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot LE, Baskin B, Ong SH, Tong J, Pawson T, Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 2001;106:745–757. doi: 10.1016/s0092-8674(01)00496-2. [DOI] [PubMed] [Google Scholar]
- York RD, Yao H, Dillon T, Ellig CL, Eckert SP, McCleskey EW, Stork PJ. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zwang Y, Yarden Y. Systems biology of growth factor-induced receptor endocytosis. Traffic. 2009;10:349–363. doi: 10.1111/j.1600-0854.2008.00870.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. RTK mutations in diseases.
Locations of gain-of-function (green arrows) and loss of function (red arrows) mutations in epidermal growth factor receptor (EGFR) (left), KIT (middle), and fibroblast growth factor receptor (FGFR) (right). Receptors are depicted as in Figure 1.
EGFR Mutations in the EGFR tyrosine kinase domain (TKD) relieve autoinhibitory interactions and constitutively elevate kinase activity. These mutations were identified in patients with non-small cell lung carcinoma (Sharma et al., 2007). Additional activating mutations are found in the juxtamembrane region (Red Brewer et al., 2009). Point mutations in Domains I, II and III of the extracellular region were also identified in glioblastoma patients (Libermann et al., 1985; Pedersen et al., 2001). In addition, variant III (vIII) or Δ2–7 EGFR, which lacks Domain I and the majority of Domain II was identified in glioblastoma patients.
KIT Gain-of-function mutations in KIT have been identified in gastrointestinal stromal tumors (GIST), acute myeloid leukemia (AML) and in melanoma. Activating mutations in the juxtamembrane region release the autoinhibitory constraints and constitutively elevate tyrosine kinase activity. The oncogenic mutations in the KIT D5 extracellular region enhance contacts between neighboring KIT molecules, resulting in enhanced tyrosine kinase activity. Loss-of-function mutations in KIT give rise to the disorder Piebaldism. Examples include point mutations in the catalytic domain that impair tyrosine kinase activity and point mutations in D2 of the extracellular region that compromise stem cell factor binding.
FGFRs Gain-of-function FGFR1, FGFR2 and FGFR3 have been identified in severe skeletal dysplasias such as Crouzon, Jackson-Weiss, Apert and Pfeiffer Syndromes. Similar activating mutations were identified in bladder cancer, cervical carcinoma and in multiple myeloma among other cancers. Activating mutations in the extracellular region are primarily seen in D3 of FGFR2; these result in aberrantly disulfide-linked and activated FGFR2 dimers. Other point mutations in the FGFR2 extracellular region enhance fibroblast growth factor (FGF) binding or alter FGF selectivity. Activating mutations in the cytoplasmic domain enhance tyrosine kinase activity of FGFR2 and FGFR3. Loss-of-function FGFR2 mutations in the TKD were identified in Lacrimo-Auriculo-Dento-Digital Syndrome. A loss-of-function point mutation in D3 of FGFR1C that impairs its ability to bind FGF8 was also shown to be responsible for the autosomal dominant Kallmann Syndrome.
Supplementary Figure 1. Receptor tyrosine kinase families.
The scheme depicts all 58 human RTKs. Key domain types are immunoglobulin-like and fibronectin-III domains (blue and orange rectangles respectively), Frizzled-related (Fz) domains (mauve rectangles), solenoid domains (green rectangles containing coils), and cysteine-rich domains (yellow rectangles). Ryk also contains a WIF domain (gray oval). The extracellular region of each receptor is connected by a single transmembrane helix (black) to the cytoplasmic region (red) that contains the tyrosine kinase domain together with regulatory regions. Splits in the intracellular kinase domains represent the kinase-insert domain. The four members of the ErbB receptor family are EGFR, ErbB2, ErbB3, and ErbB4. The insulin receptor (InsR) family has three members: InsR, IGF1R and the InsR-related receptor (InsRR). The PDGF receptor family has 5 members: PDGFRα, PDGFRβ, CSF1R/Fms, KIT/SCFR, and Flt3/Flk2. The three VEGF receptors are VEGFR1/Flt1, VEGFR2/KDR and VEGFR3/Flt4. The four FGF receptors are FGFR1–4, the three Trks are TrkA, TrkB, and TrkC, and the two Ror receptors (for orphan receptor tyrosine kinase) are Ror1 and Ror2. The Met family has two members (Met and Ron), and Axl, Mer and Tyro3 make up the three Axl family RTKs. Tie1 and Tie2 make up the Tie family, and the Eph receptors have fourteen members, named EphA1–8, EphA10, EphB1–4 and Eph6. The DDR (for discoidin domain receptor) family has two members – DDR1 and DDR2, and the LMR family (for lemur) has three members (LMR1–3). Finally, ALK and LTK make up the two members of the ALK (Anaplastic lymphoma kinase) family.