Abstract
Purpose
Initial studies indicate that the newly developed hCMEC/D3 cell line may prove to be a useful model for studying the physiology of the human blood-brain barrier (BBB) endothelium. The purpose of this study was to assess the mRNA expression of several ABC and SLC transporters, with an emphasis on the Proton-Coupled Oligopeptide Transporter Superfamily (POT) transporters in this immortalized BBB cell model. The transport kinetics of POT-substrates was also evaluated.
Methods
The hCMEC/D3 cell line was maintained in a modified EGM-2 medium in collagenated culture flasks and passaged every 3–4 days at approximately 85%–95% confluence. Messenger RNA (mRNA) expression of a variety of ABC and SLC transporters was evaluated using qRT-PCR arrays, while additional qRT-PCR primers were designed to assess the expression of POT members. The transport kinetics of mannitol and urea were utilized to quantitatively estimate the intercellular pore radius, while POT substrate transport was also determined to assess the suitability of the cell model from a drug screening perspective. Optimization of the cell line was attempted by culturing with on laminin and fibronectin enhanced collagen and in the presence of excess Ca2+.
Results
HCMEC/D3 cells express both hPHT1 and hPHT2, while little to no expression of either hPepT1 or hPepT2 was observed. The relative expression of other ABC and SLC transporters is discussed. While POT substrate transport does suggest suitability for BBB drug permeation screening, the relative intercellular pore radius was estimated at 19Å, significantly larger than that approximated in vivo. Culturing with extracellular matrix proteins did not alter mannitol permeability.
Conclusion
These studies characterized this relevant human hCMEC/D3 BBB cell line with respect to both the relative mRNA expression of various ABC and SLC transporters, and its potential utility as an in vitro screening tool for brain permeation. Additional studies are required to adequately determine the potential to establish an in vivo correlation.
Introduction
Despite favorable central nervous system (CNS) pharmacological properties of various natural and synthetic peptides and peptidomimetics, the potential for clinical development of these compounds and their active analogues into effective drug products remains a major challenge.[1] Part of the difficulty in translating these compounds into new pharmaceutical treatments lies in our current lack of understanding of the basic permeation pathways by which compounds traverse into the CNS, particularly with respect to the underlying transport physiology of the Blood Brain Barrier (BBB). The primary barrier mediating access to the brain, the BBB is primarily composed of the endothelial cells lining the perfusive capillaries.
Cerebral capillary endothelial cells differ from other mammalian capillary endothelial cells in that they exhibit significantly fewer cytoplasmic vesicles, more mitochondria, and a larger number of tight junctional complexes between overlapping cells.[2] Commensurate with its protective role, the BBB is essential to maintaining an optimal chemical environment for proper neural function. The barrier functionality of the BBB is composed of a single layer of endothelial cells, however several anatomical layers exist between the blood and the brain; namely capillary endothelial cells, the basement membrane, consisting of the extracellular matrix proteins collagen, laminin and fibronectin, pericytes embedded in the basement membrane and astrocyte processes that surround the basement membrane.[2] Although the mechanistic pathways to barrier permeation remain very similar to other endothelium, these subtle differences result in a highly specialized membrane that exhibits less endocytotic and pinocytotic activity. As such, a detailed knowledge of the chemical nature of the peptide transport systems present at the BBB is not only important for understanding their potential roles as mediators of essential nutrient transfer, but also as potential mediators of neuroactive/ neurotoxic agents.
In general, it is quite difficult to determine the underlying mechanisms of brain permeation using current experimental models, such as whole animal brain perfusion.[3,4] While a number of human-derived brain endothelial cell lines have been previously developed and described in the literature, these cell lines all exhibit significant shortcomings ranging from failure to exhibit BBB phenotype, to genetic instability.[3,4] Recently a novel, immortalized brain endothelial cell line, hCMEC/D3, derived from a primary cell culture of human origin has been developed and characterized.[5] The hCMEC/D3 cell line is a lentivirus-mediated co-transfect of hTERT (human telomerase catalytic unit) and the SV40 T antigen of primary isolated human brain endothelial cells. This cell line is truly unique in that unlike other brain endothelial cell cultures, it retains much of the morphological and functional characteristics of brain endothelial cells, even without glial cell co-culture. As such, it has been proposed that the hCMEC/D3 cell line may constitute a reliable in vitro model of the human BBB.[5–7] hCMEC/D3 cells have been demonstrated to functionally express three ATP-Binding Cassette (ABC) efflux transporters (P-gp, MRP1 and BCRP), each of which is known to be expressed in the human BBB.[8] In stark contrast however, little is known concerning the relative expression and function of potential influx transporters that may facilitate xenobiotic flux into the brain.
A clearer understanding of peptide transport mechanisms at the BBB may also lead to new avenues of therapeutic utilization of agents to promote treatment of neurological conditions. However, the BBB does exhibit cellular polarization due to the presence of tight junctional complexes, not only limiting potential paracellular diffusion,[9] but also producing an asymmetry of transporter expression between the luminal (blood facing) and abluminal (CNS facing) membranes. These factors highlight the importance of understanding the relationship between a transporter's tissue and cellular expression with it basal physiological function. This understanding is also imperative to effectively delineate strategies to circumvent endogenous drug resistance mechanisms to effectively deliver compounds to their site of therapeutic action. To this end, one of the long-term goals of our laboratory has been to delineate the mechanisms by which oligopeptides and peptide-based pharmaceuticals traverse biological membranes, specifically with respect to the functional activity of members of the Proton-Coupled Oligopeptide Transporter Superfamily (POT; SLC15A).
While the net observable transport of amino acids and di- and tripeptides is mediated by a number of different transporter families, it is generally accepted that the bulk of oligopeptide transport is attributable to the activity of members of the SLC15A superfamily. To date, four mammalian members of the SLC15A superfamily have been identified and functionally described, including Peptide Transporters 1 and 2 [SLC15A1 (PepT1) and SLC15A2 (PepT2), respectively] and the relatively recently identified Peptide/Histidine Transporters 1 and 2 [SLC15A4 (PHT1) and SLC15A3 (PHT2), respectively]. Numerous comprehensive reviews can be found describing members of this extraordinarily important transporter family.[10–16]
The primary goal of our research has been to develop cellular models to delineate the uptake and transport mechanisms of peptides through physiological barriers. To that end, much of our research has been focused on the functional activity of members of the POT superfamily. These studies aimed to assess the relative expression and functional activity of POT members in the hCMEC/D3 cell line to ascertain the validity of using this endothelial cell line to model oligopeptide transport across the BBB. These studies will serve as an initial assessment into the validity of using this model as a tool for not only screening CNS active compounds, but to mechanistically explore the potential routes of CNS permeation in an attempt to aid rational drug design strategies.
Materials and Methods
Materials
Trizol reagent for RNA extraction, the RT-PCR first strand cDNA synthesis kit, and Taq polymerase for end-point PCR reactions were obtained from Invitrogen (Carlsbad, CA). RNA isolation and SYBRII-based Mastermix kits for qRT-PCR reactions were obtained from Stratagene (La Jolla, CA). Hank's Balanced Salt Solution (HBSS), penicillin-streptomycin solution, trypsin and Phosphate Buffered Saline were obtained from Mediatech. EGM-2 growth medium was obtained from Lonza (Walkersville, MD). The West Femto Super Signal Detection kit and the BCA protein quantitation reagents were obtained from Pierce (Thermo Electron, Rockford, IL). All other chemicals, reagents and tissue culture supplies were obtained from Sigma Chemical Company (St. Louis, MO).
Cell Culture
The hCMEC/D3 cell line was kindly donated by Dr. P. Couraud from the Institut Cochin, Université René Descartes, Paris, France. The hCMEC/D3 cell line was cultured as described previously.[5] Briefly, the cells were maintained in a modification of the proprietary Endothelial Growth Medium-2 (EGM-2) from Lonza (Walkersville, MD), containing 2.5% fetal bovine serum, penicillin, streptomycin, 0.1% fibroblast growth factor, 0.01% hydrocortisone and 0.025% each of vascular endothelial growth factor, insulin-like growth factor, and endothelial growth factor, under 37°C and 5% CO2. Cells were passaged into collagenated culture flasks every 3–4 days at approximately 85%–95% confluence.
ABC and SLC Transporter Characterization by Quantitative RT-PCR
Quantitative expression of various ABC and SLC transporters was determined using the RT2 Pathway-Focused Profiler™ Array from SA Biosciences (catalog PAHS-070; Frederick, MD), according to the manufacturer's recommended protocol. The RT2 Profiler™ Array consists of a 96-well panel of pre-validated qRT-PCR primer sets for relevant ABC and SLC drug transporters and suitable housekeeping genes (Table 1). The RT2 Profiler™ system affords the combined benefits of the simplicity and sensitivity of SYBR-based qRT-PCR detection, with the throughput of array-based profiling.
Table 1.
ABC and SLC genes included on the RT2 Profiler™ Array and their relative hCMEC/D3 Expression.
| Unigene | GeneBank | Symbol | Description | Gene Name | hCMEC/D3 Relative Expression [Mean ΔCt × 10−3 ± (S.D.)] |
|---|---|---|---|---|---|
| Hs.429294 | NM_005502 | ABCA1 | ABC, sub-family A (ABC1), member 1 | ABC-1/ABC1 | 4.829 (0.34) |
| Hs.134585 | NM_173076 | ABCA12 | ABC, sub-family A (ABC1), member 12 | DKFZp434G232/ICR2B | 0.069 (0.04) |
| Hs.226568 | NM_152701 | ABCA13 | ABC, sub-family A (ABC1), member 13 | DKFZp313D2411 | 0.005 (0.00) |
| Hs.421202 | NM_001606 | ABCA2 | ABC, sub-family A (ABC1), member 2 | ABC2 | 2.916 (0.16) |
| Hs.26630 | NM_001089 | ABCA3 | ABC, sub-family A (ABC1), member 3 | ABC-C/ABC3 | 7.465 (0.94) |
| Hs.416707 | NM_000350 | ABCA4 | ABC, sub-family A (ABC1), member 4 | ABC10/ABCR | 0.321 (0.08) |
| Hs.131686 | NM_080283 | ABCA9 | ABC, sub-family A (ABC1), member 9 | DKFZp686F2450/EST640918 | 0.195 (0.05) |
| Hs.489033 | NM_000927 | ABCB1 | ABC, sub-family B (MDR/TAP), member 1 | ABC20/CD243 | 58.029 (7.96) |
| Hs.658439 | NM_003742 | ABCB11 | ABC, sub-family B (MDR/TAP), member 11 | ABC16/BRIC2 | 0.039 (0.01) |
| Hs.654403 | NM_000443 | ABCB4 | ABC, sub-family B (MDR/TAP), member 4 | ABC21/GBD1 | 0.059 (0.01) |
| Hs.658821 | NM_178559 | ABCB5 | ABC, sub-family B (MDR/TAP), member 5 | ABCB5alpha/ABCB5beta | No Ct |
| Hs.107911 | NM_005689 | ABCB6 | ABC, sub-family B (MDR/TAP), member 6 | ABC/ABC14 | 8.131 (1.00) |
| Hs.709181 | NM_004996 | ABCC1 | ABC, sub-family C (CFTR/MRP), member 1 | ABC29/ABCC | 31.571 (6.02) |
| Hs.55879 | NM_033450 | ABCC10 | ABC, sub-family C (CFTR/MRP), member 10 | EST182763/MRP7 | 3.562 (0.31) |
| Hs.652267 | NM_032583 | ABCC11 | ABC, sub-family C (CFTR/MRP), member 11 | EWWD/MRP8 | No Ct |
| Hs.410111 | NM_033226 | ABCC12 | ABC, sub-family C (CFTR/MRP), member 12 | MRP9 | 0.031 (0.00) |
| Hs.368243 | NM_000392 | ABCC2 | ABC, sub-family C (CFTR/MRP), member 2 | ABC30/CMOAT | 0.420 (0.05) |
| Hs.463421 | NM_003786 | ABCC3 | ABC, sub-family C (CFTR/MRP), member 3 | ABC31/DKFZp686E22157 | 4.840 (0.78) |
| Hs.508423 | NM_005845 | ABCC4 | ABC, sub-family C (CFTR/MRP), member 4 | EST170205/MOAT-B | 26.918 (4.02) |
| Hs.368563 | NM_005688 | ABCC5 | ABC, sub-family C (CFTR/MRP), member 5 | ABC33/DKFZp686C1782 | 9.798 (0.07) |
|
| |||||
| Hs.442182 | NM_001171 | ABCC6 | ABC, sub-family C (CFTR/MRP), member 6 | ABC34/ARA | 0.090 (0.03) |
| Hs.159546 | NM_000033 | ABCD1 | ABC, sub-family D (ALD), member 1 | ABC42/ALD | 2.971 (0.51) |
| Hs.700576 | NM_002858 | ABCD3 | ABC, sub-family D (ALD), member 3 | ABC43/PMP70 | 64.739 (7.87) |
| Hs.94395 | NM_005050 | ABCD4 | ABC, sub-family D (ALD), member 4 | ABC41/EST352188 | 0.250 (0.10) |
| Hs.655285 | NM_001090 | ABCF1 | ABC, sub-family F (GCN20), member 1 | ABC27/ABC50 | 46.211 (9.71) |
| Hs.480218 | NM_004827 | ABCG2 | ABC, sub-family G (WHITE), member 2 | ABC15/ABCP | 2.401 (0.52) |
| Hs.413931 | NM_022437 | ABCG8 | ABC, sub-family G (WHITE), member 8 | GBD4/STSL | 0.063 (0.02) |
| Hs.76152 | NM_198098 | AQP1 | Aquaporin 1 (Colton blood group) | AQP-CHIP/CHIP28 | 0.136 (0.04) |
| Hs.455323 | NM_001170 | AQP7 | Aquaporin 7 | AQP7L/AQP9 | 0.087 (0.01) |
| Hs.104624 | NM_020980 | AQP9 | Aquaporin 9 | HsT17287/SSC1 | No Ct |
|
| |||||
| Hs.389107 | NM_001694 | ATP6V0C | ATPase, H+ transporting, lysosomal 16kDa, V0 sub c | ATP6C/ATP6L | 83.087 (16.93) |
| Hs.496414 | NM_000052 | ATP7A | ATPase, Cu++ transporting, alpha polypeptide | MK/MNK | 8.677 (0.46) |
| Hs.492280 | NM_000053 | ATP7B | ATPase, Cu++ transporting, beta polypeptide | PWD/WC1 | 1.794 (0.30) |
| Hs.632177 | NM_017458 | MVP | Major vault protein | LRP/VAULT1 | 63.929 (9.04) |
| Hs.952 | NM_003049 | SLC10A1 | SLC 10 (sodium/bile acid cotransporter family), member 1 | NTCP | 0.161 (0.14) |
| Hs.194783 | NM_000452 | SLC10A2 | SLC 10 (sodium/bile acid cotransporter family), member 2 | ASBT/ISBT | No Ct |
| Hs.436893 | NM_005073 | SLC15A1 | SLC 15 (oligopeptide transporter), member 1 | HPECT1/HPEPT1 | 0.052 (0.00) |
| Hs.518089 | NM_021082 | SLC15A2 | SLC 15 (H+/peptide transporter), member 2 | PEPT2 | 0.088 (0.05) |
| Hs.75231 | NM_003051 | SLC16A1 | SLC 16, member 1 (monocarboxylic acid transporter 1) | HHF7/MCT | 10.736 (0.67) |
| Hs.75317 | NM_006517 | SLC16A2 | SLC 16, member 2 (monocarboxylic acid transporter 8) | AHDS/DXS128 | 0.067 (0.01) |
|
| |||||
| Hs.500761 | NM_004207 | SLC16A3 | SLC 16, member 3 (monocarboxylic acid transporter 4) | MCT3/MCT4 | 36.966 (7.17) |
| Hs.84190 | NM_194255 | SLC19A1 | SLC 19 (folate transporter), member 1 | CHMD/FOLT | 3.046 (0.16) |
| Hs.30246 | NM_006996 | SLC19A2 | SLC 19 (thiamine transporter), member 2 | TC1/THT1 | 10.741 (0.77) |
| Hs.221597 | NM_025243 | SLC19A3 | SLC 19, member 3 | THTR2 | 0.026 (0.00) |
| Hs.117367 | NM_003057 | SLC22A1 | SLC 22 (organic cation transporter), member 1 | HOCT1/OCT1 | 0.103 (0.11) |
| Hs.436385 | NM_003058 | SLC22A2 | SLC 22 (organic cation transporter), member 2 | OCT2 | No Ct |
| Hs.567337 | NM_021977 | SLC22A3 | SLC 22 (extraneuronal monoamine transporter), member 3 | EMT/EMTH | 0.784 (0.24) |
| Hs.369252 | NM_004790 | SLC22A6 | SLC 22 (organic anion transporter), member 6 | HOAT1/OAT1 | No Ct |
| Hs.485438 | NM_006672 | SLC22A7 | SLC 22 (organic anion transporter), member 7 | NLT/OAT2 | No Ct |
| Hs.266223 | NM_004254 | SLC22A8 | SLC 22 (organic anion transporter), member 8 | OAT3 | No Ct |
|
| |||||
| Hs.502772 | NM_080866 | SLC22A9 | SLC 22 (organic anion transporter), member 9 | HOAT4/OAT4 | No Ct |
| Hs.459187 | NM_004213 | SLC28A1 | SLC 28 (sodium-coupled nucleoside transporter), member 1 | CNT1/HCNT1 | No Ct |
| Hs.367833 | NM_004212 | SLC28A2 | SLC 28 (sodium-coupled nucleoside transporter), member 2 | CNT2/HCNT2 | 0.082 (0.03) |
| Hs.591877 | NM_022127 | SLC28A3 | SLC 28 (sodium-coupled nucleoside transporter), member 3 | CNT3 | No Ct |
| Hs.25450 | NM_004955 | SLC29A1 | SLC 29 (nucleoside transporters), member 1 | ENT1 | 40.233 (11.21) |
| Hs.569017 | NM_001532 | SLC29A2 | SLC 29 (nucleoside transporters), member 2 | DER12/ENT2 | 1.101 (0.33) |
| Hs.473721 | NM_006516 | SLC2A1 | SLC 2 (facilitated glucose transporter), member 1 | DYT17/DYT18 | 89.189 (12.62) |
| Hs.167584 | NM_000340 | SLC2A2 | SLC 2 (facilitated glucose transporter), member 2 | GLUT2 | 0.044 (0.00) |
| Hs.419240 | NM_006931 | SLC2A3 | SLC 2 (facilitated glucose transporter), member 3 | GLUT3 | 7.200 (0.77) |
| Hs.532315 | NM_001859 | SLC31A1 | SLC 31 (copper transporters), member 1 | COPT1/CTR1 | 37.374 (3.81) |
|
| |||||
| Hs.221847 | NM_018976 | SLC38A2 | SLC 38, member 2 | ATA2/PRO1068 | 259.569 (43.26) |
| Hs.195155 | NM_033518 | SLC38A5 | SLC 38, member 5 | JM24/SN2 | 5.471 (0.41) |
| Hs.112916 | NM_000341 | SLC3A1 | SLC 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 | ATR1/CSNU1 | 32.043 (2.43) |
| Hs.502769 | NM_002394 | SLC3A2 | SLC 3 (activators of dibasic and neutral amino acid transport), member 2 | 4F2/4F2HC | 128.694 (16.80) |
| Hs.1964 | NM_000343 | SLC5A1 | SLC 5 (sodium/glucose cotransporter), member 1 | D22S675/NAGT | 0.025 (0.00) |
| Hs.130101 | NM_014227 | SLC5A4 | SLC 5 (low affinity glucose cotransporter), member 4 | SAAT1/SGLT3 | 0.229 (0.01) |
| Hs.489190 | NM_014251 | SLC25A13 | SLC 25, member 13 (citrin) | ARALAR2/CITRIN | 19.461 (2.26) |
| Hs.390594 | NM_014331 | SLC7A11 | SLC 7, (cationic amino acid transporter, y+ system) member 11 | CCBR1/xCT | 12.157 (0.64) |
| Hs.513797 | NM_003486 | SLC7A5 | SLC 7 (cationic amino acid transporter, y+ system), member 5 | 4F2LC/CD98 | 29.632 (3.41) |
| Hs.653193 | NM_003983 | SLC7A6 | SLC 7 (cationic amino acid transporter, y+ system), member 6 | DKFZp686K15246/LAT-2 | 26.364 (5.09) |
|
| |||||
| Hs.513147 | NM_003982 | SLC7A7 | SLC 7 (cationic amino acid transporter, y+ system), member 7 | LAT3/LPI | 0.879 (0.24) |
| Hs.632348 | NM_182728 | SLC7A8 | SLC 7 (cationic amino acid transporter, y+ system), member 8 | LAT2/LPI-PC1 | 0.092 (0.02) |
| Hs.408567 | NM_014270 | SLC7A9 | SLC 7 (cationic amino acid transporter, y+ system), member 9 | BAT1/CSNU3 | 0.031 (0.00) |
| Hs.46440 | NM_005075 | SLCO1A2 | Solute carrier organic anion transporter family, member 1A2 | OATP/OATP-A | No Ct |
| Hs.449738 | NM_006446 | SLCO1B1 | Solute carrier organic anion transporter family, member 1B1 | LST-1/LST1 | No Ct |
| Hs.504966 | NM_019844 | SLCO1B3 | Solute carrier organic anion transporter family, member 1B3 | LST-3TM13/LST3 | No Ct |
| Hs.518270 | NM_005630 | SLCO2A1 | Solute carrier organic anion transporter family, member 2A1 | OATP2A1/PGT | 14.307 (1.98) |
| Hs.7884 | NM_007256 | SLCO2B1 | Solute carrier organic anion transporter family, member 2B1 | DKFZp686E0517/OATP-B | 0.318 (0.08) |
| Hs.311187 | NM_013272 | SLCO3A1 | Solute carrier organic anion transporter family, member 3A1 | OATP-D/OATP3A1 | 0.756 (0.19) |
| Hs.235782 | NM_016354 | SLCO4A1 | Solute carrier organic anion transporter family, member 4A1 | OATP-E/OATP1 | 6.245 (1.07) |
|
| |||||
| Hs.352018 | NM_000593 | TAP1 | Transporter 1, ABC, sub-family B (MDR/TAP) | ABC17/ABCB2 | 5.281 (0.70) |
| Hs.502 | NM_000544 | TAP2 | Transporter 2, ABC, sub-family B (MDR/TAP) | ABC18/ABCB3 | 48.582 (6.78) |
| Hs.519320 | NM_003374 | VDAC1 | Voltage-dependent anion channel 1 | PORIN/PORIN-31-HL | 163.857 (5.33) |
| Hs.355927 | NM_003375 | VDAC2 | Voltage-dependent anion channel 2 | FLJ23841 | 304.554 (30.63) |
| Hs.534255 | NM_004048 | B2M | Beta-2-microglobulin | B2M | Housekeeper 1 |
| Hs.412707 | NM_000194 | HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | HGPRT/HPRT | Housekeeper 2 |
| Hs.523185 | NM_012423 | RPL13A | Ribosomal protein L13a | RPL13A | Housekeeper 3 |
| Hs.592355 | NM_002046 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | G3PD/GAPD | Housekeeper 4 |
| Hs.520640 | NM_001101 | ACTB | Actin, beta | PS1TP5BP1 | Housekeeper 5 |
| N/A | SA_00105 | HGDC | Human Genomic DNA Contamination | HIGX1A | N/A |
|
| |||||
| N/A | SA_00104 | RTC | Reverse Transcription Control | RTC | N/A |
| N/A | SA_00104 | RTC | Reverse Transcription Control | RTC | N/A |
| N/A | SA_00104 | RTC | Reverse Transcription Control | RTC | N/A |
| N/A | SA_00103 | PPC | Positive PCR Control | PPC | N/A |
| N/A | SA_00103 | PPC | Positive PCR Control | PPC | N/A |
| N/A | SA_00103 | PPC | Positive PCR Control | PPC | N/A |
In short, qRT-PCR-quality total RNA was extracted from hCMEC/D3 cells seeded at 2 × 105 cells/cm2 onto collagenated 6-well plates at passage 38, using the Absolutely RNA® isolation kit (Stratagene, Inc.). Growth medium was changed the day after seeding and RNA extraction was performed on day 2 post-seeding. The seeding and growth protocol is meant to mimic that used for Permeability studies, as outlined below. After verifying RNA integrity by agarose separation, first strand cDNA synthesis was accomplished using the SA Biosciences recommended cDNA Synthesis kit (C-03), according to the manufacturer's instructions. PCR reactions were performed on a Stratagene MX3000P Thermocycler, using Stratagene Brilliant II PCR Mastermix (La Jolla, CA), using the optimized conditions defined by SA Biosciences. The raw data was analyzed using the SA Bioscience online RT2 Profiler™ analysis software to determine the relative limit of detection, based on DNA-contaminant controls included on the array. Results are reported as the mean ΔCt ± standard deviation of three replicates (arrays), normalized to the mean Ct of the five housekeeping genes.
Since our laboratory is also keenly interested in characterizing the relative expression and function of all POT members in the cell line, qRT-PCR primers for hPHT1 and hPHT2 were designed separately using Primer3 software (Table 2), as these genes were not included on the validated array. After primer set optimization, PCR experiments were run as above, where relative expression was determined as the mean ΔCt ± standard deviation of three replicates, normalized to the mean Ct of the housekeeping gene GAPDH.
Table 2.
Primers utilized for PHT qRT-PCR analysis and their relative hCMEC/D3 expression.
| Amplicon | Genebank | qRT-PCR Primers | hCMEC/D3 Relative Expression [Mean ΔCt × 10−3 ± (S.D.)] | |
|---|---|---|---|---|
| hPHT1 | NM_145648 | Sense: | CCAACATCACGCCCTTCG | 0.764 (0.05)† |
| Antisense: | ACAGTGGGGATCGCATAA | |||
|
| ||||
| hPHT2 | NM_016582 | Sense: | TGCTGGTGGTGGCGTTTATTCA | 1.021 (0.34)† |
| Antisense: | TGGAAGTTGGCGATGTCCTCTT | |||
|
| ||||
| GAPDH | NM_002046 | Sense: | TTCGACAGTCAGCCGCATCTTCTT | N/A |
| Antisense: | GCCCAATACGACCAAATCCGTTGA | |||
Relative expression of SLC15A4 and SLC15A3 were determined separate from the SA Biosciences RT2 Profiler™ Array, as they are not included on the validated array. Relative expression reported is the mean ΔCt of three individual replicates, normalized to the housekeeping gene GAPDH.
Permeability Studies
All transport studies were carried out for 2 hours in triplicate to ascertain the transport characteristics of known POT substrates in wild type hCMEC/D3 cells. Studies were performed in 12 mm tissue culture treated, collagen-coated polyester membranes (0.4 μm pore size Transwells, Corning Costar). Per previous methodology,[5] cells were seeded at a confluent density of 2 × 105 cells/cm2. Growth medium was changed 24 hours post-seeding and transport experiments were conducted on day 2 post-seeding. On the day of the study, the culture medium was removed and the cells were washed twice in pre-warmed transport buffer [Hank's Balanced Salt Solution (HBSS), pH 7.4, 37°C]. The cells were then equilibrated in transport buffer for 15 minutes prior to study initiation. A working buffer solution consisting of 1μCi/mL substrate in pre-warmed HBSS was then added to the appropriate chamber for each respective study and the transport characteristics of known POT substrates (glycylsarcosine, carnosine, histidine and valacyclovir) were determined. The cells were maintained on a rocker platform at 37°C. The transport kinetics of various POT model substrates were determined at 15, 30, 45, 60, 90 and 120 minutes. [14C] Mannitol and transepithelial electrical resistance (TEER) were used to monitor cellular monolayer integrity. Apparent permeability coefficients were determined using the following equation:
| (1) |
Where dQ/dt is the steady state appearance rate in the receiver compartment, C0 is the initial concentration in the donor compartment and A is the surface area of exposed membrane (cm2). Sink conditions were maintained throughout each study.
In order to control for bulk flow differences and the effects of permeability resistances arising from the study system, the apparent permeability of each compound across collagenated filter supports without cells were also determined. These permeabilities were used to to focus the transport kinetics on the compound permeability across the hCMEC/D3 monolayer itself, per Equation 2, below:
| (2) |
Where Papp is the apparent permeability across a cellular monolayer as calculated by (1), PM is the permeability across the monolayer itself and PSYS is the permeability contribution derived from the study system, such as the aqueous boundary and collagen layers, the filter support, etc.
Effective Intercellular Pore Radius
The intercellular pore radius was determined using the Renkin molecular sieving function, as previously described.[9,17,18] Briefly, the transport characteristics of mannitol and urea, two paracellular markers, were determined in the wild type cells. Using the effective permeability coefficients as calculated above (Equation 1, as corrected per Equation 2), and the molecular radii, as calculated per Stokes-Einstein equation for equivalent spheres, the effective pore radius can be determined by:
| (3) |
| (4) |
(1) Assuming a single pore model, the dimensionless Renkin molecular sieving function compares the molecular radius (r) and the cylindrical pore radius (R) and takes values of 0 < F(r/R) < 1. The aqueous pore radius was calculated from (Equation 4) using the ratio of the paracellular permeabilities of [14C]-Mannitol and [14C]-Urea.
Given the previous difficulties with the relative leakiness of this cellular model, mannitol permeability was also determined for cells grown on a combination of the extracellular basement membrane (ECM) proteins, laminin and fibronectin (Sigma Chemical Co.). The ECM studies were performed in an attempt to decrease the intercellular pore radius by altering cellular growth characteristics, using the estimated in vivo concentrations.
Results
ABC and SLC Transporter Expression
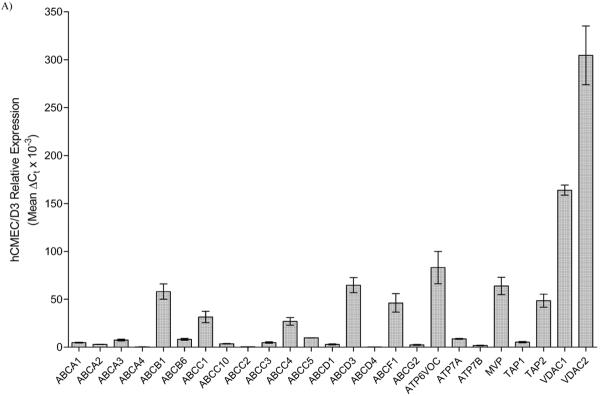
In order to discern the net contribution of any individual transporter to net substrate flux across cultured hCMEC/D3 cells, we first had to ascertain the transporter population expression. Here, we utilized the RT2 Drug Transporter Profiler™ array from SA Biosciences to quantitate transporter expression, relative to various housekeeping genes. Results presented numerically in Table 1 and graphically in Figure 1, indicate expression of various ABC transporters, with the highest expression observed for ABCB1 (MDR1/P-gp), ABCC1 (MRP1), ABCC4 (MRP4) and ABCC5 (MRP5). While ABCG2 (BCRP) is expressed in the cell line, the expression of other MDR and MRP members demonstrate appreciably higher expression, and thus may be more significant with respect to BBB permeation. Also interesting is the relatively high expression of Major Vault Protein (MVP/LRP), which has been demonstrated to act in multidrug resistance, as well as regulation of a number of other important cell signaling pathways.[19]
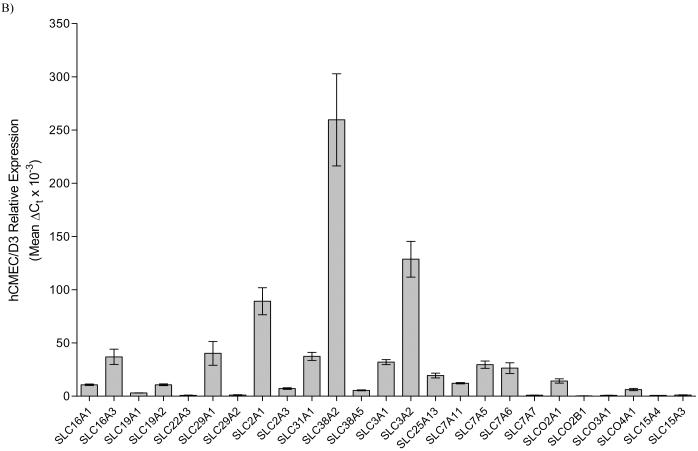
Figure 1.
Relative expression of various ABC (panel A) and SLC (panel B) transporters in hCMEC/D3 cells as determined by qRT-PCR. Relative expression was determined using the ΔCt method, normalized to five housekeeping genes. In the case of SLC15A4 and SLC15A3, expression was determined using our primer sets specific for each gene, normalized to GAPDH. A relative expression level cutoff was arbitrarily set at ΔCt greater than 0.25 for this illustration.
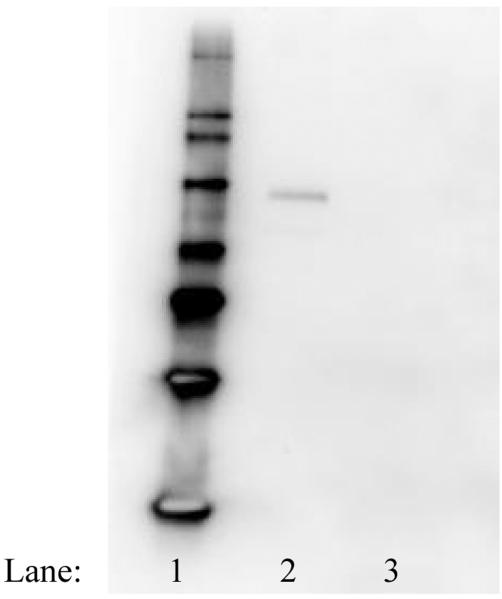
Since our laboratory is predominantly interested in members of the SLC15A family, we also designed primers specific to SLC15A4 and SLC15A3, as these were not included on the validated array. Although not standardized to multiple housekeeping genes as with the array data, comparisons can be made between the relative expressions as the raw Ct values for the housekeeper chosen (GAPDH) were the same (data not shown). That being said, array results indicate little to no expression of either SLC15A1 (PepT1) and SLC15A2 (PepT2), while our separate studies indicate relatively high expression of both SLC15A4 (PHT1) and SLC15A3 (PHT2) (Table 2), each of which was consistent across multiple passages (data not shown). These results are consistent to those of our preliminary end point assessment of POT expression in this cell line (data not shown), as well as those obtained in vivo in the human blood brain barrier.[20,21] Moreover, Western blotting using an antibody specific to hPHT1 did demonstrate protein expression in this cell line (Figure 2).
Figure 2.
Western blot analysis demonstrating hPHT1 expression in hCMEC/D3 wild type cells.Lane 1, molecular weight standard, Lane 2, hCMEC/D3 protein lysate, Lane 3, BSA blank. Studies using the primary antibody for these studies have been previously reported.[36] Briefly, whole protein lysates were obtained using a modified RIPA buffer and 40μg total protein was electrophoretically separated and blotted onto PVDF membrane. After blocking, the blot was probed with a rabbit anti-hPHT1 polyclonal (1:500 dilution), followed by a commercially available goat anti-rabbit-HRP conjugated secondary antibody (1:5000 dilution; Sigma Chemical Co.). After washing, blots were incubated using the West Femto Supersignal Chemiluminescent dection kit (Pierce Chemical Company) and the resultant immunoreactions were visualized using a Biorad Chemidoc XRS equipped with a 12 bit peltier cooled CCD camera.
Of the other SLC systems evaluated, appreciable expression of the monocarboxylate transporters (MCT) SLC16A1 and SLC16A3 were observed, while little to no expression of SLC16A2 is noted, indicative of concerted influx into the brain for MCT substrates.[22] Interestingly, relatively high expression of the equilibrative nucleoside transporters (SLC29) was observed, while little to no expression of the concentrative nucleoside transporters (SLC28) is noted. This finding is somewhat surprising considering these two families typically function in concert,[22] but also that SLC28 substrates are precursors for brain phosphatide synthesis, and while SLC29 members can transport these compounds, their respective capacity largely precludes them as the primary transporters of these important substrates.[23] Not very surprising is the relatively high complement of amino acid and facilitated glucose transporter members (SLC3, SLC38, SLC7 and SLC2). Of the organic anion/ cation transporter families (SLC22 and SLCO), only SLC22A3, SLCO2A1, SLCO3A1 and SLCO4A1 show any demonstrable expression.
Permeability Assessment
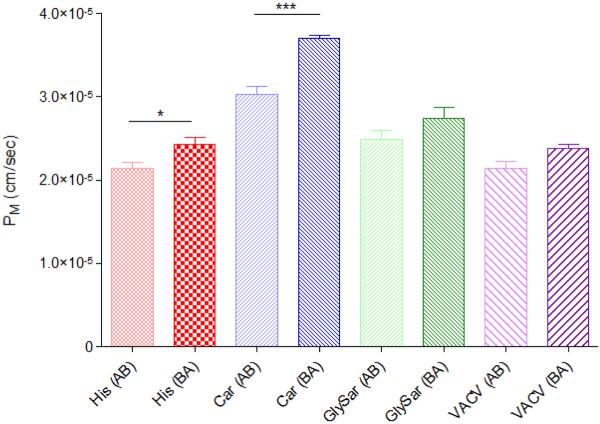
Figure 3 demonstrates the corrected membrane permeabilities (PM per Equation 2) of various established POT substrates across hCMEC/D3 cells. The transport characteristics of POT substrates were determined after 15, 30, 45, 60, 90 and 120 minutes in both the apical-basolateral (A-B) and basolateral-apical (B-A) directions. Interestingly, no differences between the A-B and B-A relative permeabilities (PM) were noted for either glycylsarcosine, or valacyclovir, when corrected for system-derived permeability resistances (PSYS). Both histidine and carnosine exhibited increased transport in the B-A direction comparative to the A-B direction. While indicative of net efflux (out of the brain and into the blood-facing compartment), a distinction must be made that this is probably not due to the influence of active transport systems such as Pgp, or MRPs, due to substrate affinities. The in vivo relevance of these findings is currently unknown.
Figure 3.
Effective permeability coefficients (PM) for a number of POT substrates across hCMEC/D3 cells. Cells were seeded at 2 × 105 cells/cm2 on membranous filters (0.4μm, polycarbonate Transwells) and permitted to grow for 2 days in whole medium. On the day of the experiment, the cells were washed twice in pre-warmed transport buffer and permitted to equilibrate in transport buffer for 20 minutes at 37°C/ 5% CO2. After equilibration, 1μCi/mL [3H] radiolabeled compound (His: histidine; Car: carnosine; GlySar: glycyclsarcosine; VACV: valaciclovir) in pre-warmed transport buffer was applied to the cells. Samples were removed after 15, 30, 45, 60, 90 and 120 minutes and an equal aliquot of pre-warmed transport buffer was added back to the system. Both the apical-basolateral (A-B) and basolateral-apical (B-A) transport was determined. [14C]Mannitol (0.25μCi/mL) transport was duplexed with each donor well to monitor the cellular integrity through the course of the experiment (data not presented). Results are presented as mean ± SD of 3 replicates. * p < 0.05, ** p < 0.01; *** p < 0.001 as determined by one-way ANOVA using Bonferroni post-hoc analysis.
Intercellular Pore Radius
The A-B transport characteristics of [14C] mannitol and [14C] urea were utilized to determine the relative intercellular pore radius of the hCMEC/D3 cell monolayer. Based on a single pore model, the ratio of the relative permeabilities of two compounds of similar charge, yet different molecular radii can be utilized to determine the effective intercellular pore radius, assuming a single pore average.[17,18] As presented in Table 3 the hCMEC/D3 effective intercellular pore radius was calculated to be 19.39 Å ± 0.84 Å, under these study conditions.
Table 3.
Intercellular pore radius determination for hCMEC/D3 wild type cells.
| Compound | MW | Molecular Radius (Å)† | PM(× 10−5 cm/sec) | Pore Radius (Å) |
|---|---|---|---|---|
| [14C] Mannitol | 182 | 4.10 | 2.47 ± 0.02 | 19.39 ± 0.84 |
| [14C] Urea | 60 | 2.67 | 5.65 ± 0.08 |
molecular radii calculated from Stokes-Einstein equation for equivalent spheres.[17]
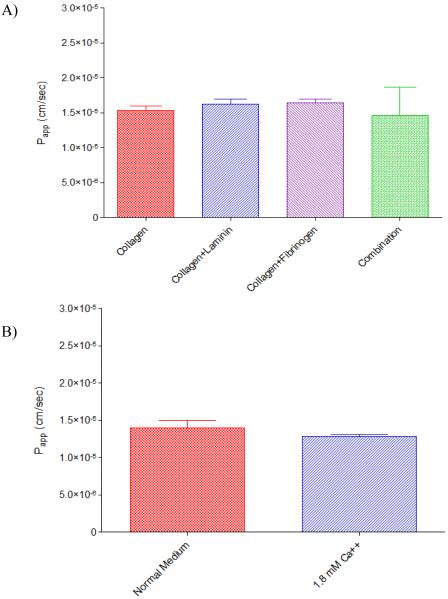
Given the previously reported relative leakiness of the hCMEC/D3 monolayer, studies were also conducted to determine the net effect of using additional extracellular basement membrane proteins during culture. In addition to Type I rat tail collagen, Transwell® inserts were coated with either laminin, derived from Engelbreth-Holm-Swarm murine sarcoma basement membrane, at 2 μg/cm2 per well, fibronectin from human plasma, at 5 μg/cm2 per well, or the combination of 2 μg/cm2 laminin and 5 μg/cm2 fibronectin. As illustrated in Figure 4, no differences in [14C] mannitol transport were observed indicating no net observable effect on the intercellular pore radius for a compound of this size.
Figure 4.
Apparent permeability coefficients (Papp) for [14C] mannitol across hCMEC/D3 cells grown on Transwell© inserts: A) coated with either collagen, collagen and laminin, collagen and fibronectin, or the combination of collagen, laminin and fibronectin and B) cells grown with normal medium, or medium further supplemented with 1.8mM Ca++. Cells were seeded at 2 × 105 cells/cm2 on membranous filters (0.4μm, polycarbonate) and permitted to grow for 2 days in whole medium. On the day of the experiment, the cells were washed twice in pre-warmed transport buffer and permitted to equilibrate in transport buffer for 20 minutes at 37°C/ 5% CO2. After equilibration, 0.25μCi/mL [14C] mannitol in pre-warmed transport buffer was applied to the cells. Samples were removed after 15, 30, 45, 60, 90 and 120 minutes and an equal aliquot of pre-warmed transport buffer was added back to the system. Transport was determined in the AB direction only. Results are presented as mean ± SD of 3 replicates. Statistical significance as determined by one-way ANOVA using Bonferroni post-hoc analysis in A), or by student's t-test in B).
Discussion
The inherent potential of using an immortalized human endothelial cell line to model the BBB gives instant relevance to characterizing the basal physiology of the hCMEC/D3 line. Initial studies conducted in Couraud's laboratory at the time of development demonstrated stable expression of various efflux transporters including P-gp, MRP-1, BCRP, and MRP-5,[5] which is consistent with our findings here and others.[8] There are a number of options to potentially formulate around gastrointestinal efflux transporters, however current studies demonstrate very little success in bypassing these transporters in the BBB.[24,25] There is also arguably greater significance in delineating the factors that contribute to influx transport through these cells, particularly with respect to POT members, as they could potentially serve as targets for effective CNS delivery.
With respect to the potential for POT-mediated transport, our characterization demonstrates expression of both hPHT1 and hPHT2. These results also demonstrate little to no expression of both hPepT1 and hPepT2, analogous to the human BBB.[20,21] As such, the cell line could serve as a surrogate model to evaluate POT substrate transport across the BBB, potentially filling a large void in pharmaceutical screening capabilities.[26] While in situ brain perfusion remains the most accurate model to monitor compound flux across the BBB, this in vivo testing method has not gained widespread utility due to the relative difficulty in establishing the technique and lack of practicality.[26] Additionally, in vivo testing methods are not generally regarded as high throughput. As such, characterizing a potential immortalized cell line as a surrogate model of human BBB transport and correlating in vitro transport kinetics with those observed in vivo could enable broad utility of this cell model in drug discovery and development.
The human hCMEC/D3 line was originally cloned due to a lack of sufficient stable in vitro models for investigating the molecular and physiological characteristics of the human BBB. Previous BBB models include co-culture systems of primary endothelial cultures with astrocytes and/or glial cells, porcine monocultures of brain endothelial cells differentiated with glucocorticoids (GC), or hTERT (catalytic unit of telomerase) transgenic cultures.[5] A number of limitations preclude the widespread utility of these models: the monoculture systems are primary cultures and thus, highly reliant on harvesting and culture conditions and confounded by pharmacogenetic variations; the co-culture systems are extraordinarily difficult to maintain, although they do exhibit a good model of in vivo physiology; finally, past transgenic cultures have either dedifferentiated into a senescent state, losing their BBB phenotype, or failed to demonstrate functional tight junctions. In stark contrast, the hCMEC/D3 cell line exhibits a stable BBB endothelial phenotype, including stable expression of tight junctional complexes.[5] Furthermore, Couraud's group demonstrated that hCMEC/D3 cells also constitutively express ICAM-1, ICAM-2, PECAM-1, and CD40 and upon cytokine activation, ICAM-1 and CD40 are up-regulated, while expression of VCAM-1 is induced.[5] The observations that hCMEC/D3 cells express a large number of chemokine receptors confirms the utility of this cell model for studying the mechanisms of human leukocyte infiltration into the CNS, which is a marker of brain injury such as ischemia, infection and traumatic brain injury.[27]
The potential utility of hCMEC/D3 cells as a useful model for studying the pathophysiology of the human brain endothelium during neuroinflammation or infectious disease states has been the thrust of further investigations by others.[28,29] To this end, recent investigations have detailed the effects of GC on the barrier functionality of these cells,[29] where a strong argument was made that the resultant increase in mRNA and protein expression of occludin and claudin-5 due to GC treatment resulted in a more selective barrier functionality. However, closer examination of the results of this and their initial assessment indicate that in spite of the presence of the tight junctional complexes, hCMEC/D3 cells may not convey as significant a barrier functionality as that observed in vivo.[5,29] Additional characterization studies focused on efflux transport mechanisms in this cell line also support this finding.[30]
Studies conducted in our laboratory also demonstrate that the cells may not present a significant barrier to paracellular permeation, although the barrier does appear to be slightly more restrictive under our growth conditions. Using [14C] radiolabelled mannitol and urea, we were also able to approximate the intercellular pore radius by means of the Renkin Molecular Sieving Function (using Equations 1 and 3 above),[31] as previously performed by our laboratory and others.[9,17,18,32] Using the reported molecular radii of 2.67Å and 4.10Å for urea and mannitol, respectively, the effective intercellular pore radius was calculated to be 19.39Å ± 0.84Å; significantly larger than those previously determined for Caco-2 cells (5.2Å ± 0.12Å), or BBMEC (14.4Å ± 2.8Å), using this methodology.[9,18] Additionally, this is almost three times that reported by Fenstermacher and Johnson for the in vivo rabbit BBB (7Å − 9Å).[33]
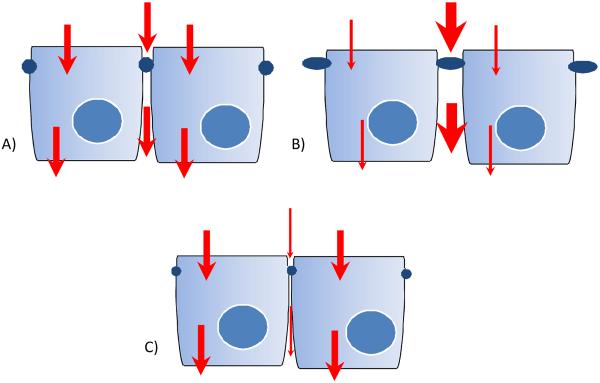
The real difficulty in numerically defining the intercellular pore radius is attempting to determine limitations for the suitability of the model for its intended use. Our intentions for characterizing the intercellular pore radius were in terms of understanding the potential for the paracellular transport component to obfuscate the additive function of the total transcellular transport, as described previously.[9] As in Figure 5, if the intercellular pore radius is too large, the relative contribution of transcellular transport to overall substrate flux will be negligible and can then be discounted, possibly undetectable. Likewise, if the pore radius is too restrictive, the model may not be a good predictor of the relative paracellular contribution and would not give accurate mechanistic information. These factors could limit the broader utility of the model from a drug screening perspective, but also in terms of its utility towards studying BBB physiology. As such, our initial compound screening included known POT substrates, with known limited paracellular transport in other screening models. While these studies do demonstrate that transcellular flux could be distinguished from paracellular, as evidenced by higher B-A transport for both histidine and carnosine, it is clear that the model does require further optimization.
Figure 5.
This is an idealized schematic illustrating the potential transport pathways that may contribute to permeability of substrates. Arrow thickness denotes relative contribution to overall permeability. A) A “normal” intercellular pore radius would mimic in vivo permeation and allow for accurate mechanistic study of compound flux. B) With a larger than “normal” pore radius, the paracellular contribution would be large enough to obscure the transcellular contribution to compound flux. C) With a smaller than “normal” pore radius, the paracellular contribution would be so restrictive it would not allow an accurate portrayal of in vivo permeability.
Towards this end, preliminary studies were conducted to determine the relative effectiveness of the extracellular matrix proteins laminin and fibronectin, components of the in vivo endogenous basement membrane, on decreasing the intercellular pore radius. This strategy has been used in the past to decrease porosity of the BBMECs [9] and has been demonstrated to be integral to the culturing of the BBMEC primary culture system. One of the immediate benefits of using this methodology is that it does not directly affect the cellular physiology, but utilizes the endogenous cellular physiology to increase the cell's natural barrier functionality through well-characterized integrin and catenin signaling. As evidenced in Figure 3 however, the addition of either laminin, or fibronectin, or the combination of the two together to the collagen matrix solution did not alter the apparent A-B mannitol permeability, indicating no net observable effect of these two matrix proteins. Interestingly, the addition of human serum to the growth medium was demonstrated to decrease the permeability of the paracellular marker sucrose.[30] While an exciting finding in its own right, given that the serum was obtained from a blood donor bank, and therefore subject to source-derived variations, it is likely that enabling such a protocol would introduce additional lab-to-lab variabilities.
Recognizing that the low calcium ion concentration in the growth medium could also affect the paracellular permeability, we also studied the effect of calcium supplementation on mannitol A-B permeability (Figure 4). It is well known that calcium sequestration through the use of EGTA or other calcium chelators can open tight junctions, leading to much higher paracellular transport.[9,18,34] Here, the addition of 1.8 mM Ca++ did not affect A-B mannitol permeability, indicating that the calcium content in the medium is sufficient to mediate tight junction contacts. This is not to say however, that calcium does not play a significant role in controlling monolayer integrity in vivo. Anatomically, the astrocytic processes surround the endothelial capillaries (analogous to hCMEC/D3 cells), and were once considered the primary barrier to brain transport.[2] While this hypothesis has been dismissed in the past, recent evidence suggests that astrocytes secrete a number of factors, including calcium, that could modulate the functional barrier activity of the BBB.[35] As such, there seems to be growing sentiment that it is the cooperative coordination of the capillary endothelial cells, the undifferentiated pericytes and the astrocytes that all synergistically compose the BBB.
While attempts to optimize the hCMEC/D3 cell line for drug screening are still early in progress, it is clear the cell line can be used to monitor both A-B and B-A transport processes in a human-derived, functionally relevant cell line. These studies have also demonstrated that transport studies can be used to discern the net flux of substrates across the cells, as evidenced by the higher B-A transport for both histidine and carnosine (analogous to brain efflux). Having been corrected for system derived resistances, these studies clearly demonstrate the utility of the cell line to study mechanistic BBB permeation. Unfortunately, these limited studies were not designed to and are therefore, not able to distinguish the individual predominating mechanisms by which these compounds permeate the membrane. In short, the potential for overlapping specificity of these substrates for other SLC and ABC transporters precludes us from concluding that the observed differences in transport were indeed due to POT transporter activity. That being said, our laboratory is currently in the process of investigating the contributions of individual POT members to BBB permeability of these and other POT substrates.
Primary cultured brain endothelial cell systems, such as the BBMEC in America, or Porcine Brain Endothelial Cells in Europe, have become the in vitro models of choice to monitor BBB permeation.[26] However, due to the labor intensive nature of establishing these culture models, as well as the relative experience level required to consistently and reproducibly extract the cells, primary cultures are not readily used by industry at-large and most studies are performed through contract laboratories. Additionally, the relative tightness of these cell systems also appears to be an issue, having led researchers to use Mandin-Darby Canine Kidney II (MDCKII) cells to rank order the passive brain permeation of discovery compounds.[26] To put this dilemma in perspective, the permeation across this kidney endothelial cell line was more predictive of in vivo brain permeation than primary culture models, most likely due to issues with monolayer tightness.[26] Although additional studies are required to determine the predictability of the hCMEC/D3 cell line of in vivo permeation, this immortalized in vitro model has the potential to fill a very large void in our understanding of BBB physiology, as well as serve as a surrogate model for brain permeability screening.
Acknowledgements
Financial support for these studies was kindly provided by the National Institute of General Medical Sciences (R01-GM65448).
References
- 1.Liederer BM, Phan KT, Ouyang H, Borchardt RT. Significant differences in the disposition of cyclic prodrugs of opioid peptides in rats and guinea pigs following IV administration. J. Pharm. Sci. 2005;94(12):2676–2687. doi: 10.1002/jps.20476. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins RA, O'Kane RL, Simpson IA,, Viña JR. Structure of the blood-brain barrier and its role in the transport of amino acids. J. Nutr. 2006;136(1 Suppl):218S–226S. doi: 10.1093/jn/136.1.218S. [DOI] [PubMed] [Google Scholar]
- 3.Nicolazzo JA, Charman SA, Charman WN. Methods to assess drug permeability across the blood-brain barrier. J. Pharm. Pharmacol. 2006;58(3):281–293. doi: 10.1211/jpp.58.3.0001. [DOI] [PubMed] [Google Scholar]
- 4.Jeffrey P, Summerfield SG. Challenges for blood-brain barrier (BBB) screening. Xenobiotica. 2007;37(10–11):1135–1151. doi: 10.1080/00498250701570285. [DOI] [PubMed] [Google Scholar]
- 5.Weksler BB, Subileau EA, Perrière N, Charneau P, Holloway K, Leveque M, Tricoire-Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19(13):1872–1904. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- 6.Poller B, Gutmann H, Krähenbühl S, Weksler B, Romero I, Couraud PO, Tuffin G, Drewe J, Huwyler J. The human brain endothelial cell line hCMEC/D3 as a human blood-brain barrier model for drug transport studies. J. Neurochem. 2008;107(5):1358–1368. doi: 10.1111/j.1471-4159.2008.05730.x. [DOI] [PubMed] [Google Scholar]
- 7.Dauchy S, Miller F, Couraud PO, Weaver RJ, Weksler B, Romero IA, Scherrmann JM,, De Waziers I, Declèves X. Expression and transcriptional regulation of ABC transporters and cytochromes P450 in hCMEC/D3 human cerebral microvascular endothelial cells. Biochem. Pharmacol. 2009;77(5):897–909. doi: 10.1016/j.bcp.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Nicolazzo JA, Katneni K. Drug transport across the blood-brain barrier and the impact of breast cancer resistance protein (ABCG2) Curr. Top. Med. Chem. 2009;9(2):130–147. doi: 10.2174/156802609787521580. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen M, Steenberg B, Knipp GT, Wang W, Steffansen B, Frokjaer S, Borchardt RT. The effect of beta-turn structure on the permeation of peptides across monolayers of bovine brain microvessel endothelial cells. Pharm. Res. 1997;14(10):1341–1348. doi: 10.1023/a:1012104301773. [DOI] [PubMed] [Google Scholar]
- 10.Graul RC, Sadée W. Sequence alignments of the H(+)-dependent oligopeptide transporter family PTR: Inferences on structure and function of the intestinal PEPT1 transporter. Pharm. Res. 1997;14:388–400. doi: 10.1023/a:1012070726480. [DOI] [PubMed] [Google Scholar]
- 11.Nussberger S, Steel A, Hediger M. Structure and pharmacology of proton-linked peptide transporters. J. Control. Rel. 1997;46:31–38. [Google Scholar]
- 12.Yang CY, Dantzing AH, Pidgeon C. Intestinal peptide transport of systems and oral drug availability. Pharm. Res. 1999;16:1331–1343. doi: 10.1023/a:1018982505021. [DOI] [PubMed] [Google Scholar]
- 13.Meredith D, Boyd CAR. Structure and function of eukaryotic peptide transporters. Cell. Mol. Life Sci. 2000;57:754–778. doi: 10.1007/s000180050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubio-Aliaga I, Daniel H. Mammalian peptide transporters as targets for drug delivery. Trends Pharm. Sci. 2002;23(9):434–440. doi: 10.1016/s0165-6147(02)02072-2. [DOI] [PubMed] [Google Scholar]
- 15.Herrera-Ruiz D, Knipp GT. Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharm. Sci. 2003;92(4):691–714. doi: 10.1002/jps.10303. [DOI] [PubMed] [Google Scholar]
- 16.Carl SM, Herrera-Ruiz D, Bhardwaj RK, Gudmundsson O, Knipp GT. Mammalian Oligopeptide Transporters. In: You G, Morris ME, editors. Drug Transporters: Molecular Characterization and Role in Drug Disposition. John Wiley & Sons, Inc.; Hoboken, NJ: 2007. pp. 105–145. ISBN: 978-0-471-78491-3. [Google Scholar]
- 17.Adson A, Burton PS, Raub TJ, Barsuhn CL, Audus KL, Ho NF. Passive diffusion of weak organic electrolytes across Caco-2 cell monolayers: uncoupling the contributions of hydrodynamic, transcellular, and paracellular barriers. J. Pharm. Sci. 1995;84(10):1197–1204. doi: 10.1002/jps.2600841011. [DOI] [PubMed] [Google Scholar]
- 18.Knipp GT, Ho NF, Barsuhn CL, Borchardt RT. Paracellular diffusion in Caco-2 cell monolayers: effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. J. Pharm. Sci. 1997;86(10):1105–1110. doi: 10.1021/js9700309. [DOI] [PubMed] [Google Scholar]
- 19.Steiner E, Holzmann K, Elbling L, Micksche M, Berger W. Cellular functions of vaults and their involvement in multidrug resistance. Curr. Drug Targets. 2006;7(8):923–934. doi: 10.2174/138945006778019345. [DOI] [PubMed] [Google Scholar]
- 20.Berger UV, Hediger MA. Distribution of peptide transporter PEPT2 mRNA in the rat nervous system. Anat. Embryol. (Berl) 1999;199(5):439–449. doi: 10.1007/s004290050242. [DOI] [PubMed] [Google Scholar]
- 21.Smith DE, Johanson CE, Keep RF. Peptide and peptide analog transport systems at the blood-CSF barrier. Adv. Drug Deliv. Rev. 2004;56(12):1765–1791. doi: 10.1016/j.addr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Bhardwaj RK, Herrera-Ruiz DR, Xu Y, Carl SM, Cook TJ, Vorsa N, Knipp GT. Drug Transporters and Absorption. In: Krishna R, Yu L, Skelly J, Agarwal V, editors. In Biopharmaceutics Applications in Drug Development. Kluwer Press; pp. 175–261. [Google Scholar]
- 23.Cansev M. Uridine and cytidine in the brain: their transport and utilization. Brain Res. Rev. 2006;52(2):389–397. doi: 10.1016/j.brainresrev.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Chikhale EG, Ng KY, Burton PS, Borchardt RT. Hydrogen bonding potential as a determinant of the in vitro and in situ blood-brain barrier permeability of peptides. Pharm. Res. 1994;11(3):412–419. doi: 10.1023/a:1018969222130. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Yang JZ, Andersen R, Nielsen LH, Borchardt RT. Evaluation of the permeation characteristics of a model opioid peptide, H-Tyr-D-Ala-Gly-Phe-D-Leu-OH (DADLE), and its cyclic prodrugs across the blood-brain barrier using an in situ perfused rat brain model. J. Pharmacol. Exp. Ther. 2002;303(2):849–857. doi: 10.1124/jpet.102.037143. [DOI] [PubMed] [Google Scholar]
- 26.Reichel A. The role of blood-brain barrier studies in the pharmaceutical industry. Curr. Drug Metab. 2006;7(2):183–203. doi: 10.2174/138920006775541525. [DOI] [PubMed] [Google Scholar]
- 27.Shaftel SS, Carlson TJ, Olschowka JA, Kyrkanides S, Matousek SB, O'Banion MK. Chronic interleukin-1beta expression in mouse brain leads to leukocyte infiltration and neutrophil-independent blood brain barrier permeability without overt neurodegeneration. J. Neurosci. 2007;27(35):9301–9309. doi: 10.1523/JNEUROSCI.1418-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cucullo L, Couraud PO, Weksler B, Romero IA, Hossain M, Rapp E, Janigro D. Immortalized human brain endothelial cells and flow-based vascular modeling: a marriage of convenience for rational neurovascular studies. J. Cereb. Blood Flow Metab. 2008;28(2):312–328. doi: 10.1038/sj.jcbfm.9600525. [DOI] [PubMed] [Google Scholar]
- 29.Foerster C, Burek M, Romero IA, Weksler B, Couraud PO, Drenckhahn D. Differential effects of hydrocortisone and TNF{alpha} on tight junction proteins in an in vitro model of the human blood-brain barrier. J. Physiol. 2008 Feb 7; doi: 10.1113/jphysiol.2007.146852. DOI: 10.1113/jphysiol.2007.146852 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poller B, Gutmann H, Krähenbühl S, Weksler B, Romero I, Couraud PO, Tuffin G, Drewe J, Huwyler J. The human brain endothelial cell line hCMEC/D3 as a human blood-brain barrier model for drug transport studies. J. Neurochem. 2008;107(5):1358–1368. doi: 10.1111/j.1471-4159.2008.05730.x. [DOI] [PubMed] [Google Scholar]
- 31.Renkin EM. Filtration, diffusion and molecular sieving through porous cellulose membranes. J. Gen. Physiol. 1954;38(2):225–243. [PMC free article] [PubMed] [Google Scholar]
- 32.Seki T, Kanbayashi H, Nagao T, Chono S, Tabata Y, Morimoto K. Effect of cationized gelatins on the paracellular transport of drugs through caco-2 cell monolayers. J. Pharm. Sci. 2006;95(6):1393–1401. doi: 10.1002/jps.20616. [DOI] [PubMed] [Google Scholar]
- 33.Fenstermacher JD, Johnson JA. Filtration and reflection coefficients of the rabbit blood-brain barrier. Am. J. Physiol. 1966;211(2):341–346. doi: 10.1152/ajplegacy.1966.211.2.341. [DOI] [PubMed] [Google Scholar]
- 34.Knipp GT, Vander Velde DG, Siahaan TJ, Borchardt RT. The effect of beta-turn structure on the passive diffusion of peptides across Caco-2 cell monolayers. Pharm. Res. 1997;14(10):1332–1340. doi: 10.1023/a:1012152117703. [DOI] [PubMed] [Google Scholar]
- 35.Kuchibhotla KV, Lattarulo CR, Hyman BT, Bacskai BJ. Synchronous hyperactivity and intercellular calcium waves in astrocytes in Alzheimer mice. Science. 2009;323(5918):1211–1215. doi: 10.1126/science.1169096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhardwaj RK, Herrera-Ruiz DR, Eltoukhy N, Saad M, Knipp GT. The Functional Evaluation of Human Peptide/Histidine Transporter 1 (HPHT1) in Transiently Transfected COS-7 Cells. Eur. J. Pharm. Sci. 2006;27:533–542. doi: 10.1016/j.ejps.2005.09.014. [DOI] [PubMed] [Google Scholar]