Abstract
Fatty acid transport protein (FATP) 4 is one of a family of six FATPs that facilitate long- and very long-chain fatty acid uptake. Mice lacking FATP4 are born with tight, thick skin and a defective epidermal barrier; they die neonatally due to dehydration and restricted movements. Both the skin phenotype and the lethality are rescued by transgene-driven expression of FATP4 solely in suprabasal keratinocytes. Here we show that Fatp4 mutants exhibit epidermal hyperplasia resulting from an increased number of proliferating suprabasal cells. In addition, barrier formation initiates precociously but never progresses to completion. To investigate possible mechanisms whereby Fatp4 influences skin development, we identified misregulated genes in Fatp4 mutants. Remarkably, three members of the epidermal growth factor (EGF) family (Ereg, Areg, and Epgn) showed increased expression that was associated with elevated epidermal activation of the EGF receptor (EGFR) and STAT3, a downstream effector of EGFR signaling. Both Tyrphostin AG1478, an EGFR tyrosine kinase inhibitor, and curcumin, an inhibitor of both STAT3 and EGFR, attenuated STAT3 activation/nuclear translocation, reduced skin thickening, and partially suppressed the barrier abnormalities. These data identify FATP4 activity as negatively influencing EGFR activation and the resulting STAT3 signaling during normal skin development. These findings have important implications for understanding the pathogenesis of ichthyosis prematurity syndrome, a disease recently shown to be caused by FATP4 mutations.
Keywords: Epiregulin, amphiregulin, epithelial mitogen, PPAR, skin barrier, epidermal hyperplasia
Introduction
The mature mammalian skin is a stratified epithelium derived from the embryonic ectoderm; it is the first line of defense against mechanical and chemical trauma. The skin also functions as a barrier to block the entry of microorganisms and to prevent the loss of body hydration. During skin morphogenesis, several signaling pathways participate in a series of inductive events that guide the separation of the hair lineage from the epidermal lineage and promote epidermal differentiation and stratification (Fuchs and Raghavan, 2002; Millar, 2002).
Epidermal stratification begins when proliferative basal cells migrate suprabasally and exit the cell cycle, beginning a series of steps culminating in cornification (Elias and Jackson, 1996). The first event is keratinization, the switch in intermediate filaments from a keratins 5/14-containing network to a keratins 1/10-containing network, permitting cells in the spinous layer to form a rigid cytoskeleton. Secondly, keratohyalin granules containing profilaggrin are synthesized to facilitate the assembly of keratin bundles, forming the granular layer. Thirdly, involucrin (IVL), loricrin, and other proteins are cross-linked by transglutaminase, forming an insoluble cornified envelope beneath the cell membrane. Finally, lipid-enriched lamellar bodies secrete their contents into the intercellular spaces of the cornified layer, completing the formation of a water-resistant barrier. This progression from a proliferating basal cell to a terminally differentiated, cornified squamous cell occurs continuously throughout life, as skin is a constantly regenerating organ (Fuchs and Raghavan, 2002).
We previously identified an autosomal recessive mouse mutation called wrinkle free (Moulson et al., 2003). Homozygous mutants are born with taut and shiny skin, a thickened epidermis, a defective skin barrier, and sparse hair follicles; neonates die due to dehydration and restricted movements. By positional cloning, we found the mutation to be caused by a spontaneous retrotransposon insertion into Slc27a4, the gene encoding fatty acid transport protein (FATP) 4. One targeted mutation in Slc27a4 (here referred to as Fatp4 for simplicity) shows an identical phenotype (Herrmann et al., 2003), and another was reported to cause very early embryonic lethality (Gimeno et al., 2003).
FATP4 is one of a family of six transmembrane proteins that facilitate long- and very long-chain fatty acid uptake. FATP4 exhibits acyl-CoA synthetase (ACS) activity and has been proposed to facilitate uptake of fatty acids indirectly by mediating their esterification to CoA (Hall et al., 2005; Herrmann et al., 2001), in contrast to the direct fatty acid transport functions identified for other FATPs, such as FATP1 (Richards et al., 2006; Schaffer and Lodish, 1994). FATP4 is widely expressed, suggesting roles in many organs (Herrmann et al., 2001; Moulson et al., 2003). In skin FATP4 is normally detected in basal and suprabasal keratinocytes (MHL and JHM, unpublished data), with the strongest expression in the granular layer of the epidermis (Moulson et al., 2007). Suprabasal keratinocyte expression of a FATP4 transgene in the epidermis rescues the neonatal lethality and ameliorates the skin phenotype of Fapt4 mutant mice, indicating crucial, skin-intrinsic roles for FATP4 in the development of skin and its appendages (Moulson et al., 2007).
The recent identification of Fatp4 mutations in patients with ichthyosis prematurity syndrome (Klar et al., 2009) makes an understanding of the mechanism whereby the absence of FATP4 causes the wrinkle free phenotype in mice an especially important goal. Here we characterized the skin abnormalities in Fatp4−/− mice at the cellular level and found an increased number of proliferating keratinocytes in the suprabasal layer. To identify possible mechanisms that link the lack of FATP4 to specific defects in the development of skin and its appendages, we carried out microarray analyses with embryonic skin RNAs and found upregulation of several EGF family ligands in Fatp4 mutant skin, together with increased activation of STAT3, a downstream effector of the EGF receptor (EGFR) signaling pathway. Furthermore, pharmacological blockade of EGFR and STAT3 activation suppressed epidermal hyperplasia in Fatp4 mutants, and this correlated with reduced hyperproliferation. These data indicate that the lack of FATP4 creates an environment, presumably via direct effects on lipid metabolism and homeostasis, that promotes epidermal proliferation via overactivation of the EGFR and the downstream STAT3 signaling pathways.
Materials and methods
Mice and skin barrier assays
Fatp4 mutant and transgenic mice have been previously described (Moulson et al., 2007; Moulson et al., 2003). Embryonic day (E) 15.5 to E17.5 embryos were dissected from pregnant females, with the morning when the copulation plug was observed considered E0.5. For inward permeability assays, embryos were stained in the dark at 37°C overnight in X-Gal solution (1 mg/ml X-Gal, 3 mM K4Fe(CN)6, 3 mM K3Fe(CN)6, 1.3 mM MgCl2, 0.1 M NaH2PO4) at pH 4.5 as described (Hardman et al., 1998). In some experiments embryos were incubated in a series of ascending and then descending concentrations of methanol, equilibrated in phosphate buffered saline (PBS), and stained briefly in 1% toluidine blue in water followed by destaining in PBS (Hardman et al., 1998). Stained samples were fixed in 4% paraformaldehyde in PBS at room temperature for 1 hour to overnight. For outward transepidermal water loss (TEWL) assays, embryos were rinsed in PBS, blotted gently with a Kimwipe, and air-dried for 5 min. The water loss through the dorsal or lateral skin was measured using a Vapometer (Delfin Technologies, Kuopio, Finland) with the sensor chamber attached to a nail adaptor.
Immunohistochemistry
Embryos were fixed at room temperature for 2 to 3 hours in 4% paraformaldehyde in PBS. To increase the penetration of fixative, E15.5 or older embryos were decapitated, and the abdominal cavity was exposed. Fixed embryos at E14.5 were cut into halves along the dorsal midline, embedded in paraffin, and sectioned parasagittally at 5 μm. For fixed embryos at older stages, the dorsal skin was collected for paraffin embedding.
Immunohistochemical analyses were performed using the peroxidase Vectastain ABC kit (Vector Laboratories, Burlingame, CA) with DAB as a chromogen (Pierce, Rockford, IL) as described (Lin et al., 2000), with some modifications. To retrieve antigens, rehydrated sections were boiled in 10 mM citrate buffer (pH 6), Trilogy (Cell Marque, Rocklin, CA), or 1 mM EDTA (pH 8). To block endogenous biotin and avidin, the Avidin/Biotin blocking kit (Vector Laboratories) was used before adding primary antibodies. The antigen retrieval method and dilutions of primary antibodies used were as follows: EDTA and 1:50 for phospho-STAT3 (Tyr705); EDTA and 1:100 with tyramide amplification for phospho-EGFR (Tyr1068); citrate and 1:100 for phospho-p44/42 MAP kinase (Thr202/Tyr204); citrate and 1:100 for phospho-p38 MAPK (Thr180/Tyr182); citrate and 1:100 for phospho-SAPK/JNK (Thr183/Tyr185) (all antibodies were from Cell Signaling Technology, Beverly, MA); Trilogy and 1:100 with tyramide amplification for JAK2 (phospho Y1007+Y1008) (Abcam, Cambridge, UK); 1:1,000 for keratin 6 (Covance, Princeton, NJ). Sections were counterstained with hematoxylin and mounted.
Double immunofluorescence analysis of Ki67 (Novocastra Laboratories, Newcastle upon Tyne, UK) and laminin γ1 (Millipore, Danvers, MA) was performed on paraformaldehyde-fixed, frozen sections of tissue embedded in OCT (Sakura Finetek, Torrance, CA). Both primary antibodies were applied at 1:1,000, and signals were detected using Alexa 488-conjugated anti-rabbit and Alexa 594-conjugated anti-rat, respectively, with Hoechst 33258 as a counterstain. The number of Ki67-positive suprabasal cells present in a field encompassing 100 Ki67-positive basal cells was used as a measurement of suprabasal cell hyperproliferation (Hansen et al., 2000). A total of at least 100 Ki67-positive basal cells were counted in each sample. Proliferation of basal cells was assayed by determining the percentage of Ki67-positive cells in the basal layer in a population of at least 100 Hoechst 33258-positive basal cells for each sample.
In situ hybridization
Embryos at various stages were fixed and paraffin-embedded as described above, except that the fixation was performed at 4°C overnight. In situ hybridization with digoxigenin-UTP labeled riboprobes was performed as described (Moulson et al., 2007), with hybridization temperatures ranging from 55 to 60°C.
Riboprobes were synthesized from either linearized plasmids or PCR amplicons using DIG RNA labeling mix (Roche, Mannheim, Germany) and RNA polymerase Plus (Ambion, Austin, TX), and purified by NucAway spin columns (Ambion) following the manufacturers' instructions. For some of the genes examined, both anti-sense and sense riboprobes were tested on samples to confirm their specificities. The plasmids contained cDNAs from: Areg (clone ID 3597695), Ereg (clone ID 5325124), Epgn (clone ID 8734042), Mmp3 (clone ID 3962288), Ccl12 (clone ID 1548072), Ifi202b (clone ID 4945974; all above are IMAGE clones obtained from OpenBiosystesm, Huntsville, AL), and Il24 (clone ID RZPDp981C12223D; from imaGenes, Berlin, Germany). The PCR amplicons were generated using the cDNAs reverse-transcribed from RNA with Superscript III reverse transcriptase (Invitrogen). Primers used to make PCR amplicons were as follows, with T7 and T3 promoter sequence underlined: Klk7: 5′ TATAATACGACTCACTATAGGGGAGTGCAAGAAGGTGTACAAG 3′ and 5′ GAAATTAACCCTCACTAAAGGG1TGGAGGAAAGGTAAAGCCAG 3′, Dusp6: 5′ TATAATACGACTCACTATAGGGGGATCACTGGAGCCAAAAC 3′ and 5′ GAAATTAACCCTCACTAAAGGGGGAACTGAAGGAATGGGGAC 3′.
RNA isolation and microarray analyses
Total RNAs were isolated from the dorsal skin of embryos at E15.5 using a tissue homogenizer (Powergen 125, Fisher Scientific, Pittsburgh, PA) and the RNeasy Fibrous Tissue Mini kit with an in-column DNase treatment (Qiagen, Chatsworth, CA) following the manufacturer's instructions. Purified RNAs were qualitatively assessed by RNA LabChip (Agilent, Palo Alto, CA) and quantified spectrophotometrically in NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). To make analytical duplicates, equal amounts of two RNA samples of the same genotype were pooled and split into two tubes. The preparation of biotin-labeled, antisense cRNA targets and microarray hybridization were performed in the Siteman Cancer Center Multiplexed Gene Analysis Core using standard protocols supplied by the manufacturer (Affymetrix, Santa Clara, CA). Double-stranded cDNAs were generated from 5 μg of total RNA with T7-T24 primers (Genset, Paris, France), Superscript II reverse transcriptase, E. coli ligase, and E. coli polymerase I (Invitrogen, Carlsbad, CA). Biotin-labeled, antisense cRNA targets were generated from the synthesized cDNAs using the Enzo BioArray High Yield RNA Transcript Labeling kit (Enzo Biochemical, New York, NY). Ten micrograms of each biotinylated cRNA preparation were fragmented and hybridized to the mouse genome array MOE430A2 for 16 hours in Hybridization Oven 640. Microarrays were then washed in Fluidics Station using the standard “Eukaryotic GE Wash 2” protocol followed by signal detection with antibody-mediated signal amplification. The processed arrays were scanned by GeneChip Scanner 3000 to generate image files (.dat files) and cell intensity files (.cel files).
Analyses of microarray data
Scanned images were processed using Affymetrix Microarray Suite (MAS 5.0) and exported as text files containing both qualitative and quantitative analyses for each probe set. Cell intensity files were normalized based on total intensity using affy package (RMA method) from BioConductor (http://www.bioconductor.org/, Biostatistics Unit of Dana Farber Cancer Institute, Harvard Medical School) and exported as text files for numerical analyses. Fatp4 mutant embryos (experimental samples as the numerator) were compared to their heterozygous littermates (control samples as the denominator). Data from analytical duplicates were analyzed statistically by multtest package (Resampling-based multiple hypothesis testing, BioConductor), and those with adjusted P-value less than 0.05 were selected. To filter false positives, data with increased fold changes but with absent calls in the experimental samples were eliminated. Likewise, data with decreased fold changes but with absent calls in the control samples were also eliminated. A cut-off value of 1.5-fold change was used to identify genes that showed the most significant variation in expression. To biologically annotate genes that showed differential expression, GenMAPP was applied to grouping genes along Gene-Ontology (GO) categories or biological pathways (http://www.genmapp.org/, Gladstone Institutes, University of California at San Francisco) (Doniger et al., 2003). Significance of differential gene expression in a specific GO was determined by Z scores, an indicator to show whether in a specific GO, there was a difference in the ratio of genes changed as compared to the background ratio in the entire GO. A positive Z score indicates there are more genes changed in a GO term than would be expected by random chance, and vice versa.
RT- quantitative PCR (RT-qPCR)
Total RNAs were isolated from E15.5 dorsal skin or from E16.5 dorsal skin epidermis as described above. To isolate the epidermis, the dorsal skin was incubated in dispase II (5 mg/ml; Roche) at 4°C for 4 hours, and the epidermis was peeled from the skin. Complementary DNA (cDNA) was generated from 0.2 μg of total RNA using Superscript III reverse transcriptase with oligo(dT)12-18 (Invitrogen) in a 30 μl reaction incubated at 50°C for 1h. Real-time PCR amplification was performed in triplicate using the fast mode (annealing and extending at 60°C) of the 7900 HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). Each replicate contained cDNA transcribed from 3 ng of RNA, a primer pair at optimized concentration (100 or 30 nM), and Fast SYBR Green Master Mix in a 10 μl reaction (Applied Biosystems). In triplicates with a standard deviation greater than 0.3, the outlier Ct was excluded. The comparative Ct (threshold cycle) method (2−ΔCt) was used for relative quantification of gene expression (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008), with Tbp as a reference gene (Minner and Poumay, 2009). To validate the comparative Ct method (Bustin et al., 2009), cDNA derived from 1 μg of total RNA in a 30 μl RT reaction was five-fold serially diluted over a 3 log10 range and subjected to real-time PCR with all primer pairs. The amplification efficiency (Ex) of each primer pair was calculated as described by the manufacturer from the slope of its linear regression line of Ct versus log10 cDNA concentrations using the equation Ex =10(−1/slope)−1, with Ct larger than 35 excluded from the calculation. Except for Il24, Cst6, and Junb, whose expression levels were too low to be assayed in a 3 order of magnitude range, all target genes showed Ex values similar to the reference gene Tbp (within a range of ±10%), with close correlation among dilutions (squared correlation coefficients R2> 0.99). Genomic DNA contamination was verified to be minimal for all target genes of interest using RT reactions without reverse transcriptase.
Most primer pairs were designed to anneal either to the 3′ untranslated region or to different exons of genes using Primer Express (Applied Biosystems). The specificity of each primer was verified by BLAST screen against the mouse genome and transcript database, by a distinct single peak during the dissociation curve analysis, and by a single band of the expected size on an electrophoretic gel. The primer sequences used are listed in Supplementary Table 1.
Tyrphostin AG1478 and Curcumin treatment
Tyrphostin AG1478 (purity >99%; LC laboratories, Woburn, MA) was prepared as a 160 mM working solution in DMSO and stored at −20°C. Curcumin (purity >80%; Sigma-Aldrich, St. Louis, MO) was prepared freshly as a 50 mM working solution in DMSO. Pregnant females at 14.5 days of gestation were injected i.p. in the morning for 2 days with AG1478, curcumin, or vehicle only at 100 μl per 35 g of body weight. Embryos were then removed at E16.5.
Statistical analysis
Two-tailed, unpaired Student's t-tests were used to determine statistical difference in TEWL and hyperproliferation assays. Two-way mixed model ANOVA tests were used to evaluate statistical difference in gene expression assays by RT-qPCR. Differences were considered significant when the P value was <0.05.
Results
Epidermal hyperproliferation in Fatp4 mutants
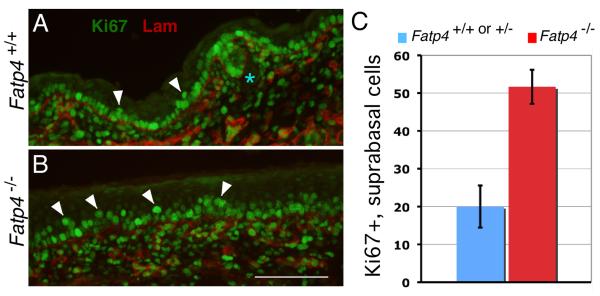
Fatp4−/− embryos display epidermal hyperplasia and hyperkeratosis from E15.5 onwards. At E15.5, when the cornified layer in control animals is barely visible, mutants already possess a distinct cornified layer. Moreover, expression of keratin 6 in mutant epidermis indicates an altered proliferation or differentiation program (Moulson et al., 2003). To directly examine whether the epidermal hyperplasia was associated with hyperproliferation of keratinocytes, the skin of Fatp4 mutant embryos was analyzed for Ki67 immunoreactivity to identify proliferating cells. From E15.5 onwards, mutant embryos exhibited many more Ki67-positive cells in the suprabasal layers compared to controls (E16.5 shown in Fig. 1A,B). For example, at E16.5 Fatp4 mutant embryos displayed 2.6 fold as many proliferating cells as did controls (P<0.01; Fig. 1C). A significant increase in suprabasal proliferating cells was also observed in Fatp4 mutants as late as E18.5 (data not shown). These results show that the epidermal hyperplasia of Fatp4 mutants is associated with an increased number of suprabasal proliferating cells.
Figure 1. Epidermal hyperplasia in Fatp4 mutants is associated with hyperproliferation of suprabasal keratinocytes.
(A and B) Dorsal skin sections from E16.5 Fatp4−/− (B) and control littermates (A) were subjected to double immunofluorescence analyses for Ki67 (green) and laminin γ1 (red). Laminin γ1 demarcates the dermo-epidermal boundary. Mutants exhibited an increased number of Ki67-positive cells (arrowheads) in the suprabasal layer compared to controls. Hair follicle progenitors are indicated by the asterisk. Scale bar is 100 μm. (C) Fatp4 mutants at E16.5 showed 2.6 times as many Ki67-positive suprabasal cells as did controls (51.7±4.5, n=3 mutants, vs. 20.0±5.6, n=3 controls; P<0.01). The Y axis indicates numbers of Ki67-positive suprabasal cells per 100 Ki67-positive basal cells. Error bars indicate standard deviation.
Abnormal skin barrier formation in Fatp4 mutants
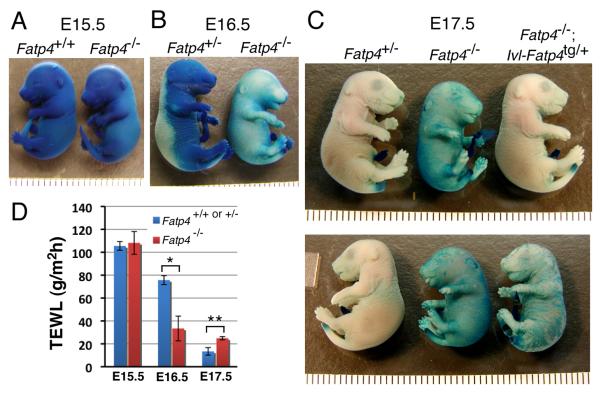
We previously reported that Fatp4 mutants exhibit a defective skin barrier at late fetal stages, as assayed by inward permeability of X-gal at low pH (Moulson et al., 2003). Because the epidermal hyperproliferation was observed from E15.5 onwards, when embryos are still in the uterus and not challenged by the postnatal environment, it is unlikely that the hyperproliferation shown above is a compensatory response to a defective skin barrier in the manner shown previously (Proksch et al., 1991). To investigate the barrier defect in more detail, we reexamined it at multiple stages. At E17.5, barrier formation was complete in control embryos but incomplete in Fatp4 mutants, as indicated by staining of the body wall (Fig. 2C). At E15.5, neither controls nor Fatp4 mutants manifested a skin barrier (Fig. 2A). At E16.5, some of the controls had just begun to establish a barrier dorsally, but the Fatp4 mutants had already established a substantial but imperfect skin barrier throughout the body wall (Fig. 2B), comparable to that observed in E17.5 mutants (compare to Fig. 2C). This premature barrier formation in Fatp4 mutants was confirmed by an alternative skin permeability assay using toluidine blue (data not shown). The precocious barrier did not, however, progress into a complete barrier at later stages. It remains to be determined whether premature barrier formation exhibits the typical dorsal to ventral wave.
Figure 2. Abnormal skin barrier formation in Fatp4 mutants.
Embryos at E15.5 to E17.5 were tested for barrier integrity by (A-C) X-Gal permeability assays and (D) TEWL assays. (A) At E15.5 both controls and mutants were permeable to X-Gal, as indicated by the blue staining throughout the body wall. (B) At E16.5 the controls had just begun to establish a skin barrier dorsally, while Fatp4 mutants had already formed a substantial skin barrier throughout the body wall. (C) Whereas Fatp4 mutants showed incomplete barrier formation at E17.5, Fatp4−/−; Ivl-Fatp4tg/+ fetuses displayed a remedied barrier, comparable to that of normal Fatp4+/− embryos (upper panel). The barrier defect at E17.5 was only partially rescued in female Fatp4 mutants carrying a hemizygous X-linked Ivl-Fatp4 transgene (lower panel). Each caliber in A-C is 1 mm. (D) At E15.5 Fatp4 mutants showed little change in TEWL compared to controls (108.2±9.9 g/m2h, n=2 vs. 105.5±3.8 g/m2h, n=5; P>0.05). At E16.5 Fatp4 mutants displayed reduced TEWL compared to controls (33.4±10.8 g/m2h, n=3 vs. 75.7±3.9 g/m2h, n=11; *P<0.05), indicating premature barrier establishment. However, the premature barrier did not progress to completion by E17.5, when control skin had done so (24.5±1.4 g/m2h, n=3 mutants vs. 13.4±3.3 g/m2h, n=3 controls; **P<0.01).
In addition to abnormalities in inward permeability of the skin, Fatp4 mutants also showed defects in outward transepidermal water loss (TEWL) from E16.5 onwards (Fig. 2D). At E15.5, Fatp4 mutants showed little difference in TEWL compared to controls (P>0.05). In contrast, at E16.5 Fatp4 mutants displayed a dramatic reduction in water evaporation rate compared to control littermates (*P<0.05), indicating premature skin barrier formation. However, the premature barrier of Fatp4 mutants did not progress into a complete barrier by E17.5, when control embryos had formed one (**P<0.01).
We have shown that transgenic expression of FATP4 in Fatp4 mutant epidermis using the involucrin (Ivl) promoter rescues the neonatal lethality and ameliorates the skin phenotypes (Moulson et al., 2007); here we show that an effective barrier was established in Fatp4−/−; Ivl-Fatp4tg/+ mice carrying an autosomal Ivl-Fatp4 transgene (upper panel in Fig. 2C). In contrast, the abnormalities in barrier formation were only partially rescued in female Fatp4 mutants carrying a hemizygous X-linked Ivl-Fatp4 transgene (lower panel in Fig. 2C), which is consistent with a mosaic pattern of transgene expression due to random X-inactivation in females. Together, these studies show crucial, skin-intrinsic roles for FATP4 in the development of skin.
Gene expression analysis of Fatp4 mutant skin
To identify the possible mechanisms accounting for the abnormal development of skin and its appendages in Fatp4 mutants, we carried out microarray analyses with skin RNAs from E15.5 Fatp4+/− and Fatp4−/− embryos. E15.5 was chosen because it is the age at which the skin phenotype first becomes apparent, increasing the likelihood that observed changes in gene expression would be causative of, rather than a result of, the phenotype. Total skin RNAs were isolated and hybridized in analytical duplicates to Affymetrix oligonucleotide microarrays. Both statistical and nonstatistical criteria were applied to filter the array data, as described in Materials and Methods. In comparisons of Fatp4 mutant samples to heterozygous littermates, 531 and 213 probe sets were found to show at least a 1.5-fold increase or decrease in expression, respectively. Out of those probe sets, 338 and 148 genes, respectively, were mapped to Gene-Ontology (GO) terms (supplementary Tables 2 and 3).
Table 1 lists a subset of genes that showed differential expression in Fatp4 mutants. Given the impact of FATP4 loss on proliferation and differentiation in the epidermis, we focused our analysis on gene classes known to be involved in regulating the formation of skin and its appendages. Genes encoding EGF family ligands and signal transducer and activator of transcription 3 (STAT3) were significantly upregulated, along with genes regulating cell proliferation, such as the Myc, Fos, Jun, and Junb oncogenes. Additionally, there was significant upregulation of the genes encoding proteins involved in keratinocyte differentiation and barrier formation, such as Ivl, small proline-rich proteins (Sprr's), and S100 calcium binding proteins (S100a's). This pattern is consistent with the premature barrier formation observed in Fatp4 mutants (Fig. 2).
Table 1.
Genes changed in the skin at E15.5 in Fatp4−/− embryos, compared to controls.
| GenBank accession |
Gene symbol |
Gene name | Fold change by arrays^ |
Fold change by RT-qPCR# |
|---|---|---|---|---|
| Immune response | ||||
| U50712 | Ccl12 | chemokine (C-C motif) ligand 12 | 7.3 | 6.1*** |
| NM_011940 | Ifi202b | interferon activated gene 202B | 10.4 | 8.3*** |
| AF333251 | Il24 | interleukin 24 | 4.7 | ↑ |
| NM_011019 | Osmr | oncostatin M receptor | 5.1 | 2.6** |
| NM_021367 | Tslp | thymic stromal lymphopoietin | 14.6 | |
| Signal transduction | ||||
| NM_009704 | Areg | amphiregulin | 4.1 | 11.2*** |
| AV239587 | Bmp2 | bone morphogenetic protein 2 | −1.6 | |
| NM_053087 | Epgn | epithelial mitogen | 44.6 | 30.9*** |
| NM_007950 | Ereg | epiregulin | 9.0 | 3.3** |
| AU023352 | Jak2 | Janus kinase 2 | 1.8 | |
| BB241535 | Socs3 | suppressor of cytokine signaling 3 | 9.8 | 2.9** |
| AI325183 | Stat3 | signal transducer and activator of transcription 3 | 2.1 | 1.8** |
| U65016 | Tgfa | transforming growth factor alpha | 3.9 | 1.1 |
| Transcription | ||||
| AV231755 | hr | hairless | 3.7 | 2.6** |
| NM_010703 | Lef1 | lymphoid enhancer binding factor 1 | −2.3 | −2.1** |
| Cell proliferation | ||||
| AV026617 | Fos | FBJ osteosarcoma oncogene | 1.6 | |
| NM_010235 | Fosl1 | fos-like antigen 1 | 4.4 | 19.8*** |
| NM_010591 | Jun | Jun oncogene | 1.9 | |
| NM_008416 | Junb | Jun-B oncogene | 7.3 | 7.0*** |
| BC006728 | Myc | myelocytomatosis oncogene | 1.5 | 1.6** |
| AI462015 | Nfkbia | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha |
2.0 | |
| NM_018754 | Sfn | stratifin | 2.1 | |
| Cell adhesion | ||||
| BC008626 | Icam1 | intercellular adhesion molecule | 3.4 | 2.2*** |
| L04678 | Itgb4 | integrin beta 4 | 1.7 | |
| Keratinocyte differentiation | ||||
| NM_008412 | Ivl | involucrin | 20.9 | |
| NM_011313 | S100a6 | S100 calcium binding protein A6 (calcyclin) | 1.6 | |
| NM_013650 | S100a8 | S100 calcium binding protein A8 (calgranulin A) | 9.2 | 8.0** |
| NM_009114 | S100a9 | S100 calcium binding protein A9 (calgranulin B) | 21.4 | 23.5*** |
| NM_009264 | Sprr1a | small proline-rich protein 1A | 9.9 | 10.7*** |
| NM_009265 | Sprr1b | small proline-rich protein 1B | 18.7 | |
| NM_011468 | Sprr2a | small proline-rich protein 2A | 20.2 | |
| NM_011470 | Sprr2d | small proline-rich protein 2D | 19.4 | |
| NM_011472 | Sprr2f | small proline-rich protein 2F | 8.1 | |
| BC026422 | Tgm1 | transglutaminase 1, K polypeptide | 2.4 | |
| Protein modification | ||||
| NM_026268 | Dusp6 | dual specificity phosphatase 6 | 3.5 | 2.6*** |
| BE136125 | Dusp7 | dual specificity phosphatase 7 | 3.0 | |
| BB261602 | Map2k6 | mitogen activated protein kinase kinase 6 | −2.0 | |
| Intermediate filament cytoskeleton | ||||
| NM_008470 | Krt16 (Krt1-16) | keratin 16 (keratin complex 1, acidic, gene 16) | 70.2 | |
| AA798563 | Krt17 (Krt1-17) | keratin 17 (keratin complex 1, acidic, gene 17) | 8.3 | |
| NM_008476 | Krt6a (Krt2-6a) | keratin 6A (keratin complex 2, basic, gene 6a) | 4.5 | |
| NM_010669 | Krt6b (Krt2-6g) | keratin 6B (keratin complex 2, basic, gene 6g) | 166.9 | |
| Intercellular junction | ||||
| AV239646 | Gjb2 | gap junction membrane channel protein beta 2 | 3.9 | 3.0* |
| Peptidase activity | ||||
| NM_007403 | Adam8 | a disintegrin and metallopeptidase domain 8 | 5.4 | 6.6*** |
| NM_011177 | Klk6 (Prss18) | kallikrein related-peptidase 6 (protease, serine, 18) | 12.0 | |
| BB283507 | Klk7 | kallikrein related-peptidase 7 | 20.7 | 64.2*** |
| NM_010809 | Mmp3 | matrix metalloproteinase 3 | 20.1 | 39.0*** |
| Peptidase inhibitor activity | ||||
| NM_011111 | Serpinb2 | serine (or cysteine) proteinase inhibitor, clade B, member 2 |
7.8 | 5.4*** |
| AK003744 | Cst6 | cystatin E/M | 9.9 | 8.3*** |
Data indicate gene expression changes in Fatp4 mutant compared to control skin by array analyses in analytical duplicates, as described in Materials and Methods.
26 genes were tested by RT-qPCR, using RNA from three separate litters (5 controls and 6 Fatp4 mutants) different from those used for the arrays. Gene expression was quantified as described in Materials and Methods and analyzed by two-way mixed model ANOVA tests.
Changes in 25 of the 26 genes were verified, and all except Il24 are indicated with fold change and statistical significance P < 0.05;
P <0.01;
P<0.001.
The fold change and statistical significance of Il24 upregulation were unable to be determined because expression of Il24 was undetectable in four out of the five control RNAs assayed.
Ectopic activation of EGFR signaling in Fatp4 mutant epidermis
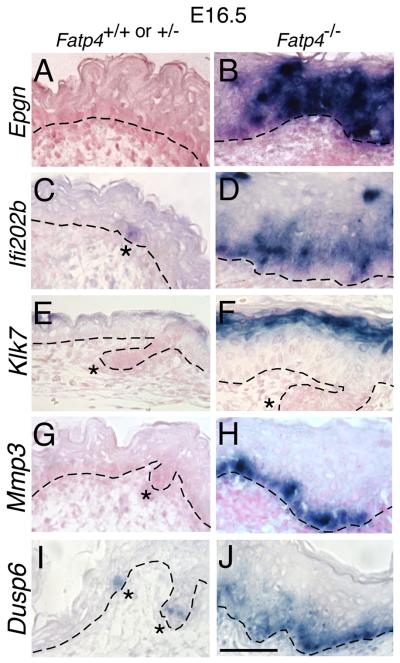
To further investigate the expression of a subset of candidate genes identified in the arrays, RT- quantitative PCR (qPCR) was performed for relative quantification of gene expression using TATA box binding protein (Tbp) as an internal control (Minner and Poumay, 2009). Of the 26 genes tested, all except transforming growth factor alpha (Tgfa) were found to be differentially expressed in Fatp4 mutants compared to controls (Table 1). One group of verified genes encodes proteases, including kallikrein 7 (KLK7), a serine protease involved in desquamation (Komatsu et al., 2003) and matrix metalloproteinase 3 (MMP3); their upregulation was also confirmed by in situ hybridization (Fig. 3 E-H). Another group of genes verified by RT-qPCR encodes EGF family ligands involved in the regulation of keratinocyte functions: amphiregulin (Areg), epiregulin (Ereg), and epithelial mitogen/epigen (Epgn). These three genes are clustered within 110 kb on mouse chromosome 5 (Harris et al., 2003). By in situ hybridization Epgn transcripts were found throughout the epidermis in Fatp4 mutants, whereas no expression was detected in controls (Fig. 3A,B). Upregulation of Areg and Ereg was also validated by in situ hybridization (data not shown). Concomitant with this, Fatp4 mutants displayed ectopic suprabasal activation of the EGFR, indicated by the expanded domain of phosphorylated EGFR (pEGFR), from E15.5 onwards (Fig. 4A,B and Fig. 5M,N). Suprabasal pEGFR localization is an indicator of epidermal hyperproliferation (Fuchs and Raghavan, 2002; Weiss et al., 1984).
Figure 3. Abnormal gene expression in Fatp4 mutant skin.
Dorsal skin sections from Fatp4−/− and control littermates at E16.5 were subjected to in situ hybridization with the indicated riboprobes. Sections A to H were counterstained with nuclear fast red. (A and B) Epgn was ectopically expressed throughout the epidermis in mutants at E16.5. (C and D) Ifi202b was ectopically activated in mutants in interfollicular epidermis but was detected only in hair placodes in controls (asterisk in C). (E and F) Klk7 was induced in the granular and upper spinous layers in mutants. (G and H) Induction of Mmp3 in mutants was detected mostly in the basal layer. (I and J) Dusp6 was induced in mutants in interfollicular epidermis (J) but was expressed only in hair follicle progenitors in controls (asterisks in I). Dashed lines demarcate the dermo-epidermal boundary. Hair follicle progenitors are indicated by asterisks. Scale bar is 50 μm.
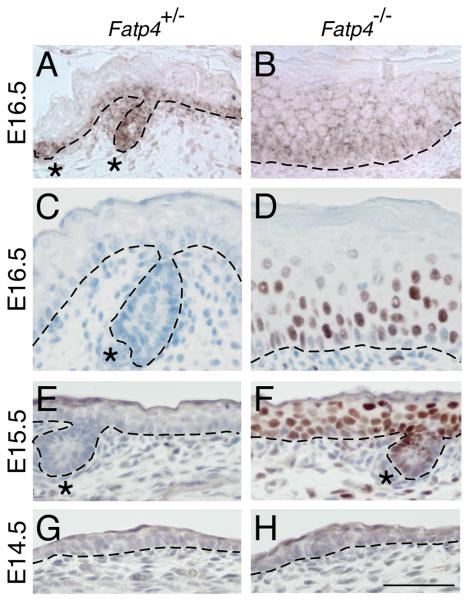
Figure 4. Activation of EGFR and STAT3 in Fatp4 mutant skin.
Dorsal skin sections from Fatp4+/− and Fatp4−/− littermates at the indicated ages were analyzed by immunohistochemistry with antibodies against pEGFR (A and B) or pSTAT3 (C to H; counterstained with hematoxylin). (A and B) Whereas pEGFR signals were detected only in basal keratinocytes in controls, ectopic signals were detected in suprabasal keratinocytes in mutants. (C-H) At E15.5 and E16.5, only mutants showed distinct nuclear pSTAT3 signals in basal and suprabasal layers. At E14.5, neither controls nor mutants showed epidermal pSTAT3 signals. Hair follicle progenitors are indicated by asterisks. Dashed lines demarcate the dermo-epidermal boundary. Scale bar is 50 μm.
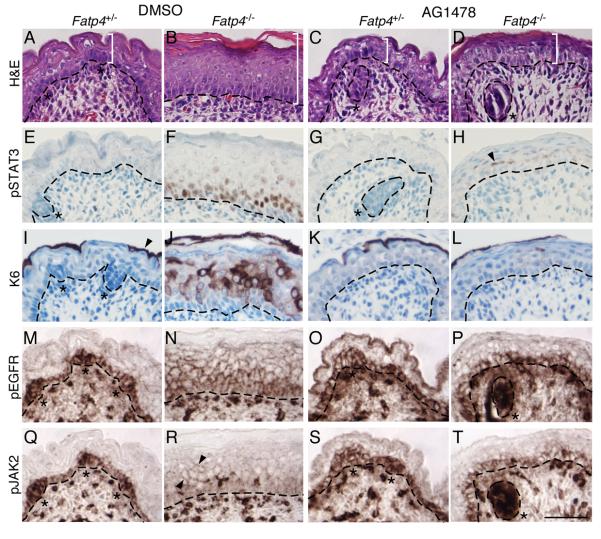
Figure. 5. AG1478 treatment suppresses STAT3 activation and epidermal hyperplasia.
Embryos were dissected at E16.5 from pregnant females injected i.p. with AG1478 or DMSO vehicle daily for 2 days. Dorsal skin sections of Fatp4−/− and control littermates were then subjected to hematoxylin and eosin staining (A-D) and immunohistochemical analyses (E-T). Some sections were counterstained with hematoxylin (E-L). (A-D) AG1478 treatment suppressed the thickened epidermis phenotype (compare white brackets in B and D) but not the flat skin phenotype (compare C to D). (E-H) Nuclear localization of pSTAT3 in mutants was dramatically reduced by AG1478 (compare F to H; arrowhead in H marks residual nuclear signal). (I-L) AG1478 treatment dramatically reduced the ectopic expression of keratin 6 (compare J to L), which is normally expressed only in periderm (arrowhead in I). (M-P) Ectopic activation of the EGFR in the suprabasal layer of mutants was greatly reduced by AG1478 (compare N to P). (Q-T) AG1478 treatment also reduced ectopic JAK2 activation in mutant suprabasal layers (arrowhead in R; compare R to T). Note the pEGFR and pJAK2 signals in dermal blood vessels in both controls and mutants. Dashed lines demarcate the dermo-epidermal boundary. Hair follicle progenitors are indicated by asterisks. Scale bar is 50 μm.
EGFR signaling can be exerted through multiple downstream pathways, including MAPK superfamily cascades (ERK, JNK, p38), the PI3K/Akt pathway (Dent et al., 2003), and the STAT3 pathway (Buettner et al., 2002). For example, Ras/ERK-mediated EGFR signaling can regulate cell growth and transformation (Howe et al., 1992). Although Fatp4 mutants exhibited ectopic activation of EGFR in suprabasal keratinocytes, they showed reduced activation of ERK in the epidermis at E15.5 (supplementary Fig. 1). Because ERK activity reflects the opposing actions of upstream activator kinases (MEK) and inhibitory MAPK phosphatases, reduced ERK activity could result from the observed induction of its inhibitory phosphatase Dusp6, shown by RT-qPCR (Table 1) and in situ hybridization (Fig. 3I,J). Activation of p38 MAPK was not altered in Fatp4 mutant epidermis, as detected by immunostaining for phospho-p38 (supplementary Fig. 1). In contrast, activation of JNK appeared reduced in Fatp4 mutant epidermis, as determined by immunostaining for phospho-JNK (supplementary Fig. 1). It is not clear whether the reduction in JNK activation is related to the skin abnormalities in Fatp4 mutants. Nevertheless, these results suggest that the ectopic EGFR signaling was not exerted via activation of MAPK superfamily cascades, but rather via activation of other pathways.
Activation of STAT3 signaling in Fatp4 mutant epidermis
STAT3 is a signaling molecule activated by both cytokines and growth factors, and STAT3 activation is required for EGFR-mediated cell growth in vitro (e.g., Grandis et al., 1998) and in vivo (Chan et al., 2004). Activation of STAT3 involves phosphorylation by receptor tyrosine kinases or Janus kinases (JAKs), dimerization, and translocation into nuclei, where it serves as a transcription factor to drive expression of downstream target genes (Buettner et al., 2002). Stat3 mRNA was modestly upregulated in Fatp4 mutants (Table 1), but to examine whether the observed ectopic EGFR signaling (Fig. 4A,B and Fig. 5M,N) correlated with ectopic STAT3 activation, skin samples were subjected to immunohistochemical analyses. At E14.5, phosphorylated STAT3 (pSTAT3) was neither detected in controls nor in Fatp4 mutants (Fig. 4G,H). At E15.5, however, Fatp4 mutants showed distinct signals in the nuclei of basal and suprabasal keratinocytes (Fig. 4F), indicating STAT3 activation. Activation of STAT3 signaling in the mutant epidermis continued at E16.5 (Fig. 4D) and was evident at least through E18.5 (data not shown). These data suggest that STAT3 signaling could be an effector of the ectopic EGFR activation, and perhaps other signaling pathways, in Fatp4 mutants.
Besides receptor tyrosine kinases such as EGFR, cytokine receptors are the other major initiators of STAT signaling. As cytokine receptors lack intrinsic tyrosine kinase activity, cytokine-induced activation of STAT is mediated by receptor-associated tyrosine kinases such as JAK2 (Buettner et al., 2002). Interestingly, Fatp4 mutants exhibited ectopic activation of JAK2 in suprabasal layers (Fig. 5R). Furthermore, the array data showed a robust immune response signature in Fatp4 mutants (Table S1). Many cytokines have been found to be expressed in keratinocytes, either constitutively or in pathological states (Grone, 2002). Upregulation of several cytokines in the Fatp4 mutant skin was verified by RT-qPCR and in situ hybridization. For example, whereas Il24 expression was not detectable by RT-qPCR in four of the five control embryos examined, it was induced in all six mutants (Table 1). Induction of Il24 was also validated by in situ hybridization (data not shown). Similarly, upregulation of Ccl12, encoding a chemokine, and Ifi202b, a gene induced by interferons in autoimmune patients (Mondini et al., 2007), was detected in Fatp4 mutants by RT-qPCR (Table 1) and by in situ hybridization (Fig. 3C,D and data not shown). Upregulation of Tslp, encoding thymic stromal lymphopoietin, has been previously verified in Fatp4 mutants by the presence of elevated serum TSLP levels at birth (Demehri et al., 2008). These results suggest that in addition to EGFR-mediated activation, cytokine-mediated STAT3 activation could also account for the skin abnormalities in Fatp4 mutants.
Inhibition of EGFR/STAT3 activation suppresses hyperproliferation
To investigate whether EGFR activation was related mechanistically to the skin abnormalities of Fatp4 mutants, we inhibited EGFR activation using Tyrphostin AG1478, an EGFR-specific tyrosine kinase inhibitor (El-Abaseri et al., 2005; Gazit et al., 1989). Because the skin abnormalities in Fatp4 mutants were not observed until E15.5, pregnant females were injected with AG1478 solubilized in DMSO or with DMSO only at E14.5 and E15.5 and then sacrificed at E16.5. The epidermal hyperplasia normally observed in Fatp4 mutants was suppressed by AG1478 but not by vehicle (DMSO) (compare Fig. 5B to D); the flat skin and sparse hair phenotypes, however, were not remedied. AG1478 treatment did not appear to adversely affect control littermates (compare Fig. 5A to C).
To correlate suppression of epidermal hyperplasia with suppression of EGFR and perhaps also STAT3 activation, immunohistochemical analyses were performed. Upon AG1478 treatment, both suprabasal activation of EGFR and nuclear localization of pSTAT3 were dramatically reduced in Fatp4 mutant skin, compared to vehicle treatment. For example, nuclear pSTAT3 was detected in about 25% of the epidermal cells in the AG1478-treated mutant skin, compared to about 50% in the vehicle-treated mutant skin (compare Fig. 5F to H). Similarly, the AG1478-treated mutants showed only a few pEGFR-positive cells in the spinous layer, whereas the vehicle-treated mutants showed positive cells throughout the spinous layer (compare Fig. 5N to P), as observed with no treatment (Fig. 4B). AG1478 treatment also dramatically reduced the expression of K6 in Fatp4 mutants (compare Fig. 5J to L). We next examined whether the attenuation of STAT3 signaling was correlated with suppressed activation of JAK2, a molecule involved in both EGFR-mediated and cytokine-mediated STAT3 activation. With AG1478 treatment, the activation of JAK2 in the suprabasal layer was significantly reduced as compared to mutants treated with DMSO (compare Fig. 5R to T), suggesting that inhibition of EGFR signaling is sufficient to block JAK2 activation regardless of the presence of cytokines. Alternatively, administration of AG1478 may inhibit JAK2 activation by suppressing cytokine expression.
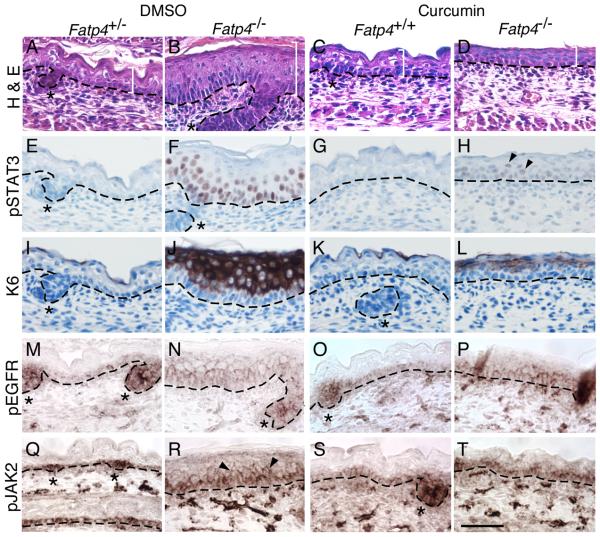
To provide evidence that the effects of AG1478 were not nonspecific, we inhibited EGFR and STAT3 activation in a completely different fashion, using curcumin. Curcumin is an Indian spice isolated from the turmeric plant that is a documented inhibitor of STAT3 activation; it suppresses both phosphorylation and nuclear translocation of STAT3 (Bharti et al., 2003). It also inhibits phosphorylation of EGFR (Korutla and Kumar, 1994) and JAK2 (Natarajan and Bright, 2002) and inhibits the tyrosine kinase activities of the EGFR (Korutla and Kumar, 1994) and Src (Reddy and Aggarwal, 1994), both of which can phosphorylate STAT3. Indeed, the epidermal hyperplasia of Fatp4 mutants was suppressed by curcumin, accompanied by a dramatic reduction of nuclear pSTAT3 (compare Fig. 6B to D, 6F to H). In contrast to AG1478, curcumin only moderately reduced the ectopic activation of EGFR and JAK2 in the suprabasal layer of mutants (compare Fig. 6N to P, 6R to T). But as with AG1478 treatment, the flat skin and sparse hair phenotypes were not ameliorated by curcumin. Taken together, these results suggest that hyperplasia in Fatp4 mutants is mediated by EGFR/STAT3 activation.
Figure. 6. Curcumin treatment suppresses STAT3 activation and epidermal hyperplasia.
E16.5 embryos were dissected from pregnant females injected i.p. with curcumin or vehicle daily for 2 days. Dorsal skin sections from Fatp4−/− and control littermates were then subjected to hematoxylin and eosin staining (A-D) or immunohistochemical analyses (E-T). Some sections were counterstained with hematoxylin (E-L). (A-D) Curcumin treatment suppressed the thickened epidermis phenotype (compare white brackets in B and D) but not the flat skin phenotype (compare C to D). (E-H) Nuclear localization of pSTAT3 was dramatically reduced by curcumin (compare F to H; arrowheads in H mark residual nuclear signals). (I-L) Curcumin treatment drastically reduced the ectopic activation of keratin 6 (compare J to L). (M-P) Ectopic EGFR activation in mutant suprabasal cells was moderately suppressed by curcumin (compare N to P). (Q-T) Curcumin treatment also moderately reduced ectopic JAK2 activation in mutant suprabasal layers (arrowheads in R; compare R to T). Dashed lines demarcate the dermo-epidermal boundary. Hair follicle progenitors are indicated by asterisks. Scale bar is 50 μm.
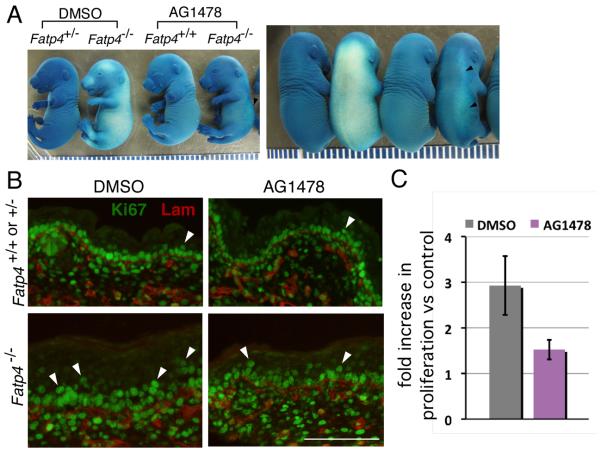
In addition to suppression of hyperplasia, administration of AG1478 (Fig. 7A) and curcumin (data not shown) also inhibited precocious barrier formation in Fatp4 mutants. At E16.5 the vehicle-treated mutants displayed a widespread precocious skin barrier, whereas AG1478-treated mutants had acquired only a slight barrier in the dorso-lateral region (arrowheads in Fig. 7A). Nevertheless, AG1478 treatment did not remedy the incomplete skin barrier observed in mutants from E17.5 onwards (data not shown; curcumin has not been tested in this context). These results suggest that the precocious barrier formation in Fatp4 mutants is due to increased EGFR/STAT3 activation.
Figure. 7. AG1478 treatment suppresses precocious barrier formation and hyperproliferation in Fatp4 mutants.
Embryos were dissected at E16.5 from pregnant females injected i.p. with AG1478 or vehicle (DMSO) daily for 2 days and tested for barrier integrity by X-Gal permeability assays (A) and for hyperproliferation by immunofluorescence analyses (B and C). (A) Both lateral (left panel) and dorsal (right panel) views of embryos are shown. Whereas DMSO-treated Fatp4 mutants displayed a precocious skin barrier over the entire body, AG1478-treated mutants acquired only a slight barrier in the dorso-lateral region (arrowheads). Each caliber is 1 mm. (B) Dorsal skin sections of DMSO-treated mutant embryos exhibited an increased number of Ki67-positive cells (green; arrowheads) in the suprabasal layer. Upon AG1478 treatment, hyperproliferation in the mutant skin was reduced. Laminin γ1 (red) demarcates the dermo-epidermal boundary. Scale bar is 100 μm. (C) Upon AG1478 treatment, Fatp4 mutant skin displayed a statistically significant reduction in the fold increase in proliferation, as compared to DMSO treatment (P<0.01). The Y axis indicates fold increase in epidermal proliferation in Fatp4 mutants versus control littermates.
To determine whether the AG1478-mediated reduction of epidermal hyperplasia was due to prevention of suprabasal keratinocyte hyperproliferation, the skin of vehicle- and drug-treated embryos was analyzed for Ki67 immunoreactivity to identify proliferating cells (Fig. 7B,C). Upon DMSO treatment, Fatp4 mutant skin displayed 2.9 times as many proliferating cells in the suprabasal layer as did control skin (52.5±22.1, n=4 mutants vs. 18.0±5.8, n=4 controls; P<0.05) (Fig. 7C), similar to the results with no treatment (Fig. 1C). In contrast, upon AG1478 treatment, Fatp4 mutant skin displayed only 1.5 times as many proliferating cells in the suprabasal layer vs. control (32.3±7.3, n=4 mutants vs. 21.0±2.9, n=4 controls), a more modest but still statistically significant difference (P<0.05) (Fig. 7C). Nevertheless, the AG1478-mediated reduction in the proliferation increase observed in mutants (the 1.5- vs. 2.9-fold increase in proliferation over controls) was significant (P<0.01) (Fig. 7C). The AG1478-mediated remedy of suprabasal cell hyperproliferation was not associated with a reduction of proliferation in the basal layer, as DMSO- and AG1478-treated mutant epidermis showed similar proliferation in the basal layer (99%, P>0.05).
Discussion
FATP4 and epidermal hyperproliferation
Newborn mice lacking FATP4 display a dramatically thickened, flat epidermis as well as a defective skin barrier. Here we investigated the cellular and molecular mechanisms for the epidermal hyperplasia and barrier defects and made several important observations. 1) The epidermal hyperplasia manifested by Fatp4 mutants was associated with an increased number of proliferating cells in the suprabasal layer of the epidermis. 2) Expression of three EGF family ligands (Areg, Ereg, and Epgn) was increased in Fatp4 mutant epidermis. 3) Phosphorylation of the EGF receptor, an indicator of EGFR activation, was detected ectopically in Fatp4 mutant suprabasal layers. 4) Nuclear localization of phosphorylated STAT3, a downstream effector of EGFR signaling, was detected suprabasally in the mutant but not in the control epidermis. 5) Pharmacological inhibition of EGFR tyrosine kinase activity partially suppressed phospho-STAT3 nuclear translocation, suprabasal keratinocyte proliferation, skin thickening, and precocious skin barrier formation. Together, these findings strongly suggest that the absence of FATP4 during the early stages of skin development leads to an upregulation of the expression of EGF family ligands, and this sets into motion the events described above, leading to epidermal hyperproliferation and skin thickening. The mechanisms whereby the lack of FATP4 causes changes in expression of EGF family member genes remain to be determined.
Involvement of EGFR and STAT3 activation in the epidermis
EGFR signaling functions in diverse aspects of skin biology and pathology, including keratinocyte proliferation, hair follicle morphogenesis, wound healing, psoriasis, and tumorigenesis (Schneider et al., 2008). EGFR signaling is normally confined to the basal layer of the epidermis where proliferating cells reside. Mice carrying a null mutation in Egfr display a thin epidermis and abnormal differentiation of epidermis and hair follicles (Miettinen et al., 1995). Transgenic mice overexpressing TGFα (an EGFR ligand) in the basal layer of the epidermis exhibit scaly skin and epidermal hyperplasia (Vassar and Fuchs, 1991), similar to Fatp4 mutants. However, in contrast to the tight and wrinkle-free skin of Fatp4 mutants, TGFα transgenic mice show increased skin wrinkling. This discrepancy may result from induction and activation of the EGFR suprabasally in the Fatp4 mutant skin, but not in TGFα transgenic mice.
STAT3, an effector molecule for EGFR signaling, has previously been implicated in remodeling of the skin. Mice lacking STAT3 in epidermal keratinocytes display severe defects in wound healing and hair cycling (Sano et al., 1999). In contrast, mice overexpressing STAT3 in epidermis develop psoriasis-like phenotypes spontaneously or in response to wounding (Sano et al., 2005). Along with ectopic activation of STAT3 in suprabasal keratinocytes, Fatp4 mutant skin exhibited upregulation of Icam1, Myc, Socs3, Adam8 (Table 1), and Bcl3 (data not shown), five direct targets of STAT3 (Caldenhoven et al., 1995; Kiuchi et al., 1999; Snyder et al., 2008). However, although AG1478 suppressed both STAT3 activation and epidermal hyperproliferation, upregulation of these STAT3 targets was not suppressed, as assayed by RT-qPCR (data not shown). It is therefore likely that the drug-mediated remedy of epidermal hyperproliferation (and premature skin barrier formation) was exerted through suppression of other STAT3 target genes or through other EGFR effectors. Nevertheless, our results suggest that FATP4 plays a role in controlling keratinocyte proliferation in normal embryos by negatively regulating EGFR/STAT3 signaling (Fig. 8).
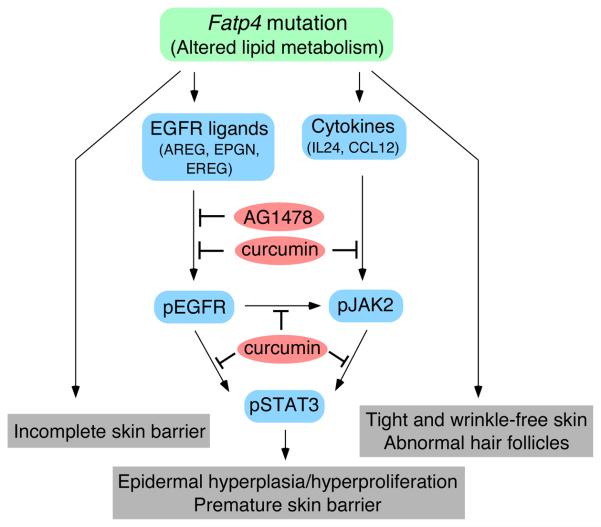
Figure. 8. Mechanisms by which Fatp4 mutation affects the development of skin.
Our data suggest discrete mechanisms for the distinct skin phenotypes of Fatp4 mutants. The incomplete barrier in Fatp4 mutants may be due to direct effects of the lack of FATP4 on the lipid-enriched lamellar membrane that seals the intercellular spaces of the cornified layer. Activation of STAT3 through pathways involving EGF family ligands and cytokines likely accounts for the epidermal hyperplasia/hyperproliferation and premature barrier formation in Fatp4 mutants, though other downstream effectors of EGFR activation may also be involved. Blockage of EGFR/STAT3 activation by either AG1478 or curcumin remedied epidermal hyperplasia/hyperproliferation and suppressed premature barrier formation. However, neither AG1478 nor curcumin treatment remedied the tight and wrinkle-free skin and sparse hair phenotypes, which may be mediated by other pathways. It remains unclear how altered lipid metabolism in Fatp4 mutants initiates the observed alterations in epidermal signaling pathways.
Whereas AG1478-mediated suppression of STAT3 activation was associated with drastic reductions in EGFR and JAK2 activation (Fig. 5), curcumin only moderately reduced EGFR and JAK2 activation (Fig. 6). Despite suppressing hyperplasia, neither AG1478 nor curcumin treatment remedied the flat skin and sparse hair aspects of the wrinkle free phenotype, implying that those abnormalities may be attributable to deregulated expression of other signaling molecules (Fig. 8). This also demonstrates the multipronged nature of the phenotype.
Members of the interleukin 20 (IL-20) subfamily of cytokines, including IL-19, -20, -22, and -24, have been shown to be upregulated in human psoriatic skin and to be involved in the pathogenesis of psoriasis (Sabat et al., 2007); they are able to induce expression of psoriasis-associated proteins, activation of STAT3, hyperproliferation, and abnormal differentiation of epidermal keratinocytes both in vivo (Blumberg et al., 2001) and in vitro (Sa et al., 2007). Most of those phenotypes are also found in Fatp4 mutants. Consistent with activation of STAT3 by IL24 in human keratinocytes (Wang et al., 2002), our data showed upregulation of IL24 and activation of STAT3 in mutant suprabasal keratinocytes (Fig. 3 and 4). Taken together, our data indicate roles for cytokines (in addition to the EGFR) in STAT3-mediated skin abnormalities in Fatp4 mutants (Fig. 8).
FATP4 and barrier formation
In addition to its effects on proliferation, the absence of FATP4 also led to precocious establishment of a skin barrier. The early onset of barrier formation reflected premature expression of barrier-relevant genes, including members of the epidermal differentiation complex. Nevertheless, the barrier never progressed to completion. Interestingly, we did not find upregulated expression of Klf4, which encodes a transcription factor that positively regulates the expression of barrier genes in suprabasal keratinocytes (Jaubert et al., 2003; Patel et al., 2006; Segre et al., 1999). This demonstrates that the lack of FATP4 initiates premature barrier formation independent of increased Klf4 expression. On the other hand, the cellular environment created by the absence of FATP4 may lead to premature activation of KLF4-mediated transcription, perhaps due to increased expression of a required cofactor.
Suppression of EGFR and STAT3 activation by AG1478 and curcumin inhibited precocious barrier formation in Fatp4 mutants (Fig. 7 and data not shown), suggesting that FATP4 somehow negatively regulates EGFR/STAT3 signaling to temporally control barrier initiation in normal embryos. However, that AG1478 treatment did not remedy the incomplete barrier phenotype at later stages suggests that there are EGFR/STAT3-independent roles for FATP4 (Fig. 8). In support of this, Fatp4−/− epidermis exhibits reduced levels of ceramides containing very long chain fatty acids (Herrmann et al., 2003; Moulson et al., 2007), which are crucial for normal barrier integrity (Feingold, 2007). The observed inability of the precocious barrier to progress to a complete barrier is likely due directly to the effects of the lack of FATP4 on the formation or composition of the lipid-enriched lamellar membrane that seals the intercellular spaces of the cornified layer.
How does FATP4 activity influence keratinocyte proliferation and differentiation?
The analysis presented here has provided important new insights into the mechanisms leading to the wrinkle free skin phenotype. But why the absence of FATP4 initiates the cascade of events that causes the phenotype remains to be determined. FATP4 has been reported to exhibit ACS activity (Hall et al., 2005; Herrmann et al., 2001; Jia et al., 2007; Milger et al., 2006), and we recently showed that FATP4 with mutations in its ACS domain fail to rescue the wrinkle free phenotype (Moulson et al., 2007). Furthermore, Fatp4−/− skin fibroblasts show an impaired ability to esterify C24:0 to CoA (Jia et al., 2007). It is therefore possible that the lack of FATP4 activity results in accumulation of free, unesterified fatty acids that serve as ligands to activate signaling pathways disruptive to normal skin development. Free fatty acids are ligands for peroxisome proliferator-activated receptors (PPARs), whose activation promotes keratinocyte differentiation and barrier formation (Di-Poi et al., 2004; Komuves et al., 1998; Schmuth et al., 2004). To test if the lack of FATP4 leads to aberrant keratinocyte differentiation via overactivation of PPAR signaling, we generated mice lacking both FATP4 and either PPARα or PPARβ/δ (Lee et al., 1995; Peters et al., 2000). However, Fatp4 mutants lacking either PPARα or PPARβ/δ still died perinatally and did not show any changes in their wrinkle free phenotype (MHL and JHM, unpublished results). These results suggest that the Fatp4 phenotype is not due to overactivation of PPAR signaling by an elevated level of free fatty acids, though it would be interesting to determine whether elimination of both PPARα and PPARβ/δ activation would ameliorate the skin defects.
That there are already extensive changes in keratinocyte gene expression in Fatp4−/− embryos by E15.5 suggests that FATP4 must have crucial functions at even earlier stages of development. FATP4 also has later roles in promoting barrier formation and hair and sebaceous gland maturation and function (Moulson et al., 2007; MHL and JHM, unpublished observations). Thus, FATP4 has multiple, separable roles in the development and function of skin and its appendages, presumably via direct effects on lipid metabolism and homeostasis that we are currently investigating. A more complete understanding of FATP4 biology will provide important insights into the pathophysiology of ichthyosis prematurity syndrome, an autosomal recessive congenital disorder caused by FATP4 mutations that leads to premature birth, respiratory distress, and edematous skin with severe ichthyotic scaling (Klar et al., 2009). It will be interesting to determine whether these patients show defects in epidermal proliferation and lipid metabolism that are similar to those observed in Fatp4 mutant mice.
Supplementary Material
Acknowledgments
We thank Cheng-Hwang Perng, Bioinformatics Center, National Yang-Ming University for quality check and statistical analyses of the array data. We are grateful to: Seth Blackshaw for important suggestions on the in situ hybridization protocol; Darlene Stewart and Teresa Tolley of the Pulmonary Morphology Core (supported by NIH P01HL029594) for assistance with histology; Jennifer Richardson for genotyping mice; the Mouse Genetics Core for husbandry services; the Renal Disease Models Core (NIH P30DK079333) for qPCR and imaging support; and Raphael Kopan for critical comments on the manuscript. This work was supported by NIH R01AR049269 (to JHM) and by NSC93-3112-B-010-001 (to SCL). Mice were housed in a facility supported by NIH C06RR015502.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J. Immunol. 2003;171:3863–3871. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, Haugen H, Jelinek L, Kelly JD, Madden K, Maurer MF, Parrish-Novak J, Prunkard D, Sexson S, Sprecher C, Waggie K, West J, Whitmore TE, Yao L, Kuechle MK, Dale BA, Chandrasekher YA. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Caldenhoven E, van Dijk T, Raaijmakers JA, Lammers JW, Koenderman L, De Groot RP. Activation of the STAT3/acute phase response factor transcription factor by interleukin-5. J. Biol. Chem. 1995;270:25778–25784. doi: 10.1074/jbc.270.43.25778. [DOI] [PubMed] [Google Scholar]
- Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R. Notch-deficient skin induces a lethal systemic Blymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent P, Yacoub A, Contessa J, Caron R, Amorino G, Valerie K, Hagan MP, Grant S, Schmidt-Ullrich R. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat. Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:sariao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Di-Poi N, Michalik L, Desvergne B, Wahli W. Functions of peroxisome proliferator-activated receptors (PPAR) in skin homeostasis. Lipids. 2004;39:1093–1099. doi: 10.1007/s11745-004-1335-y. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Abaseri TB, Fuhrman J, Trempus C, Shendrik I, Tennant RW, Hansen LA. Chemoprevention of UV light-induced skin tumorigenesis by inhibition of the epidermal growth factor receptor. Cancer Res. 2005;65:3958–3965. doi: 10.1158/0008-5472.CAN-04-2204. [DOI] [PubMed] [Google Scholar]
- Elias PM, Jackson SM. What does normal skin do? In: Arndt KA, et al., editors. Cutaneous Medicine and Surgery: An Integrated Program in Dermatology. Vol. 1. W.B. Saunders; Philadelphia: 1996. pp. 46–57. [Google Scholar]
- Feingold KR. Thematic review series: skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J. Lipid Res. 2007;48:2531–2546. doi: 10.1194/jlr.R700013-JLR200. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- Gazit A, Yaish P, Gilon C, Levitzki A. Tyrphostins I: synthesis and biological activity of protein tyrosine kinase inhibitors. J. Med. Chem. 1989;32:2344–2352. doi: 10.1021/jm00130a020. [DOI] [PubMed] [Google Scholar]
- Gimeno RE, Hirsch DJ, Punreddy S, Sun Y, Ortegon AM, Wu H, Daniels T, Stricker-Krongrad A, Lodish HF, Stahl A. Targeted deletion of fatty acid transport protein-4 results in early embryonic lethality. J. Biol. Chem. 2003;278:49512–49516. doi: 10.1074/jbc.M309759200. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth In vitro. J. Clin. Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grone A. Keratinocytes and cytokines. Vet. Immunol. Immunopathol. 2002;88:1–12. doi: 10.1016/s0165-2427(02)00136-8. [DOI] [PubMed] [Google Scholar]
- Hall AM, Wiczer BM, Herrmann T, Stremmel W, Bernlohr DA. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J. Biol. Chem. 2005;280:11948–11954. doi: 10.1074/jbc.M412629200. [DOI] [PubMed] [Google Scholar]
- Hansen LA, Woodson RL, 2nd, Holbus S, Strain K, Lo YC, Yuspa SH. The epidermal growth factor receptor is required to maintain the proliferative population in the basal compartment of epidermal tumors. Cancer Res. 2000;60:3328–3332. [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp. Cell Res. 2003;284:2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Buchkremer F, Gosch I, Hall AM, Bernlohr DA, Stremmel W. Mouse fatty acid transport protein 4 (FATP4): characterization of the gene and functional assessment as a very long chain acyl-CoA synthetase. Gene. 2001;270:31–40. doi: 10.1016/s0378-1119(01)00489-9. [DOI] [PubMed] [Google Scholar]
- Herrmann T, van der Hoeven F, Grone HJ, Stewart AF, Langbein L, Kaiser I, Liebisch G, Gosch I, Buchkremer F, Drobnik W, Schmitz G, Stremmel W. Mice with targeted disruption of the fatty acid transport protein 4 (Fatp 4, Slc27a4) gene show features of lethal restrictive dermopathy. J. Cell Biol. 2003;161:1105–1115. doi: 10.1083/jcb.200207080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe LR, Leevers SJ, Gomez N, Nakielny S, Cohen P, Marshall CJ. Activation of the MAP kinase pathway by the protein kinase raf. Cell. 1992;71:335–342. doi: 10.1016/0092-8674(92)90361-f. [DOI] [PubMed] [Google Scholar]
- Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- Jia Z, Moulson CL, Pei Z, Miner JH, Watkins PA. Fatty acid transport protein 4 is the principal very long chain fatty acyl-CoA synthetase in skin fibroblasts. J. Biol. Chem. 2007;282:20573–20583. doi: 10.1074/jbc.M700568200. [DOI] [PubMed] [Google Scholar]
- Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, Hibi M, Hirano T. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar J, Schweiger M, Zimmerman R, Zechner R, Li H, Torma H, Vahlquist A, Bouadjar B, Dahl N, Fischer J. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am. J. Hum. Genet. 2009;85:248–253. doi: 10.1016/j.ajhg.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, Saijoh K. Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J. Invest. Dermatol. 2003;121:542–549. doi: 10.1046/j.1523-1747.2003.12363.x. [DOI] [PubMed] [Google Scholar]
- Komuves LG, Hanley K, Jiang Y, Elias PM, Williams ML, Feingold KR. Ligands and activators of nuclear hormone receptors regulate epidermal differentiation during fetal rat skin development. J. Invest. Dermatol. 1998;111:429–433. doi: 10.1046/j.1523-1747.1998.00296.x. [DOI] [PubMed] [Google Scholar]
- Korutla L, Kumar R. Inhibitory effect of curcumin on epidermal growth factor receptor kinase activity in A431 cells. Biochim. Biophys. Acta. 1994;1224:597–600. doi: 10.1016/0167-4889(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MH, Leimeister C, Gessler M, Kopan R. Activation of the Notch pathway in the hair cortex leads to aberrant differentiation of the adjacent hair-shaft layers. Development. 2000;127:2421–2432. doi: 10.1242/dev.127.11.2421. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Milger K, Herrmann T, Becker C, Gotthardt D, Zickwolf J, Ehehalt R, Watkins PA, Stremmel W, Fullekrug J. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J. Cell Sci. 2006;119:4678–4688. doi: 10.1242/jcs.03280. [DOI] [PubMed] [Google Scholar]
- Millar SE. Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- Minner F, Poumay Y. Candidate housekeeping genes require evaluation before their selection for studies of human epidermal keratinocytes. J. Invest. Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- Mondini M, Vidali M, Airo P, De Andrea M, Riboldi P, Meroni PL, Gariglio M, Landolfo S. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann. N. Y. Acad. Sci. 2007;1110:47–56. doi: 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- Moulson CL, Lin MH, White JM, Newberry EP, Davidson NO, Miner JH. Keratinocyte-specific expression of fatty acid transport protein 4 rescues the wrinkle-free phenotype in Slc27a4/Fatp4 mutant mice. J. Biol. Chem. 2007;282:15912–15920. doi: 10.1074/jbc.M701779200. [DOI] [PubMed] [Google Scholar]
- Moulson CL, Martin DR, Lugus JJ, Schaffer JE, Lind AC, Miner JH. Cloning of wrinkle-free, a previously uncharacterized mouse mutation, reveals crucial roles for fatty acid transport protein 4 in skin and hair development. Proc. Natl. Acad. Sci. U S A. 2003;100:5274–5279. doi: 10.1073/pnas.0431186100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Bright JJ. Curcumin inhibits experimental allergic encephalomyelitis by blocking IL-12 signaling through Janus kinase-STAT pathway in T lymphocytes. J. Immunol. 2002;168:6506–6013. doi: 10.4049/jimmunol.168.12.6506. [DOI] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc. Natl. Acad. Sci. USA. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol. Cell. Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proksch E, Feingold KR, Man MQ, Elias PM. Barrier function regulates epidermal DNA synthesis. J. Clin. Invest. 1991;87:1668–1673. doi: 10.1172/JCI115183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Aggarwal BB. Curcumin is a non-competitive and selective inhibitor of phosphorylase kinase. FEBS Lett. 1994;341:19–22. doi: 10.1016/0014-5793(94)80232-7. [DOI] [PubMed] [Google Scholar]
- Richards MR, Harp JD, Ory DS, Schaffer JE. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J. Lipid Res. 2006;47:665–672. doi: 10.1194/jlr.M500514-JLR200. [DOI] [PubMed] [Google Scholar]
- Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, Kasman I, Winer J, Modrusan Z, Danilenko DM, Ouyang W. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J. Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- Sabat R, Philipp S, Hoflich C, Kreutzer S, Wallace E, Asadullah K, Volk HD, Sterry W, Wolk K. Immunopathogenesis of psoriasis. Exp. Dermatol. 2007;16:779–798. doi: 10.1111/j.1600-0625.2007.00629.x. [DOI] [PubMed] [Google Scholar]
- Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, Itami S, Nickoloff BJ, DiGiovanni J. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer JE, Lodish HF. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 1994;79:427–436. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schmuth M, Haqq CM, Cairns WJ, Holder JC, Dorsam S, Chang S, Lau P, Fowler AJ, Chuang G, Moser AH, Brown BE, Mao-Qiang M, Uchida Y, Schoonjans K, Auwerx J, Chambon P, Willson TM, Elias PM, Feingold KR. Peroxisome proliferator-activated receptor (PPAR)-beta/delta stimulates differentiation and lipid accumulation in keratinocytes. J. Invest. Dermatol. 2004;122:971–983. doi: 10.1111/j.0022-202X.2004.22412.x. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Werner S, Paus R, Wolf E. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am. J. Pathol. 2008;173:14–24. doi: 10.2353/ajpath.2008.070942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Snyder M, Huang XY, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008;283:3791–3798. doi: 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- Vassar R, Fuchs E. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 1991;5:714–727. doi: 10.1101/gad.5.5.714. [DOI] [PubMed] [Google Scholar]
- Wang M, Tan Z, Zhang R, Kotenko SV, Liang P. Interleukin 24 (MDA-7/MOB-5) signals through two heterodimeric receptors, IL-22R1/IL-20R2 and IL-20R1/IL-20R2. J. Biol. Chem. 2002;277:7341–7347. doi: 10.1074/jbc.M106043200. [DOI] [PubMed] [Google Scholar]
- Weiss RA, Eichner R, Sun TT. Monoclonal antibody analysis of keratin expression in epidermal diseases: a 48- and 56-kdalton keratin as molecular markers for hyperproliferative keratinocytes. J. Cell Biol. 1984;98:1397–1406. doi: 10.1083/jcb.98.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.