Abstract
The complete consensus genome sequences of avian paramyxovirus (APMV) serotype 2 strains Bangor, England and Kenya were determined and compared with those of APMV-2 prototype strain Yucaipa and other paramyxoviruses. The genome lengths of APMV-2 strains Bangor, England and Kenya are 15024, 14904, 14916 nucleotides (nt), respectively, compared to 14904 nt for Yucaipa. Each genome consists of six non-overlapping genes in the order of 3′N-P/V/W-M-F-HN-L5′, with a 55-nt leader at the 3′end. The length of the trailer at the 5′ end of strain Bangor was 173 nt, compared to 154 nt for strains England, Kenya, and Yucaipa. In general, sequence comparison of APMV-2 strains England and Kenya with strain Yucaipa have 94.5 and 88% nt and 96 and 92% aggregate amino acid (aa) identity, respectively. In contrast, strain Bangor has a much lower percent nt identity (70.4, 69.4, and 70.8%) and aa identity (75.3, 76.2, and 76.3%) with strains Yucaipa, England, and Kenya, respectively. Furthermore, strain Bangor has a single basic aa residue (101TLPSAR↓F108) at the fusion protein cleavage site compared to the dibasic aa (93DKPASR↓F100) found in those of other three strains. Reciprocal cross-hemagglutination inhibition (HI) and cross-neutralization assays using post-infection chicken sera indicated that strain Bangor is antigenically related to the other APMV-2 strains, but with a 4- to 8-fold difference in homologous versus heterologous HI titer. These differences in antigenic relatedness suggests that these four APMV-2 strains represent a single serotype with two antigenic subgroups, and this is strongly supported by the dimorphism in nt and aa sequence identity.
Introduction
The family Paramyxoviridae is large and diverse and includes members that have been isolated from many species of avian, terrestrial, and aquatic animals around the world (Lamb and Parks, 2007; Wang and Eaton, 2001). Paramyxoviruses are pleomorphic, enveloped, cytoplasmic viruses with a non-segmented negative-strand RNA genome. Paramyxoviruses are divided into two subfamilies, Paramyxovirinae and Pneumovirinae, based on structure, genome organization, and sequence relatedness (Lamb et al., 2005). Subfamily Paramyxovirinae comprises five genera; Respirovirus (including Sendai virus [SeV] and human parainfluenza virus types 1 and 3 [HPIV-1 and -3]), Rubulavirus (including simian virus type 5 [SV5], mumps virus [MuV], and human parainfluenza virus types 2 and 4 [HPIV-2 and -4]), Morbillivirus (including measles [MeV] and canine distemper [CDV] viruses), Henipavirus (including Hendra [HeV] and Nipah [NiV] viruses), and Avulavirus (comprising the nine serotypes of avian paramyxoviruses [APMV-1 to -9]). Subfamily Pneumovirinae contains two genera, Pneumovirus (comprising human respiratory syncytial virus [HRSV] and its animal counterparts) and Metapneumovirus (comprising human metapneumovirus [HMPV] and its avian counterpart [AMPV].
The genome lengths of members of Paramyxoviridae range from 15 to 19 kb and contain 6–10 genes arranged in tandem (Lamb and Parks, 2007). All members of Paramyxoviridae examined to date encode a major nucleocapsid protein (N) that binds the entire length of the genomic and the replicative antigenomic RNAs, a nucleocapsid phosphoprotein (P) that is a polymerase co-factor, a large protein (L) that is the major polymerase subunit and bears catalytic domains, a matrix protein (M) that lines the inner surface of the envelope, a fusion glycoprotein (F) that is a surface antigen that mediates viral penetration and syncytium formation and a major glycoprotein (G) or hemagglutinin-neuraminidase (HN) glycoprotein that is a second surface antigen and mediates attachment.
The genome termini of members of Paramyxoviridae consist of extragenic regions, called the 3′-leader and 5′-trailer: the 3′-leader region contains the genome promoter, and the trailer encodes the 3′ end of the antigenome, which is the full-length positive-sense replicative intermediate, which contains the antigenome promoter. Each gene starts with a conserved gene start (GS) sequence and ends with a conserved gene end (GE) sequence. Transcription begins at the 3′-leader region and proceeds in a sequential manner by a start–stop mechanism that is guided by short, conserved GS and GE signals that flank each gene (Lamb & Parks, 2007). The genes are separated by non-coding intergenic sequences (IGS) that are conserved in length and sequence among the different gene junctions for some genera (Respirovirus, Morbillivirus, and Henipavirus) and are non-conserved in sequence or length for others (Rubulavirus, Avulavirus, Pneumovirus, and Metapneumovirus). For the members of subfamily Paramyxovirinae, efficient genome replication depends on the total genome nucleotide (nt) length being an even multiple of six, known as ‘rule of six’ (Kolakofsky et al., 1998), which is thought to reflect a requirement of nucleocapsid structure. Most members of subfamily Paramyxovirinae encode three different proteins, namely P, V and W (or I, in case of genus Rubulavirus), from the P/V gene due to frame-shifting into alternative open reading frames (ORFs) by RNA editing. RNA editing involves the insertion of one or more G residues at a specific motif midway along the P/V gene during transcription; yielding subpopulations of P/V mRNA have frame shifts into each of the three reading frames. In the case of genus Avulavirus, the unedited mRNA encodes the P protein. The insertion of a single G residue at the P editing site shifts the reading frame to access a downstream ORF encoding a highly conserved cysteine motif, resulting in the V protein. The V protein of subfamily Paramyxovirinae has been implicated in the regulation of viral RNA synthesis (Horikami et al., 1996; Lin et al., 2005) and in counteracting host antiviral responses (Goodbourn et al., 2000). Alternatively, the insertion of two G residues shifts the reading frame to access a third, shorter internal ORF that leads to production of the W protein, whose function is not yet understood (Steward et al., 1993).
The APMVs have been classified into nine different serotypes based on hemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays (Alexander, 2003). Newcastle disease virus (NDV) belongs to serotype 1. NDV is the most characterized member among all the APMV serotypes because it produces severe disease in chickens (Alexander, 1980). APMV-2 was first isolated in 1956 in Yucaipa, California from a diseased chicken that was also infected with infectious laryngotracheitis virus (Bankowski et al., 1960). Since then, many APMV-2 strains have been isolated from chickens, turkeys and feral birds around the world (Alexander et al., 1982; Asahara et al., 1973; Collings et al., 1975; Fleury and Alexander, 1979; Goodman and Hanson, 1988; Lang et al., 1975; Lipkind et al., 1979, 1982; Mbugua and Karstad, 1985; Nymadawa et al., 1977; Shihmanter et al., 1997; Weisman et al., 1984; Zhang et al., 2006, 2007). APMV-2 strain Bangor was isolated from a finch during a routine quarantine evaluation, and the biological and serological characterization suggested that strain Bangor might represent a separate serotype or as a subgroup within serotype 2 (McFerran et al., 1973; McFerran et al., 1974).
As noted, APMV-2 strains have been isolated from a wide variety of avian species from different parts of the world. But little is known about the serological and genetic relationships among these strains. The information is important for understanding virus evolution and epidemiology and for development of vaccines against these viruses. To date, a complete genome sequence is available only for the prototype strain Yucaipa (Subbiah et al., 2008). As a first step towards understanding the serological and genetic relationship among APMV-2 strains, we have determined the complete genome sequences of three other strains of APMV-2; Bangor, England and Kenya, isolated from a finch, a chicken and a gadwell duck, respectively, and describe comparison with the complete genome sequence of prototype strain Yucaipa and other paramyxoviruses. Our sequence and antigenic analyses suggested that APMV- 2 strains can be classified into two genetic subgroups under a single serotype.
Materials and methods
Virus and cells
APMV-2/Chicken/Yucaipa/Cal/56 (APMV-2 Yucaipa) and APMV-2/Finch/N.Ireland/Bangor/73 (APMV-2 Bangor) were received from the National Veterinary Services Laboratory, Ames, Iowa, USA and APMV-2/Chicken/England/7702/06 (APMV-2 England) and APMV-2/Gadwell/Kenya/3/80 (APMV-2 Kenya) were obtained from Veterinary Laboratories Agency, Weybridge, UK. The viruses were grown in 9-day-old embryonated, specific pathogen-free (SPF) chicken eggs. Hemagglutination (HA) titers were determined using 0.5% chicken RBC at room temperature. The ability of the viruses to replicate in cell culture was examined in two established cell lines, namely DF1 chicken fibroblast and Vero African green monkey kidney cells. Both cell lines were grown in Dulbecco's MEM containing 10% fetal bovine serum (FBS) in a 37°C incubator with 5% CO2.
Replication of viruses in cell cultures
Cell monolayers (DF1 and Vero) were infected with a 10−3 dilution of 28 HA units of egg-grown APMV-2 strains Yucaipa, Bangor, England and Kenya and, after 1 h of adsorption, the viral inoculum was replaced with maintenance medium containing 2% FBS with or without the supplementation of exogenous protease (10% allantoic fluid or 1 μg/ml trypsin). The cells were observed daily for cytopathic effects (CPE) and the supernatants of the infected cells were collected every 24 h until the fifth day post-infection (dpi). Virus titers were determined by serial end-point dilution on monolayers of DF1 cells in 96-well plates. The infected cells were immunostained using polyclonal antisera raised against each of the viruses in chickens. Virus titers (TCID50/ml) were calculated using the Reed & Muench method (Reed & Muench, 1938). The ability of the viruses to produce plaques was tested in both cell lines under various conditions, including 1% methylcellulose, 1% low melting agar, or 0.8% noble agar with or without magnesium sulfate (25 mM) and 1% diethylaminoethyl dextran (30 μg/ml), and with and without allantoic fluid. The monolayers were stained with either crystal violet or neutral red in attempts to detect plaques.
Serological analysis
Antisera against APMV-2 strains Yucaipa, Bangor, England and Kenya were prepared separately by single infection of 2-week-old chickens via the intraocular (IO) and intranasal (IN) routes, mimicking natural infection. Briefly, groups of three 2-week-old chickens per group were infected with each virus (28 HAU) at separate times to avoid cross-infection. Two weeks after infection, sera were collected and stored at −20°C. HN-specific antibody titers in the serum samples were determined by HI assay using the homologous virus and chicken RBC as described previously (Alexander, 1997). The cross-reactivity of the sera was determined by HI assay against heterologous APMV-2 strains. The ability of immunized chicken sera to cross-neutralize heterologous APMV-2 strains was determined by a focus reduction microneutralization assay using standard procedures (Borisevich et al., 2007). Briefly, different dilutions of sera were mixed with a constant titer of virus (103 TCID50/ml), incubated for 2 h at room temperature, and transferred to monolayers of DF1 cells in 96-well plates. The plates were incubated for three days at 37°C with 5% CO2. Each plate included both uninfected and infected cell controls. On the third day, the culture medium was removed and cells were fixed with methanol for 30 min and washed with PBS three times. The fixed cells were immunostained to identify virus-containing wells, and a 50% focus reduction was considered as the end point of the titration.
Pathogenicity tests
The virulence of the APMV-2 strains was determined by two standard pathogenicity tests for APMV-1: mean death time (MDT) in 9-day-old embryonated SPF chicken eggs and intracerebral pathogenicity index (ICPI) test in 1-day-old SPF chicks (Alexander 1989). Briefly, for MDT, a series of 10-fold (10-6-10-9) dilutions of fresh infective allantoic fluid in PBS was made and 0.1 ml of each dilution was inoculated into the allantoic cavities of five 9-day-old SPF embryonated chicken eggs (BEE eggs company, PA), which were incubated at 37°C. The eggs were candled 3 times a day for the next 7 days and the time of embryo death, if any, were recorded. The minimum lethal dose (MLD) is the highest virus dilution that kills all the embryos. The MDT is the mean time in hours for the MLD to kill all the inoculated embryos. The MDT has been used to classify APMV-1 strains into the following groups: velogenic strains (taking less than 60 h to kill); mesogenic strains (taking 60 -90 h to kill); and lentogenic strains (taking more than 90 h to kill).
For ICPI, 0.05 ml (1:10 dilution) of fresh infective allantoic fluid of each virus was inoculated into groups of ten 1-day-old SPF chicks via the intracerebral route. The inoculation was done using a 27-gauge needle attached to a 1 ml stepper syringe dispenser that was set to dispense 0.05 ml of inoculum per bird. The birds were inoculated by inserting the needle up to the hub into the right or left rear quadrant of the cranium. The birds were observed for clinical symptoms and mortality once every 8 h for a period of 10 days. At each observation, the birds were scored: 0, if normal, 1, if sick and 2, if dead. The ICPI is the mean score per bird over the 10-day period. Highly virulent (velogenic) viruses give values approaching 2, and avirulent (lentogenic) viruses give values close to 0.
Virus RNA isolation and complete genome sequencing
The viral RNA was isolated from the allantoic fluid of virus-infected eggs using RNeasy kit according to the manufacturer's instructions (QIAGEN, USA). Each of the APMV-2 genomes, except for the 3′ and 5′ termini, was amplified into cDNAs using primers designed from the published APMV-2 strain Yucaipa (Table 1). All primers were commercially synthesized from Integrated DNA Technologies Inc, USA. Briefly, the first-strand cDNA was synthesized from viral RNA by Superscript II kit using random hexamers according to manufacturer's instructions (Invitrogen). PCR was performed using virus specific or consensus primers and Taq polymerase (Invitrogen). The PCR fragments were cloned into TOPO TA cloning kit (Invitrogen) and the clones were sequenced using vector primers. In addition, selected PCR products were purified by agarose gel electrophoresis and sequenced directly. The DNA sequencing was carried out using BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems Inc, USA) in ABI 3130xl genetic analyzer. Every nt in the genome was sequenced at least three times and once directly from RT-PCR product without cloning, thus ensuring a consensus sequence. The sequences of the 3′ and 5′ genomic ends were determined from cDNA prepared by rapid amplification of cDNA ends (RACE) as described previously (Subbiah et al., 2008).
Table. 1.
Primers used to amplify APMV-2 genome based on previously available genome sequence of APMV-2 prototype strain Yucaipa.
| Primer name | position within APMV-2 strain Yucaipa genome | Primer sequence |
|---|---|---|
| Gene Start forward | Gene start consensus | * NNNNNNNGGGGGCGA |
| Gene End reverse | Gene end consensus | * NNNNNNNNNNNTTTTTTCTTAA |
| N Forward | 388-415 | ACATGCGAGCTCACGCAACCCTTGCAGC |
| N Reverse | 1019-1044 | GCCTGATCAAGGACGACATCTTCTTC |
| P Forward | 1758-1781 | CGAAGTCAAGGGCCCGCAAACAAC |
| P Reverse | 2464-2484 | CTGACTAATCTCATTCTTTAT |
| M Forward | 3135-3157 | CCAAAGAGTTGCAGCAGCAAATC |
| F Forward | 5217-5242 | AGTGTCACTACACCAAAAGGAGAAGG |
| HN Forward | 6698-6719 | CCAGTATGTATATCTCTCTGGG |
| L1 Forward | 8869-8890 | ATGCTAGTGAGACACACGCAGG |
| L1 Reverse | 10422-10441 | GAATACACAAAGAATGATTG |
| L2 Forward | 11967-11986 | ATATATCAGCAAATCATGCT |
| L2 Reverse | 13314-13332 | CAGCATACTTGTACCAGCT |
| L3 Forward | 14170-14186 | TCACCCTATTCGGACAG |
N=A/T/C/G
Virus genome sequence alignment and phylogenetic analyses
Sequence compilation and prediction of ORFs were carried out using the SeqMan and EditSeq programs in the Lasergene 6 (DNASTAR) software package (http://www.dnastar.com). The search for matching protein sequences in GenBank was done using the blastp program of the same package. The bootstrap values in phylogenetic tree were calculated using 1000 replicas and the construction of phylogenetic trees was performed by maximum parsimony method using MEGA 4 software (Tamura et al., 2007).
Database accession numbers
The complete genome sequences of APMV-2 strains Bangor, England and Kenya were submitted to GenBank (accession number HM159995, HM159993 and HM159994, respectively). Accession numbers for other paramyxovirus sequences used in this study were: Avulaviruses: APMV-1, AF077761; APMV-2 strain Yucaipa, EU338414; APMV-3, EU403085; APMV-4KR, EU877976; APMV-4HK, FJ177514; APMV-5, GU206351.1; APMV-6TW, NC 003043; APMV-6HK, EU622637; APMV-6FE, EF569970; APMV-7, FJ231524; APMV-8DEL, FJ215863; APMV-8WAK, FJ215864; APMV-9, EU910942. Rubulaviruses: HPIV-2, NC_003443; SV5 (also known as Parainfluenza virus 5), NC_006430; MuV, NC_002200; simian virus 41 (SV41), NC_006428. Respiroviruses: HPIV-1, NC_003461; HPIV-3, NC_001796; SeV, NC_001552, BPIV-3, NC_002161. Henipaviruses: NiV, NC_002728; HeV, NC_001906. Morbilliviruses: CDV, NC_001921; MeV, AF266288; phocine distemper virus (PDV), NC_006383; rinderpest virus (RPV), NC_006296; peste des petits ruminants virus (PPRV), NC_006383; dolphin morbillivirus (DMV), NC_005283; other paramyxovirus: Atlantic salmon paramyxovirus (ASPV), EF646380; Beilong virus (BeV), NC_007803; Fer-de-Lance virus (FDLV), NC_005084; J virus (JV), NC_007454; Menangle virus (MenV), NC_007620; Mossman (MoV), NC_005339; Tupaia paramyxovirus (TpV), NC_002199; Pneumoviruses: HRSV, NC001781; BRSV, NC001989. Metapneumoviruses: AMPV, NC007652; HMPV, NC004148.
Results
In vitro growth characteristics of APMV-2 strains Bangor, England and Kenya
APMV-2 strains Bangor, England and Kenya yielded titers of 210–212 HA units in 9-day-old embryonated SPF chickens eggs at 4 dpi. The inclusion of exogenous protease, either 10% allantoic fluid or 1 μg/ml trypsin, did not affect the efficiency of replication of these viruses in cell culture, indicating a lack of requirement of external proteases for efficient cleavage of the F protein. The viruses grew to one log higher titer in DF1 cells than in Vero cells (data not shown). Viral CPE involved rounding and detachment of the cells. The growth kinetics and the CPE of all the three strains were similar to those of APMV-2 prototype strain Yucaipa. None of the strains produced syncytia or formed plaques but caused single cell infections similar to that of APMV-2 strain Yucaipa (Fig. 1).
Fig. 1.
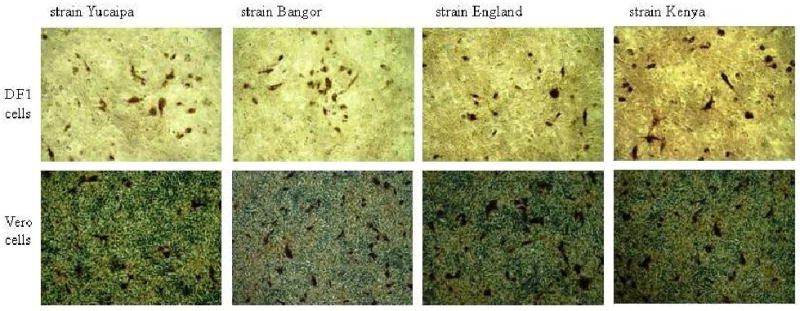
Single cell infection caused by APMV-2 strains Yucaipa, Bangor, England and Kenya in DF1 and Vero cells, three days post infection. The infected cells were immunostained using single-infection sera raised specifically against each of these strains in chickens.
Antigenic relationship among APMV-2 strains
The antigenic relationship among APMV-2 strains Yucaipa, Bangor, England and Kenya was evaluated by reciprocal HI tests using strain specific convalescent sera raised by a single infection of chickens via the IN/IO route. Each of the antiserum exhibited a 2 to 16-fold difference in HI titer between the homologous and heterologous strains (Table 2). Conversely, the HI titer of antisera specifically against strains Bangor, England and Kenya were 4, 4 and 8-fold higher against the homologous strains than against the prototype strain Yucaipa. The antiserum against strain Bangor showed 2-, 2-, and 4-fold higher HI titer against strain Bangor than against strains England, Kenya, and Yucaipa. The antiserum specific for strain England showed 4-fold higher titer against strain England and Kenya than against strains Bangor and Yucaipa. The antiserum specific for strain Kenya showed 8-, 16- and 2-fold higher titers against the homologous strain Kenya than against strains Yucaipa, Bangor, and England, respectively. The ability of antisera to neutralize homologous and heterologous APMV-2 strains was assessed by a microneutralization assay in DF1 cells. The antiserum specific for strain Yucaipa showed 4-fold higher neutralization titer against homologous strain Yucaipa and strains England and Kenya than against strain Bangor. On the contrary, antisera specific for strain Bangor showed 4-fold higher neutralization titer against homologous strain Bangor than against prototype strain Yucaipa and 2-fold higher neutralization titer against homologous strain Bangor than against strains England and Kenya. The antisera specific to strains England and Kenya showed 4-fold higher neutralization titers against their homologous strains compared to those against strains Yucaipa and Bangor, while showing 2-fold difference between either of the strains (Table 2). These reactions indicated the existence of a low level of antigenic differences among APMV-2 strains. These results suggested that the strains Yucaipa, England and Kenya represented one antigenically-distinct subgroup while strain Bangor represented a second subgroup, a distinction that was not observed in most, but not every, comparison.
Table 2.
Antigenic analyses of APMV-2 strains Yucaipa, Bangor, England and Kenya using antisera from chickens infected with the individual strains.
| APMV-2 antiserum | APMV-2 strains | Cross HI titer a | Neutralization titer b |
|---|---|---|---|
| Yucaipa | 160 | 40 | |
| strain Yucaipa | Bangor | 20 | 10 |
| England | 40 | 40 | |
| Kenya | 40 | 40 | |
| Yucaipa | 20 | 10 | |
| strain Bangor | Bangor | 80 | 40 |
| England | 40 | 20 | |
| Kenya | 40 | 20 | |
| Yucaipa | 40 | 20 | |
| strain England | Bangor | 40 | 20 |
| England | 160 | 80 | |
| Kenya | 160 | 40 | |
| Yucaipa | 80 | 20 | |
| strain Kenya | Bangor | 40 | 20 |
| England | 320 | 40 | |
| Kenya | 640 | 80 | |
Cross HI titer is the reciprocal of the highest dilution of antisera that inhibited 4 HA units of the virus.
Neutralization titer was defined as the reciprocal of highest dilution of antisera that caused 50% reduction in the number of infected wells compared to the positive control wells.
The pathogenicity of APMV-2 strains
The pathogenicity of APMV-2 strains Bangor, England and Kenya was evaluated by MDT in 9-day-old embryonated SPF chicken eggs and ICPI test in 1-day-old chicks. The MDT and ICPI values for all the three APMV-2 strains were >168 h and 0, respectively, similar to those of APMV-2 strain Yucaipa (> 168 h and 0, respectively). These results indicated that these APMV-2 strains are avirulent in chickens, similar to lentogenic NDV strains.
Determination of the complete genome sequences of APMV-2 strains Bangor, England and Kenya
We determined the complete genome sequences of APMV-2 strains Bangor, England and Kenya. A number of the initial cDNAs in this analysis was synthesized using primers derived from the published sequence of APMV-2 strain Yucaipa (Table 1). The 3′ and 5′ ends of each genome were determined by RACE procedures (Materials and Methods). Every nt in each complete sequence was confirmed in uncloned RT-PCR cDNA, providing a consensus sequence. The genome of strain England is identical in length (14904 nt) to that of strain Yucaipa, whereas the genome lengths of strains Bangor (15024 nt) and Kenya (14916 nt) are slightly larger than that of strain Yucaipa (14904 nt). The nt lengths of the genomes of all three strains are multiple of six, as in the case of the previously reported sequence for strain Yucaipa. Thus all three strains conform to the rule of six, which is a characteristic of the genome of all members of subfamily Paramyxovirinae (Kolakofsky et al., 1998). All three APMV-2 strains have the gene order of 3′N-P/V/W-M-F-HN-L5′, which is the same as previously reported for strain Yucaipa.
The complete genome and predicted proteins of strain Bangor have 70.4% nt and 75.3% aggregate aa sequence identity with those of the previously sequenced strain Yucaipa, and have 69.4% and 70.8% nt and 76.15% and 76.3% aggregate aa sequence identity with strains England and Kenya, respectively. In contrast, strains England and Kenya are much more closely related to strain Yucaipa, with nt sequence identities of 94.5% and 88%, respectively, and aggregate aa sequence identities of 96.1% and 92.4%, respectively. Thus, strains Yucaipa, England and Kenya are genetically closely related, whereas strain Bangor is somewhat distinct. This is consistent with the finding noted before that strain Bangor is distinct antigenically, and provides unequivocal evidence for dimorphism within the APMV-2 serotype.
The 3′-leader sequences of APMV-2 strains consist of 55 nt, a length that is conserved among almost all the members of the subfamily Paramyxovirinae. The nt sequences of the leader regions of strains Bangor and Yucaipa shows differences at 9 out of 55 nt positions, while those of strains England and Kenya are 100% identical to strain Yucaipa (Fig. 2A). The lengths of trailer regions of APMV-2 strains England and Kenya are 154 nt each, same as strain Yucaipa. But the length of trailer region of strain Bangor is 173 nt (Fig. 2B). This difference accounted for most of the difference in genome length between strain Bangor versus the others. The sequence of trailer region of strains England and Kenya are 100% identical to strain Yucaipa, but the sequence of strain Bangor had only 51.3% nt identity with the other three strains. The proposed GS and GE signal sequences are highly conserved among the APMV-2 strains (Table 3). In general, the conserved GS and GE sequences of all the four strains are (mRNA-sense) 5′-GGGGGCGA(A/C)(A/T) and 5′-T(T/A)(A/T)(A/G)NAAAAA respectively. In strain Bangor, the GS and GE sequences had a number of single nt variations compared to the other three strains (Table 3).
Fig. 2.
Nucleotide (nt) sequence alignment of the leader region (A) and of the 5′-terminal 60 nt of the trailer region (B) of the indicated APMV-2 strains, shown 3′ to 5′ in negative sense. Dots indicate identity with strain Yucaipa. Sequences are in negative-sense. Numbers indicate nt position.
Table. 3.
Molecular features of the genes and their deduced protein products for the four strains of APMV-2:
| Gene | Strain | mRNA features (nt) | Intergenic sequence (nt) |
Deduced protein (aa) |
||||
|---|---|---|---|---|---|---|---|---|
| Gene-start | 5′ UTR | ORF | 3′ UTR | Gene-end | ||||
| N | Yucaipa | GGGGGCGACA | 75 | 1374 | 77 | TTAAGAAAAAA | 7 | 457 |
| Bangor | GGGGGCGACA | 75 | 1374 | 77 | TTAAGAAAAAA | 7 | 457 | |
| England | GGGGGCGACA | 75 | 1374 | 77 | TTAAGAAAAAA | 7 | 457 | |
| Kenya | GGGGGCGACA | 75 | 1374 | 77 | TTAAGAAAAAA | 7 | 457 | |
| P | Yucaipa | GGGGGCGAAG | 61 | 1200 | 97 | TTAACAAAAAA | 7 | 399 |
| Bangor | GGGGGCGAAT | 61 | 1200 | 97 | TAAGAAAAAAA | 7 | 399 | |
| England | GGGGGCGAAG | 61 | 1200 | 97 | TTAACAAAAAA | 7 | 399 | |
| Kenya | GGGGGCGAAG | 61 | 1200 | 97 | TTAACAAAAAA | 7 | 399 | |
| P/V | Yucaipa | GGGGGCGAAG | 61 | 699 | 599 | TTAACAAAAAA | - | 232 |
| Bangor | GGGGGCGAAT | 61 | 699 | 599 | TAAGAAAAAAA | - | 232 | |
| England | GGGGGCGAAG | 61 | 699 | 599 | TTAACAAAAAA | - | 232 | |
| Kenya | GGGGGCGAAG | 61 | 699 | 599 | TTAACAAAAAA | - | 232 | |
| P/W | Yucaipa | GGGGGCGAAG | 61 | 624 | 675 | TTAACAAAAAA | - | 207 |
| Bangor | GGGGGCGAAT | 61 | 462 | 837 | TAAGAAAAAAA | - | 153 | |
| England | GGGGGCGAAG | 61 | 624 | 675 | TTAACAAAAAA | - | 207 | |
| Kenya | GGGGGCGAAG | 61 | 624 | 675 | TTAACAAAAAA | - | 207 | |
| M | Yucaipa | GGGGGCGAAG | 32 | 1110 | 117 | TTAAGAAAAAA | 23 | 369 |
| Bangor | GGGGGCGAAT | 38 | 1110 | 135 | TTTAGAAAAAA | 23 | 369 | |
| England | GGGGGCGAAG | 32 | 1110 | 117 | TTAAGAAAAAA | 23 | 369 | |
| Kenya | GGGGGCGAAG | 32 | 1110 | 117 | TTAAGAAAAAA | 23 | 369 | |
| F | Yucaipa | GGGGGCGACA | 44 | 1611 | 31 | TTAAGAAAAAA | 9 | 536 |
| Bangor | GGGGGCGAAA | 20 | 1635 | 84 | TTAAGAAAAAA | 4 | 544 | |
| England | GGGGGCGACA | 44 | 1611 | 31 | TTAAGAAAAAA | 9 | 536 | |
| Kenya | GGGGGCGACA | 44 | 1611 | 31 | TTAAGAAAAAA | 9 | 536 | |
| HN | Yucaipa | GGGGGCGACA | 66 | 1743 | 69 | TTAAGAAAAAA | 3 | 580 |
| Bangor | GGGGGCGAAA | 60 | 1752 | 61 | TTAATAAAAAA | 8 | 583 | |
| England | GGGGGCGACA | 66 | 1743 | 69 | TTAAGAAAAAA | 3 | 580 | |
| Kenya | GGGGGCGACA | 72 | 1749 | 64 | TTAATAAAAAA | 8 | 582 | |
| L | Yucaipa | GGGGGCGAAT | 11 | 6729 | 73 | TTAAGAAAAAA | - | 2242 |
| Bangor | GGGGGCGAAT | 11 | 6729 | 102 | TTAAGAAAAAA | - | 2242 | |
| England | GGGGGCGAAT | 11 | 6729 | 73 | TTAAGAAAAAA | - | 2242 | |
| Kenya | GGGGGCGAAT | 11 | 6729 | 73 | TTAAGAAAAAA | - | 2242 | |
Differences relative to the most conserved sequence are underlined.
The intergenic sequences (IGS) of APMV-2 strains vary in length from 3 to 23 nt and are exactly conserved in length between the N, P, M and F genes (Table 3). The IGS sequences of strain England are 100% identical in length and sequence to strain Yucaipa, and the IGS sequences of strain Kenya are also are identical in length and sequence to strain Yucaipa except between HN and L genes. In contrast, the IGS between the F and HN in strain Bangor is only 4 nt in length compared to 9 nt in length in the other three strains, and the IGS between HN and L is 8 nt in length in strains Bangor and Kenya compared to 3 nt in length in the other two strains. In addition, the IGS sequences of strain Bangor have less than 50% nt identity with those of strain Yucaipa.
The nucleocapsid protein (N) gene
The N gene of APMV-2 strains Bangor, England and Kenya is 1547 nt in length and encodes a N protein of 457 aa (Table 3), as is the case for strain Yucaipa. The N protein of strains Bangor, England and Kenya has 90.4%, 99.3% and 94.5% aa sequence identity, respectively, with that of strain Yucaipa (Table 4). An amino acid sequence motif that is highly conserved in the N proteins of members of subfamily Paramyxovirinae and is involved in N–N self assembly, F-X4-Y-X3-φ -S-φ -A-M-G, where X represents any amino acid residue and φ represents an aromatic amino acid residue (Morgan, 1991), is present within the central domain of the N protein of each the four strains and is exactly conserved among all four strains (324FAPANFSTLYSYAMG338).
Table 4.
Percent amino acid percentage identity between APMV-2 strains Yucaipa, Bangor, England and Kenya for the indicated proteins.
| Strains | N | P | M | F | HN | L | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bangor | England | Kenya | Bangor | England | Kenya | Bangor | England | Kenya | Bangor | England | Kenya | Bangor | England | Kenya | Bangor | England | Kenya | ||
| Yucaipa Bangor England Kenya | 90.4 | 99.3 | 94.5 | 55.8 | 87.7 | 99.5 | 85.1 | 99.7 | 98.4 | 79.1 | 99.8 | 98.1 | 75 | 96 | 76.2 | 66.5 | 94.2 | 87.8 | |
| 89.9 | 89.7 | 60.8 | 55.3 | 84.8 | 85.1 | 78.9 | 77.6 | 75.2 | 85.1 | 67.4 | 68.2 | ||||||||
| 94.1 | 87.2 | 98.1 | 97.9 | 76.4 | 86.1 | ||||||||||||||
The phosphoprotein (P) gene and P/V/W editing
The P gene of APMV-2 strains Bangor, England and Kenya is 1379 nt in length and encodes a P protein of 399 aa (Table 3), as is the case for strain Yucaipa. The P protein of strains Bangor, England and Kenya has 55.8%, 87.7% and 99.5% aa sequence identity, respectively, with that of strain Yucaipa (Table 4). The P gene of all four APMV2 strains contains a putative P gene editing site (3′-UUUUUCCCC (negative-sense), located at nt position 2092–2100 in the viral RNA genome. The addition of a single G residue to the editing site would yield a predicted V protein and the addition of 2 G residues would yield a predicted W protein, as is the case with NDV (Steward et al., 1993). For all four APMV-2 strains, the predicted V protein is 232 aa in length. For all four strains, the V protein domain contains the conserved cysteine rich motif that is characteristic of most members of subfamily Paramyxovirinae (Fig. 3). This 52-aa motif was completely conserved among strains England, Kenya, and Yucaipa, whereas that of strain Bangor has a number of aa difference. The predicted W protein of strains England and Kenya is 207 aa in length, as also is the case for strain Yucaipa, while that of strain Bangor is only 153 aa in length (Table 3).
Fig. 3.
Amino acid sequence alignment of the C-terminal domain of the V proteins of the indicated APMV-2 strains. Conserved cysteine (C) residues are underlined; dots indicate identity with strain Yucaipa. Numbers indicate the amino acid position.
The matrix protein (M) gene
The M gene of APMV-2 strains England and Kenya is 1280 nt in length, as is the case for strain Yucaipa, whereas that of strain Bangor is 1304 nt in length (Table 3). The increased length found in strain Bangor is due to longer 5′and 3′ untranslated regions. The M gene of all four strains encodes a M protein of 369 aa. The M protein of strains Bangor, England and Kenya has 85.1%, 99.7% and 98.4% aa sequence identity, respectively, with that of strain Yucaipa (Table 4).
The fusion protein (F) gene
The F gene of APMV-2 strains Yucaipa, England, and Kenya is 1707 nt in length and encodes a F protein of 536 aa (Table 3), whereas that of strain Bangor is 1760 nt in length and encodes an F protein of 544 aa. The difference in length is due increased lengths of the 3′ untranslated region and ORF in strain Bangor, which are partially offset by a shorter 5′ untranslated region. The F protein of strains Bangor, England and Kenya has 79.1%, 99.8% and 98.1% aa sequence identity, respectively, with that of strain Yucaipa (Table 4). In APMV-1, the cleavage sequence of the F protein has been shown to be a critical factor for viral replication and pathogenesis. For APMV-2 strains England, Kenya and Yucaipa, the aa sequences spanning the F protein cleavage site and adjacent upstream end of the F1 subunit are identical (DKPASR↓F) and contain dibasic aa residues (Fig. 4). In contrast, in strain Bangor, the sequence of the six amino acids preceding the cleavage site differ from the other strains at four positions and contains only one basic aa residue (TLPSAR↓F). A similar difference in the number of basic amino acids at cleavage site between strains of same serotype has been reported in APMV-6 (Xiao et al., 2010). However, all the APMV-2 strains contain a phenylalanine residue at the F1 amino terminal end: this also is the case in virulent APMV-1 strains, whereas avirulent APMV-1 strains have a leucine at this position (Fig. 4) (Lamb and Parks, 2007).
Fig. 4.
Alignment of the F protein cleavage site sequences of the four APMV-2 strains with those of other APMVs. Basic amino acids (R=arginine and K=lysine) are underlined and in bold. Numbers indicate amino acid position.
The hemagglutinin-neuraminidase (HN) gene
The HN gene of APMV-2 strain England is 1899 nt long, as is the case for strain Yucaipa, while the lengths of the HN genes of strains Bangor and Kenya are 1894 nt and 1906 nt, respectively. These latter two strains have differences relative to the others and to each other in the lengths of the 5′ and 3′ untranslated regions and the ORFs. The lengths of HN protein of strains Yucaipa and England are 580 aa, while those of strains Bangor and Kenya are 583 and 582 aa, respectively (Table 3). The HN protein of strains Bangor, England and Kenya has 75%, 96% and 76.2% aa sequence identity, respectively, with that of strain Yucaipa (Table 4). In addition, all the four strains have the hexapeptide (NRKSCS) that forms part of the sialic acid binding site (Mirza et al., 1994).
The large polymerase protein (L) gene
The L gene of APMV-2 strains England and Kenya is 6834 nt long, as is the case for strain Yucaipa. The L gene of strain Bangor is 6863 in length, with the difference due to a longer 3′ untranslated region. The L genes of all the four strains encode an L protein of 2242 aa (Table 3). The L protein of strains Bangor, England and Kenya has 66.5%, 94.2% and 87.8% aa sequence identity, respectively, with that of strain Yucaipa (Table 4). In addition, all four strains have the conserved motif GDNQ in the L protein domain III, as seen in all non-segmented negative strand RNA viruses, which involved in L protein transcriptional activity (Schnell and Conzelmann, 1995).
Phylogenetic analysis
A phylogenetic tree was generated from alignments of the complete nt sequences of the genomes of APMV-2 strains Yucaipa, Bangor, England and Kenya with those of the representative members of family Paramyxoviridae (Fig. 5). This shows the APMV-2 strains clustering together on a branch that is distinct from other the paramyxoviruses, as would be expected. Also, strains Yucaipa, England and Kenya are more closely related to each other than to strain Bangor.
Fig. 5.
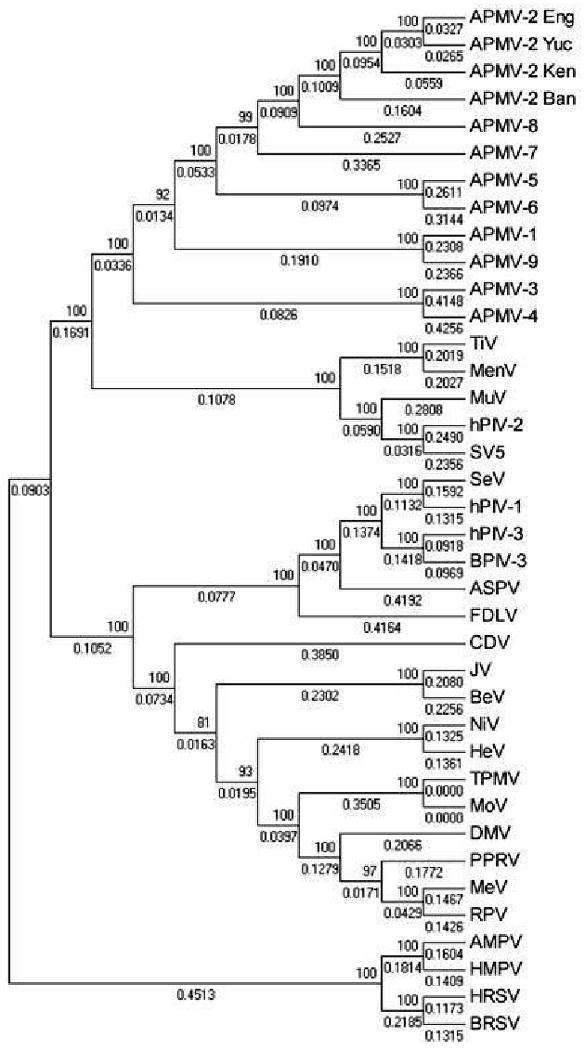
Phylogenetic tree of representative members of the family Paramyxoviridae. The phylogenetic tree of representative members of the family Paramyxoviridae was constructed with the complete genome sequences and using MEGA 4.1, Molecular Evolutionary Genetics Analysis software. The numbers at the node represent the bootstrap values among different viruses and the numbers under the lines indicate branch length.
Discussion
Avian paramyxoviruses are classified into nine serotypes based on their serological relationships in HI and NI tests (Alexander, 2003). Among these serotypes, APMV-1 causes severe disease in poultry; hence, a great deal of information is available on the antigenic and genetic relationships among APMV-1 strains isolated from different parts of the world (Alexander, 1988). Recently we and others have reported complete genome sequences for representative strains of APMV-2 to -9 (Subbiah et al., 2008; Kumar et al., 2008; Nayak et al., 2008; Samuel et al., 2010; Chang et al., 2001; Xiao et al., 2009; Paldurai et al., 2009; Samuel et al., 2009). However, very little information is available about the antigenic and genetic relationships among the strains within serotypes 2 through 9 (Alexander, 2003). In this study we have determined the antigenic and genetic relations among APMV-2 strains Yucaipa, Bangor, England and Kenya isolated from a chicken, finch, chicken and gadwell duck, respectively. Furthermore, these strains were isolated from different parts of the world and in different years. Therefore, it was interesting to know the extent of antigenic and genetic variation among these strains. The antigenic relationships among these four strains were evaluated using cross-HI and cross-serum microneutralization assays, and genetic variation was assessed by determining and comparing complete sequences for the viral genomes and predicted proteins. This information will have implications for studies in pathogenesis, epidemiology and for the development of vaccines against APMV-2.
To evaluate the antigenic relationships among the four APMV-2 strains described in the present study, we raised chicken antisera against each strain individually by respiratory infection mimicking a natural route of infection. Since serological responses tend to broaden over time, and with repeated antigenic exposure, we limited the immunization to a single infection and collected serum samples at an early time point (14 dpi). HI assays showed that, in the majority of comparisons, antigenic relatedness was greater between stains Yucaipa, England, and Kenya versus strain Bangor. Consistent with this, the results from the microneutralization tests in cell culture suggested an antigenic dimorphism that would be consistent with the existence of two antigenic subgroups within APMV-2, with strains Yucaipa, England and Kenya belonging to one antigenic subgroup and with strain Bangor belonging to the second antigenic subgroup, as seen with APMV-3 and -6 strains (Kumar et al., 2010; Xiao et al., 2010). It was previously suggested that strain Bangor be classified as a separate serotype or as a subtype of serotype 2 (McFerran et al., 1974) based on distinct differences in neuraminidase activities (Alexander et al., 1974) and cross serum neutralization tests between strains Bangor and Yucaipa. Our data support the classification of strain Bangor as a separate subgroup within serotype 2 rather than a distinct new serotype. It will be interesting to extend this analysis to additional strains to further evaluate antigenic variability among APMV-2 strains.
The genome lengths of strains Bangor, England and Kenya are 15024, 14904 and 14916 nt, respectively, compared with 14904 for strain Yucaipa. Among the APMV-1 (NDV) strains, there are three genome sizes: (1) 15,186 nt in early (>1930s) isolated strains, 2) 15,192 nt in late (>1960s) isolated strains (due to a six nt insertion in the upstream of the N gene), and (3) 15,198 nt (12 nt insertion in the P gene ORF) (Czeglédi et al., 2006). These different genome sizes of NDV strains did not relate to the viral virulence, but seem to be related to the time (year) of virus isolation with the genomes becoming progressively longer (Miller et al., 2009; Czeglédi et al., 2006). However, in APMV-2, the genome length does not seem to be decided by the year of isolation but rather by the host species. Strains Yucaipa and England were both isolated from chicken and have the same genome length (14904 nt). Despite the difference in the genome length, all the three strains follow the “rule of six” consistent with this rule being a requirement for virus replication and survival.
Comparison of the complete consensus sequences for the genomes of the four APMV strains showed that strain Bangor has 70.4, 69.4, and 70.8% nt and 75.3, 76.1, 76.8% aggregate aa sequence identity with strain Yucaipa, England, and Kenya, respectively. In contrast, strains England and Kenya are more closely related to strain Yucaipa, with a nt sequence identity of 94.5% and 88%, respectively, and an aggregate aa sequence identity of 96.1% and 92.4%, respectively. Also, strains England and Kenya have 86.1% nt and 89.9% aggregate aa sequence identity with each other. These results unequivocally show that strains Yucaipa, England and Kenya are closely related genetically, while strain Bangor is somewhat distinct. This is consistent with the proposed antigenic subgroups described above, and provides a molecular basis for this antigenic dimorphism.
Comparison of the aa sequence relatedness of cognate proteins between the APMV-2 strains revealed values ranging from 55.8 to 99.8% aa identity, with different proteins having different ranges of identity. In particular, the P and L proteins of strain Bangor were among the most divergent (55.3-60.8 and 66.5-68.2% aa identity, respectively), compared to the Yucaipa, England, and Kenya strains. However, the percent identity for these proteins was much higher among the latter three strains (87.2-99.5% for P and 86.1-94.2 for L), consistent with these three strains representing a subgroup separate from strain Bangor. The extent of variability in the APMV-2 P proteins is similar to that observed among APMV-6 strains (Xiao et al., 2010) but differs from that of the P proteins of the two subgroups of HMPV and HRSV, which are more highly conserved (85 and 90% aa identity, respectively) (Biacchesi et al., 2003). The V protein also was relatively divergent: the V protein of strain Bangor had only 56.3, 55.4 and 56.3 % aa identities, respectively with that of strains Yucaipa, England, and Kenya, whereas the V proteins of strains Yucaipa and Kenya had 100% aa identity and the V protein of both these strains had 99.1% aa identity with that of strain England. In addition, it is interesting to note that the W protein of strain Bangor was smaller in length, 153 aa compared to a length of 207 aa that was conserved for the other three strains. A similar difference in W protein size between strains of same serotype has been reported in APMV-8 (Paldurai et al., 2009). Since the role of W protein is not known, the functional significance of the W protein size difference remains to be studied. It is also interesting to find that the F and HN proteins of strain Bangor exhibited more divergence (77.6-79.1% and 75-85.1% aa identity, respectively) with those of strains Yucaipa, England, and Kenya, while the F and G proteins of the HMPV subgroups have 95% and 37% aa identity, respectively, and that of the HRSV subgroups have 89% and 55% aa identity, respectively (Biacchesi et al., 2003). Among the Yucaipa, England, and Kenya strains, Yucaipa and England were more closely related on the nt level as well as for most of the proteins. These two strains also were from the same host, namely the chicken. This was the most evident for the HN, and L proteins, for which strains Yucaipa and England were substantially more closely related to each other than either was to strain Bangor. Curiously, however, for the P protein, strains Yucaipa and Kenya were substantially more closely related than either was to strain England.
Another difference between strain Bangor and the other three strains was observed in the fusion protein cleavage site, which plays a major role in NDV pathogenesis (Lamb and Parks, 2007). Virulent NDV strains have a multiple basic aa cleavage site R-X-K/R-R↓F, which is cleaved intracellularly by ubiquitous cellular furin-like proteases, and also have a phenylalanine (F) residue at the beginning of the F1 subunit, which also may play a role in facilitating cleavage (Morrison et al., 1993). The avirulent NDV strains have one or a few basic residues at the cleavage site and do not conform to the furin motif, and have a leucine (L) residue at the first position of F1 subunit. Interestingly, the putative cleavage sites of other APMV serotypes showed that the cleavage site sequences of some serotypes are not necessarily predictive of the protease activation phenotype (Samuel et al., 2010). The putative F protein cleavage site (DKPASR↓F) of the strains England and Kenya resembled that of prototype strain Yucaipa and contained two basic residues and a phenylalanine residue at the F1 terminal end, while that of strain Bangor (TLPSAR↓F) contained only one basic amino acid. However, none of the sites conform to the preferred furin cleavage site (R-X-(K/R)-R↓). Each of these strains replicated in a trypsin-independent manner in both of the cell lines that we tested and the addition of trypsin or allantoic fluid did not substantially increase virus replication, as we previously observed for the prototype strain Yucaipa in a comparison involving nine different cell lines (Subbiah et al., 2008). Thus, on the basis of cleavage site sequence, it will be difficult to predict the virulence of these strains, unlike in the case of APMV-1 strains. Our results of MDT in chicken eggs and ICPI in day-old chicks provided evidence of an avirulent phenotype for each of these strains in chickens. APMV-2 neither produced plaques nor syncytia in tissue culture. These viruses produced single cell infection in the cell lines tested. These results indicate that APMV-2 is different from other APMV serotypes in its infection mechanism. Our result suggests that unlike other APMV serotypes, APMV-2 infection does not require cell to cell fusion. Hence, cleavage of F protein is probably not needed for APMV-2 infection. This agrees with our observation that addition of external proteases does not enhance growth of APMV-2. However, additional experiment using reverse genetics of APMV-2 is needed to confirm our hypothesis.
In conclusion, the complete genome sequences were determined for APMV-2 strains Bangor, England and Kenya. Comparison of the nt and predicted protein aa sequences among four APMV-2 strains showed the existence of divergence between strains Yucaipa, England, Kenya versus strain Bangor, suggesting that APMV-2 contains two antigenic subgroups, as reported with the APMV-3 and -6 serotypes. This grouping based on sequence relatedness and phylogenetic tree also is consistent with the antigenic analysis. This indicated that APMV-2 strains represent two APMV-2 subgroups and we propose that the prototype strain Yucaipa and strains England and Kenya represent one subgroup while strain Bangor represents a second subgroup. It will be interesting in future to look at the antigenic and genetic analyses of other APMV-2 strains isolated from different avian species.
Acknowledgments
We thank all our laboratory members for their excellent technical assistance. “This research was supported by NIAID contract no. N01A060009 (85% support) and NIAID, NIH Intramural Research Program (15% support). The views expressed herein do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. Government.”
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DJ. Comparison of the structural polypeptides of four avian paramyxoviruses. Archives of Virology. 1974;46:291–301. doi: 10.1007/BF01240071. [DOI] [PubMed] [Google Scholar]
- Alexander DJ. Avain paramyxoviruses. Vet Bull. 1980;50:737–752. [Google Scholar]
- Alexander DJ. Newcastle disease. In: Purchase HG, Arp LH, Domermuth CH, Pearson JE, editors. A laboratory manual for the isolation and identification of avian pathogens. 3rd. The American Association of Avian Pathologists, Kendall/Hunt Publishing Company; Dubuque, IA: 1989. pp. 114–120. [Google Scholar]
- Alexander DJ. Newcastle disease and other avian paramyxoviridae infections. In: Calnek BW, editor. Diseases of Poultry. Iowa State University Press; Ames: 1997. pp. 541–569. [Google Scholar]
- Alexander DJ, Allan WH, Parsons G, Collins MS. Identification of paramyxoviruses isolated from birds dying in quarantine in Great Britain. Vet Rec. 1982;111:571–574. [PubMed] [Google Scholar]
- Alexander DJ. Avian paramyxoviruses 2–9. In: Saif YM, editor. Diseases of Poultry. 11th. Iowa State University Press; Ames: 2003. pp. 88–92. [Google Scholar]
- Asahara T, Yoshimura M, Tusubaki S, Yamagamt T, Aoi T, Ide S, Masu S. Isolation in Japan of a virus similar tomyxovirus Yucaipa (MVY) Bull Azabu Vet Coll. 1973;26:67–81. [Google Scholar]
- Bankowski RA, Corstert RE, Clark CT. Isolation of an unidentified agent from the respiratory tract of chickens. Science. 1960;132:292–293. doi: 10.1126/science.132.3422.292. [DOI] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315:1–9. doi: 10.1016/s0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- Borisevich V, Nistler R, Hudman D, Yamshchikov G, Seregin A, Yamshchikov V. A highly sensitive and versatile virus titration assay in the 96-well microplate format. J Virol Methods. 2008;147:197–205. doi: 10.1016/j.jviromet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Chang PC, Hsieh ML, Shien JH, Graham DA, Lee MS, Shieh HK. Complete nucleotide sequence of avian paramyxovirus type 6 isolated from ducks. J Gen Virol. 2001;82:2157–2168. doi: 10.1099/0022-1317-82-9-2157. [DOI] [PubMed] [Google Scholar]
- Collings DF, Fitton J, Alexander DJ, Harkness JW, Pattison M. Preliminary characterization of a paramyxovirus isolated from a parrot. Res Vet Sci. 1975;19:219–221. [PubMed] [Google Scholar]
- Czeglédi A, Ujvári D, Somogyi E, Wehmann E, Werner O, Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 2006;120:36–48. doi: 10.1016/j.virusres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Fleury HJA, Alexander DJ. Isolation of twenty-three Yucaipa-like viruses from 616 wild birds in Senegal,West Africa. Avian Dis. 1979;23:742–744. [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81(Pt 10):2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Goodman BB, Hanson RP. Isolation of avian paramyxovirus-2 from domestic and wild birds in Costa Rica. Avian Dis. 1988;32:713–717. [PubMed] [Google Scholar]
- Horikami SM, Smallwood S, Moyer SA. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222(2):383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72(2):891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nayak B, Collins PL, Samal SK. Complete genome sequence of avian paramyxovirus type 3 reveals an unusually long trailer region. Virus Res. 2008;137:189–197. doi: 10.1016/j.virusres.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nayak B, Samuel AS, Xiao S, Collins PL, Samal SK. Complete genome sequence of avian paramyxovirus-3 strain Wisconsin: evidence for the existence of subgroups within the serotype. Virus Res. 2010;149:78–85. doi: 10.1016/j.virusres.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. Family Paramyxoviridae. In: Fauquet CM, editor. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Acedemic Press; 2005. pp. 655–668. [Google Scholar]
- Lamb R, Parks G. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5th. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Lang G, Gagnon A, Howell J. Occurrence of paramyxovirus Yucaipa in Canadian poultry. Can Vet J. 1975;16:233–237. [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Horvath F, Aligo JA, Wilson R, He B. The role of simian virus 5 V protein on viral RNA synthesis. Virology. 2005;338(2):270–280. doi: 10.1016/j.virol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Lipkind M, Weisman Y, Shihmanter E, Shoham D, Aronovici A. The isolation of Yucaipa-like paramyxoviruses from epizootics of a respiratory disease in turkey poultry farms in Israel. Vet Rec. 1979;105:577–578. [PubMed] [Google Scholar]
- Lipkind M, Weisman Y, Shihmanter E, Shoham D, Aronovici A. Isolation of Yucaipa-like avian paramyxovirus froma wild mallard duck (Anas platyrhinchos) wintering in Israel. Vet Rec. 1982;110:15–16. doi: 10.1136/vr.110.1.15. [DOI] [PubMed] [Google Scholar]
- Mbugua HCW, Karstad L. Isolation of avian paramyxoviruses (Yucaipa-like) from wild birds in Kenya, 1980–1982. J Wildl Dis. 1985;21:52–54. doi: 10.7589/0090-3558-21.1.52. [DOI] [PubMed] [Google Scholar]
- McFerran JB, Connor TJ, Allan GM, Purcell DAP, Young JA. Studies on a virus isolated from a finch. Research in Veterinary Science. 1973;15:116–118. [PubMed] [Google Scholar]
- McFerran JB, Connor TJ, Allan GM, Adair B. Studies on a paramyxovirus isolated from a finch. Archiv fftr die gesamte Virusforschung. 1974;46:281–290. doi: 10.1007/BF01240070. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Decanini EL, Afonso CL. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet. 2009;10:26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Mirza AM, Deng R, Iorio RM. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutininneuraminidase glycoprotein: effects on antigenic structure and function. J Virol. 1994;68:5093–5099. doi: 10.1128/jvi.68.8.5093-5099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T, McQuain C, Sergel T, McGtnnes L, Reitter J. The role of the amino terminus of F 1 of the Newcastle disease virus fusion protein in cleavage and fusion. Virology. 1993;193:997–1000. doi: 10.1006/viro.1993.1214. [DOI] [PubMed] [Google Scholar]
- Nayak B, Kumar S, Collins PL, Samal SK. Molecular characterization and complete genome sequence of avian paramyxovirus type 4 prototype strain duck/Hong Kong/D3/75. Virol J. 2008;5:124. doi: 10.1186/1743-422X-5-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymadawa P, Konstantinow-siebelist I, Schulze P, Starke G. Isolation of paramyxoviruses from free-flying birds of the order Passeriformes in German Democratic Republic. Acta Virol. 1977;56:345–351. [PubMed] [Google Scholar]
- Paldurai A, Subbiah M, Kumar S, Collins PL, Samal SK. Complete genome sequences of avian paramyxovirus type 8 strains goose/Delaware/1053/76 and pintail/Wakuya/20/78. Virus Res. 2009;142:144–153. doi: 10.1016/j.virusres.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel AS, Kumar S, Madhuri S, Collins PL, Samal SK. Complete sequence of the genome of avian paramyxovirus type 9 and comparison with other paramyxoviruses. Virus Res. 2009;142:10–18. doi: 10.1016/j.virusres.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel AS, Paldurai A, Kumar S, Collins PL, Samal SK. PLoS One. 2. Vol. 5. 2010. Feb 17, Complete genome sequence of avian paramyxovirus (APMV) serotype 5 completes the analysis of nine APMV serotypes and reveals the longest APMV genome; p. e9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell M, Conzelmann KK. Polymerase activity of in vitro mutated rabies L protein. Virology. 1995;214:522–530. doi: 10.1006/viro.1995.0063. [DOI] [PubMed] [Google Scholar]
- Shihmanter E, Weisman Y, Manwell R, Alexander DJ, Lipkind M. Mixed paramyxovirus infection of wild and domestic birds in Israel. Vet Microbiol. 1997;58:73–78. doi: 10.1016/s0378-1135(97)00147-8. [DOI] [PubMed] [Google Scholar]
- Steward M, Vipond IB, Millar NS, Emmerson PT. RNA editing in Newcastle disease virus. J Gen Virol. 1993;74(Pt 12):2539–2547. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- Subbiah M, Xiao S, Collins PL, Samal SK. Complete sequence of the genome of avian paramyxovirus type 2 (strain Yucaipa) and comparison with other paramyxoviruses. Virus Res. 2008;137:40–48. doi: 10.1016/j.virusres.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang LF, Eaton BT. Emerging paramyxoviruses. Infect Dis Rev. 2001;3:52–69. [Google Scholar]
- Weisman Y, Aronovici A, Malkinson M, Shihmanter E, Lipkind M. Isolation of paramyxoviruses from pigeons in Israel. Vet Rec. 1984;115:605. doi: 10.1136/vr.115.23.605-a. [DOI] [PubMed] [Google Scholar]
- Xiao S, Paldurai A, Nayak B, Subbiah M, Collins PL, Samal SK. Complete genome sequence of avian paramyxovirus type 7 (strain Tennessee) and comparison with other paramyxoviruses. Virus Res. 2009;145:80–91. doi: 10.1016/j.virusres.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Subbiah M, Kumar S, De Nardi R, Terregino C, Collins PL, Samal SK. Complete genome sequences of avian paramyxovirus serotype 6 prototype strain Hong Kong and a recent novel strain from Italy: evidence for the existence of subgroups within the serotype. Virus Res. 2010;150:61–72. doi: 10.1016/j.virusres.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GZ, Zhao JX, Wang HW, Yang AM, Bu CY, Wang M. Isolation, identification and comparison of four isolates of avian paramyxovirus serotype 2 in China. Avian Dis. 2006;50:386–390. doi: 10.1637/7502-010906R1.1. [DOI] [PubMed] [Google Scholar]
- Zhang GZ, Zhao JX, Wang M. Serological survey on prevalence of antibodies to avian paramyxovirus serotype 2 in China. Avian Dis. 2007;51:137–139. doi: 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]





