Abstract
Genetically-encoded protein photosensors, including the LOV (light, oxygen, voltage) domain, are promising tools for engineering optical control of cellular behavior. We are only beginning to understand how to couple these light detectors to effectors of choice. We report a method that increases the dynamic range of an artificial photoswitch based on the LOV2 domain of A. sativa phototropin1 (AsLOV2). This approach can potentially be used to improve many AsLOV2-based photoswitches.
INTRODUCTION
Photoswitchable proteins offer the unique ability to perturb living cells, tissues, and intact organisms with high spatial and temporal precision1. In particular, genetically encoded photoswitches such as the LOV and phytochrome domains can be conveniently used in many different experimental contexts2–7. The LOV2 domain of A. sativa phototropin 1 (AsLOV2) has proven especially useful for controlling functionally diverse effectors including DNA binding proteins, enzymes, and small GTPases2,3,5. To aid the optimization of these artificial LOV-effector fusions, biophysical investigations are needed to understand how these genetically-encoded light detectors are coupled both to their natural effectors and effectors of our choosing8.
AsLOV2 and other LOV domains were first identified in phototropins, a class of light-activated serine/threonine kinases that mediate phototropism and other blue light responses in plants and algae9,10. LOV domains share the mixed α/β fold common to the Per-ARNT-Sim class of environmental sensors8 and carry a flavin chromophore (either FMN or FAD), that broadly absorbs blue light. While these cofactors are non-covalently associated with the surrounding LOV domain in the dark, photoexcitation generates a covalent adduct between the flavin C4a carbon atom and an invariant cysteine residue11,12. After illumination ceases, this adduct spontaeously decays to the dark state11.
For phototropin LOV domains, the formation of this adduct is associated with kinase activation. In the dark state, the second LOV domain (LOV2) acts as a dark-state repressor of phototropin's kinase activity13. Lit-state adduct formation promotes the undocking and unfolding of the carboxy-terminal Jα helix14,15, but how structural and dynamic changes within the LOV2 domain couple these events to kinase activation has yet to be fully elucidated14,16.
Our understanding of the light-regulated undocking of the Jα helix in AsLOV2 has paved the way for its use as the input component of designed proteins2,3,5. In each of these three engineered proteins, effector activity is suppressed in the dark, concomittant with the Jα helix being in the folded and docked conformation. Although the Jα is mostly docked in the dark state and mostly undocked in the lit state, both conformations are populated in both the dark and lit states (Fig. 1a). The equilibrium between these two populations dramatically shifts between the dark and lit states, providing a large thermodynamic driving force (ΔG ≈ 4 kcal/mol) which can be used to regulate the activity of an effector17.
Figure 1.
Conformational and binding equilibria in AsLOV2 and LovTAP. (a) Photoexcitation (hν) of AsLOV2 (blue) is accompanied by displacement and unfolding of the Jα helix (dark blue). (b) In AsLOV2 (left), photoactivation shifts the equilibrium from mostly docked to mostly undocked. Black lines, dark-state free energy surfaces; dashed lines, lit-state free energy surfaces. Upon fusion with an effector (center), the equilibrium is shifted due to interactions with the effector and the helix is mostly undocked in the dark and lit states. As a consequence, the effector is mostly active in both states. Helix stabilizing mutations specifically stabilize the docked state (ΔG = – RT ln xeff), bringing the equilibrium back into a regime where the effector is mostly inactive in the dark, and photoactivation shifts the equilibrium from mostly inactive to mostly active. (c) Khelix, equlibrium constant of helix undocking for the isolated LOV2 domain. The fusion protein LovTAP is in equilibrium between an inactive conformation (left), in which the shared helix is docked on the LOV domain, and an active conformation (center), in which it is docked on the TrpR domain. Due to competition by TrpR (orange) for the helix, the equilibrium constant of the helix undocking reaction, Khelix, is increased by a factor 1 / xeff. In the active conformation, LovTAP binds DNA with an intrinsic association constant KDNA.
Despite this potentially large driving energy, initial attempts at designed AsLOV2-containing photoswitches have generally suffered from a low photoswitching dynamic range2,3,5. This is exemplified by our design of an end-to-end fusion of AsLOV2 and the E. coli trp repressor (TrpR) that selectively binds DNA when illuminated2. In our initial design, designated "LovTAP," the dynamic range is modest, with only a five-fold change in DNA affinity between the dark and lit states2. Here we present an analytical model where mutations that stabilize LOV-–Jα docking increase the dynamic range of AsLOV2-based photoswitches. We report four such mutations, and demonstrate that these increase the dynamic range of the LovTAP photoswitch to 70-fold.
RESULTS
Analytical model of photoswitching
We sought to identify mutations that improve coupling between the initiating photochemical events at the AsLOV2 chromophore and the resultant activity of an effector domain. Mutations throughout the composite signaling protein (including the FMN binding pocket, the β sheet, the Jα linker, and the effector domain) could potentially improve this coupling. However, it is not obvious what type of mutations to make, or in which regions to make them. Furthermore, because our goal is to increase the ratio of lit- to dark-state activity, mutations that preferentially stablilize either the Jα-docked conformation in the dark or the Jα-undocked conformation in the lit state would seem to offer the most benefit. Yet, the task of rationally predicting such mutations or screening for them is daunting.
To narrow the search for such mutations, we developed an analytical model of photoswitching. We focused on how fusion with an effector domain might itself restrict the dynamic range of the photoswitch. In isolated AsLOV2, the Jα helix is normally docked against the LOV core. When AsLOV2 is part of a composite signaling protein, a competition for Jα helix docking is set up between the components15. To create a molecule that signals effectively, fusion with an effector domain should introduce intra- or intermolecular interactions. These new interactions may favor the undocking of the Jα helix from the LOV core, thereby increasing the equilibrium constant of helix undocking by the factor
| Eq. 1 |
where the equilibrium constant of helix undocking is represented by Khelix, while superscripts iso and fus refer to isolated and fused AsLOV2, respectively (Fig. 1a–b). The shift in docking equilibrium, xfus, will depend on the strength of the new interactions in the fused state and on the effector's intrinsic conformational equilibrium.
Bringing the conformational equilibrium between active and inactive states into an appropriate regime (Fig. 1b) is a central challenge of engineering allosteric proteins and nucleic acids18. Photoswitching requires that the effector activity be suppressed in the dark state. However, this suppression is only achieved when the Jα helix is docked to the LOV core. The new interactions introduced by fusion likely favor undocking of the helix, and increase unwanted dark state effector activity. Potentially, the active conformation can predominate even in the dark state, such that the molecule is “always on,” and cannot be further activated by photoexcitation (Fig. 1b).
The undesirable effects of competition for the Jα helix might be mitigated using mutations that stabilize LOV–Jα docking. These mutations would change the equilibrium constant of helix undocking by the factor
| Eq. 2 |
where the superscripts wt and mut refer to wild-type and mutated species, respectively. By definition, for isolated AsLOV2, xmut = 1. For mutations that destabilize the LOV–Jα interaction (e.g. I532E15) xmut < 1. For mutations that stabilize LOV–Jα docking, and thereby reduce dark state effector activity, xmut > 1. To be independent of xfus, xmut should not depend on coupling between AsLOV2 and the effector or on the effector's intrinsic conformational equilibrium, but could depend on the photo-state.
To represent the combined effects of fusion and mutation on LOV–Jα docking, we used the parameter
| Eq. 3 |
Generally, when xeff ≪ 1, the Jα helix is undocked from the LOV domain, and the effector domain is active. Conversely, as xeff increases, LOV–Jα docking increases and effector activity is suppressed (Fig. 1c).
Intuitively, one expects that mutations which increase LOV–Jα docking equally in the lit and dark states would have no net effect on the photoswitching dynamic range (assuming that coupling between LOV–Jα and effector is the same in both states, i.e. that xfus is independent of photo-state). We questioned this supposition. In LovTAP, the DNA binding activity obeys a linked equilibrium model in which helix undocking promotes an active, binding-competent conformation (Fig. 1c). The competition for the Jα helix reduces the observed DNA binding affinity by the fraction of the population that is in the active conformation:
| Eq. 4 |
and
| Eq. 5 |
Under the assumption that xeff is the same in the lit and dark states, the photoswitching dynamic range becomes:
| Eq. 6 |
Thus, for xeff < Kdark helix < Klit helix, β ≈ 1, i.e. nearly all of the effector is active, even in the dark state. Photoexcitation may further shift the equilibrium to the active state, e.g., from 99% active to 99.9% active, but this has a negligible functional effect. On the other hand, for xeff > Klit helix > Kdark helix, the maximum photoswitching dynamic range is obtained as β ≈ Klit helix / Kdark helix. For intermediate values of xeff, however, β increases rapidly with xeff, even assuming that xeff is the same in the lit and dark states (Fig. 1c and Fig. 2). Because β is small for LovTAP (5.3)2, we expect that the protein is near the lower end of this intermediate regime and we should be able to increase β with mutations that stabilize LOV–Jα docking.
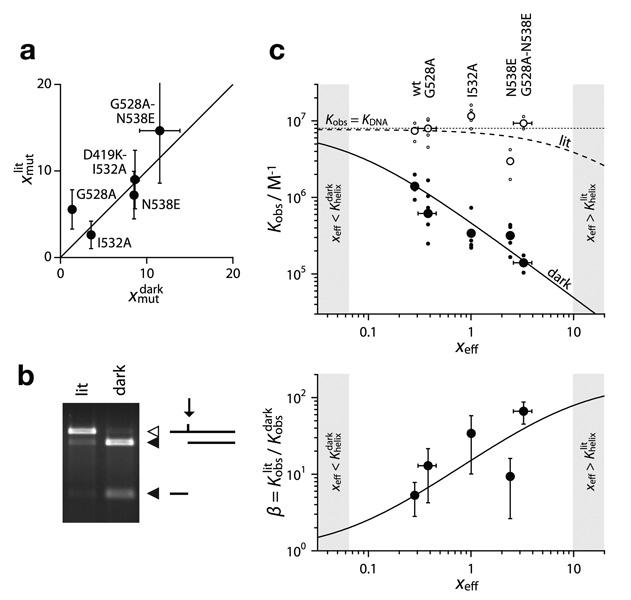
Figure 2.
Mutational stabilization LOV–Jα association and its effect on DNA binding activity in LovTAP. (a) Comparison of CD-derived xlit mut with NMR-derived xdark mut for AsLOV2 with the indicated mutations. The identity line is shown as a reference. (b) Gel image of RsaI protection assay for LovTAP(G528A, N538E). Open and closed triangles denote reactant and product bands, respectively. (c) Effects of the helix-stabilizing mutations on LovTAP. In the upper panel, observed affinity constants (Kobs) are plotted against xeff. Large data points are average Kobs values, and small points show individual measurements. The data are compared to Eqs. 4 and 5 (solid and dashed lines). For wild-type LovTAP, xeff = xfus is calculated using Eq. 3. For mutants, xmut is determined by NMR, and xeff = xmut xfus. In the lower panel, β= Klit obs / Kdark obs is plotted as a function of xeff and compared to Eq. 6. The shaded regions indicate regimes where β is relatively insensitive to changes in xeff.
Mutational stabilization of LOV–Jα docking
We used available computational tools to predict mutations that increase LOV–Jα docking either by increasing helix propensity, new tertiary interactions or increasing the electrostatic complementarity (Supplementary Fig. 1 and online methods). We quantified this stabilization (expressed as xmut in our model) for four point mutations and two tandem mutations using nuclear magnetic resonance (NMR) and circular dichrosim (CD) spectroscopy. The mutations all stabilized helix docking in both the dark and lit states (Fig. 2a, Supplementary Fig. 2 and online methods). Generally the CD-derived xlit mut values were similar to the NMR-derived xdark mut values (Fig. 2a). However, for the G528A mutant and the G528A-N538E tandem mutant, xdark mut ≪ xlit mut. This may refect experimental error in determining these quantities (online methods), or it may reveal that for some mutations xdark mut ≠ xlit mut.
Mutants that increase LOV–Jα affinity lead to improvement in photoswitching
To test how mutations that stabilize LOV–Jα interaction in the isolated AsLOV2 domain influence the dynamic range of the Lov-TAP photoswitch, we used a restriction enzyme protection assay to measure the DNA binding affinity of LovTAP (Fig. 2b)2. We found that the G528A mutation produced a ~ 2-fold decrease in dark-state DNA binding. Similarly, the I532A and N538E mutations each produced a ~ 4-fold decrease in dark state DNA binding (Table 1). Notably, the N538E mutation decreased lit-state DNA binding ~ 2-fold. This decrease may result from the introduction of a negatively charged residue near the DNA binding site of TrpR and highlights the fact that xmut and xfus may not be fully independent. When introduced together, the G528A and N538E mutations decreased dark-state DNA binding affinity by ~ 9-fold, without changing the lit-state binding, and increase β from ~ 5 to ~ 70 (Table 1).
Table 1.
Helix stabilization and DNA binding parameters
| AsLOV2 | LovTAP | ||||||
|---|---|---|---|---|---|---|---|
| pUdark | pUlit | xdarkmut | xlitmut | Kdarkobs | Klitobs | β | |
| (%) | (%) | (105 M−1) | (105 M−1) | ||||
| WT | 5.98 ± 0.08 | 91 ± 6 | 1a | 1a | 14 ± 5 | 70 ± 20 | 5 ± 3 |
| G528A | 4.5 ± 0.9 | 70 ± 10 | 1.3 ± 0.3 | 6 ± 2 | 6 ± 3 | 80 ± 30 | 13 ± 9 |
| I532A | 1.76 ± 0.03 | 80 ± 10 | 3.55 ± 0.08 | 3 ± 2 | 3 ± 2 | 110 ± 30 | 30 ± 20 |
| N538E | 0.74 ± 0.02 | 60 ± 10 | 8.5 ± 0.3 | 7 ± 3 | 3 ± 1 | 30 ± 20 | 9 ± 7 |
| G528A–N538E | N.D. | 40 ± 10 | 12 ± 2b | 15 ± 6 | 1.4 ± 0.4 | 90 ± 20 | 70 ± 20 |
| D419K–I532A | 0.73 ± 0.04 | 50 ± 10 | 8.6 ± 0.5 | 9 ± 3 | N.D. | N.D. | N.D. |
by definition
calculated from G528A and N538E single mutant data
We compared these DNA binding affinities to the prediction of our model. To determine the relationship between predicted and observed affinities for the dark state, we need a reference value of xfus (Eq. 3). In WT LovTAP2, β = 5.3 and, by Eq. 6, xfus = xeff = 0.28. We used this value of xfus as the reference for calculating values of xeff in mutant species. We found that the predicted dark-state DNA binding affinities agreed well with the observed values (Fig. 2c). According to Eq. 6, the lit-state DNA binding affinity becomes independent of xeff when xeff < 10. Consistent with this prediction, all of the mutants had lit-state DNA binding affinities that were approximately equal to the lit-state affinity of the original LovTAP design (Fig. 2c).
Discussion
Fusion of AsLOV2 with an effector introduces new interactions and a competition that depopulates the marginally-stable Jα-docked conformation in the dark state. Because suppression of effector activity requires helix docking, the competition leads to undesired activity in the dark. Conversely, mutations that stabilize the Jα-docked conformation should shift this docking equilibrium back to a regime where the helix is mostly docked in the dark state. From this condition, light can again be used to drive the protein from a largely docked state to a largely undocked state (Figs. 1c and 2c).
Based on this analysis, we identified helix-stabilizing mutations and demonstrated that the mutations generally stabilized the LOV–Jα interaction equally in the dark and lit states. As predicted by our model, these mutations increased the dynamic range of our designed DNA binding photoswitch by up to ~10-fold, to a final lit-to-dark activity ratio of β ≈ 70. Moreover, our model suggested that greater dynamic range can be achieved by further stabilization of the LOV–Jα interaction (Fig. 2c).
A systematic mutational screen of the Jα helix may uncover other stabilizing mutations. However, for xeff ≫ 10, the helix docked, inactive state predominates even in the lit state. In this regime, mutations that equally stabilize LOV–Jα interaction in the dark and lit states no longer offer an increase in photoswitching. Additional improvements require the discovery of mutations that act differentially in the dark and lit states, or the exploitation of dimeric or multimeric systems in ways that offer cooperativity (online methods).
Other designed AsLOV2-based photoswitches also rely on helix docking for dark state suppression of activity3,5. In these designs, as in LovTAP, fusion with the effector domain likely introduces competition for helix docking that increases dark state activity and limits the dynamic range of photoswitching. Because the mutations described here are in the LOV domain itself, they should reduce undesirable dark state effector activity and improve these other designs and other future AsLOV2-based photoswitches.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Munro and A. Möglich for critical reading of the manuscript, S. Crosson and T. Pan for sharing reagents and equipment, and members of the Sosnick, Gardner and Rosen labs for helpful discussions. This work was supported by grants from the NIH (R01 GM081875 to K.H.G.; GM55694 to T.R.S. and GM088668 to T.R.S. and M. Glotzer), the Robert A. Welch Foundation (I-1424 to K.H.G., I-1544 to M.K.R.), and the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust (to T.R.S., M. Glotzer and E. Weiss).
Footnotes
AUTHOR CONTRIBUTIONS:
D.S., designed and performed experiments, analyzed data and wrote the paper; X.Y., designed and performed experiments, analyzed data and wrote the paper; G.G., performed experiments and analyzed data; M.K.R., designed experiments, analyzed data and wrote the paper; K.H.G., designed experiments, analyzed data and wrote the paper; T.R.S., designed experiments, analyzed data and wrote the paper.
References
- 1.Gorostiza P, Isacoff EY. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strickland D, Moffat K, Sosnick TR. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J, et al. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Möglich A, Ayers RA, Moffat K. J. Mol. Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu YI, et al. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 8.Möglich A, Ayers RA, Moffat K. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huala E, et al. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, et al. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 11.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 12.Crosson S, Moffat K. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. Plant Cell. 2009;21:3226–3244. doi: 10.1105/tpc.109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper SM, Neil LC, Gardner KH. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 15.Harper SM, Christie JM, Gardner KH. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 16.Halavaty AS, Moffat K. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 17.Yao X, Rosen MK, Gardner KH. Nat. Chem. Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallée-Bélisle A, Ricci F, Plaxco KW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz V, Serrano L. Nat. Struct. Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 20.Selzer T, Albeck S, Schreiber G. Nat. Struct. Biol. 2000;7:537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 21.Carver JP, Richards RE. J. Magn. Reson. 1972;6:89–105. [Google Scholar]
- 22.Palmer AG, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F, et al. J. Biomol. N.M.R. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA, Blevins RA. J. Biomol. N.M.R. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 25.Loria JP, Rance M, Palmer AG. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- 26.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 27.Grzesiek S, Bax A. J. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 28.Kay LE, Xu GY, Yamazaki T. J. Mag. Resn. A. 1994;109:129–133. [Google Scholar]
- 29.Wittekind M, Mueller L. J. Mag. Resn. B. 1993;101:201–205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.