Abstract
The atonal (ato) proneural gene specifies different numbers of sensory organ precursor (SOP) cells within distinct regions of the Drosophila embryo in an epidermal growth factor-dependent manner through the activation of the rhomboid (rho) protease. How ato activates rho, and why it does so in only a limited number of sensory cells remains unclear. We previously identified a rho enhancer (RhoBAD) that is active within a subset of abdominal SOP cells to induce larval oenocytes and showed that RhoBAD is regulated by an Abdominal-A (Abd-A) Hox complex and the Senseless (Sens) transcription factor. Here, we show that ato is also required for proper RhoBAD activity and oenocyte formation. Transgenic reporter assays reveal RhoBAD contains two conserved regions that drive SOP gene expression: RhoD mediates low levels of expression in both thoracic and abdominal SOP cells, whereas RhoA drives strong expression within abdominal SOP cells. Ato indirectly stimulates both elements and enhances RhoA reporter activity by interfering with the ability of the Sens repressor to bind DNA. As RhoA is also directly regulated by Abd-A, we propose a model for how the Ato and Sens proneural factors are integrated with an abdominal Hox factor to regulate region-specific SOP gene expression.
Keywords: Atonal, proneural, Senseless, Hox, Rhomboid, EGF, enhancer
INTRODUCTION
The proneural genes encode a family of basic-Helix-Loop-Helix (bHLH) transcription factors that promote neurogenesis in the ectoderm of organisms from worms to vertebrates (Bertrand et al., 2002). In the Drosophila peripheral nervous system (PNS), for example, the atonal (ato) proneural gene promotes the formation of cells that differentiate into distinct sensory organs depending upon their location within the body plan. In the thoracic and abdominal segments, ato specifies sensory organ precursors (SOPs) that differentiate into internal stretch receptors (proprioceptors called chordotonal (ch) organs), in the larval and adult eyes ato specifies photoreceptors, and within the maxillary palp and antennal segments ato specifies olfactory receptors (Gupta and Rodrigues, 1997; Jarman et al., 1993; Jarman et al., 1994; Jarman et al., 1995). Thus, while proneural factors have the general capacity to promote neurogenesis, they are integrated with positional factors through largely unknown mechanisms to ensure the appropriate neural cell fate is adopted (Powell and Jarman, 2008).
Proneural genes not only promote neurogenesis cell autonomously, but also affect neighboring cell fates through the regulation of cell signaling pathways. In Drosophila SOP cells, proneural factors activate Delta expression to stimulate Notch signaling in adjacent cells (Heitzler et al., 1996; Hinz et al., 1994; Kunisch et al., 1994). The reception of Notch promotes epidermal fates while inhibiting neuronal development, ensuring that sensory organs, such as sensory bristles, are separated by epidermal tissue (Bray, 1998). However, some sense organs are clustered in Drosophila in spite of the Notch lateral inhibition pathway. Internal stretch receptors that function in proprioception, for example, consist of anywhere from one to 80 clustered scolopodia that together form a mature ch organ (Lai and Orgogozo, 2004; zur Lage and Jarman, 1999). Each scolopodium consists of five cells (a neuron, scolopale, cap, ligament, and attachment cell) that are derived from a single SOP cell specified by ato (Lai and Orgogozo, 2004). While ato activates the Notch-Delta lateral-inhibition pathway in all ch organ SOP cells, a subset of these cells also activate the EGF signaling pathway by up-regulating Rhomboid (Rho) proteases that cleave an EGF ligand (Spitz, Spi) to promote its secretion (Bier et al., 1990; Lage et al., 1997; Okabe and Okano, 1997; Shilo, 2005). Neighboring cells that receive Spi overcome the anti-neural effects of Notch to form additional ch organ SOP cells and/or hepatocyte-like oenocyte cells (Elstob et al., 2001; Lage et al., 1997; Okabe and Okano, 1997; Rusten et al., 2001). Thus, ato regulates cell signaling pathways that both inhibit (Notch-Delta) and stimulate (EGF) the formation of additional sensory cells.
Ch organ SOP cells activate rho in a region-specific manner to differentially pattern the Drosophila embryo. In abdominal segments, for example, a set of five Ato-positive primary (1°) SOP cells (C1-C5) activate rho, secrete Spi, and thereby induce three secondary (2°) SOP cells and a cluster of oenocytes (Elstob et al., 2001; Gebelein, 2008; Lage et al., 1997; Okabe and Okano, 1997; Rusten et al., 2001). In contrast, even though a similar set of Ato-positive 1° SOP cells forms within thoracic segments, these cells fail to up-regulate rho and 2° SOP cells and oenocytes do not form within the thorax. The differential ability of abdominal and thoracic SOP cells to activate rho is linked to the expression of a specific Hox factor, Abdominal-A (Abd-A) (Brodu et al., 2002). In the absence of Abd-A, the abdominal ch organ SOP cells fail to stimulate rho and lack Spi secretion, thereby preventing the induction of 2° SOP cells and oenocytes. What remains unclear is how the Ato proneural and Abd-A Hox pathways are integrated to stimulate gene expression within specific abdominal SOP cells.
Here, we investigate how Ato and Abd-A are integrated to activate a rho enhancer (RhoBAD) in SOP cells to specify abdomen-specific oenocytes. We show that ato is required for proper RhoBAD activity and oenocyte formation. Using transgenic reporter assays, we further show that the RhoBAD enhancer contains two conserved elements (RhoA and RhoD) that are indirectly regulated by ato to drive gene expression within SOP cells. Of these two elements, only RhoA is also regulated by the Abd-A Hox factor, and we found that, like Abd-A, Ato interferes with the DNA binding activity of the Senseless (Sens) repressor protein for RhoA. We incorporate these data with detailed expression analysis to present a model for how the Ato and Sens proneural factors are integrated with Abd-A to result in segment- and tissue-specific gene regulation of a signaling pathway.
RESULTS
RhoBAD contains separable conserved regions that drive gene expression within the embryonic PNS
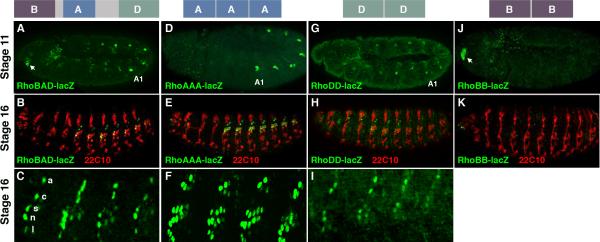
We recently identified an enhancer upstream of the rho gene (RhoBAD) that is active within a specific subset of abdominal ch organ SOP cells (the C1 SOP cells) to induce the formation of oenocytes (Li-Kroeger et al., 2008). In early embryogenesis (stage 11), the RhoBAD-lacZ reporter drives β-gal expression within the C1 SOP cells of each body segment (thorax/abdomen) with higher levels detected in the oenocyte-producing abdominal segments than in thoracic segments that do not induce this cell type (Figure 1A). Consistent with this early pattern, β-gal is detected within the C1 SOP cell lineage (ligament, neuron, scolopale, cap, and attachment cells) in abdominal but not thoracic segments of older embryos (stage 16, Figure 1B and 1C). To better understand how RhoBAD mediates SOP cell expression, we first identified three highly conserved regions through comparisons between the twelve sequenced Drosophilid genomes (RhoB, RhoA, and RhoD, Supplemental Figure 1). To determine which, if any, of these elements can drive SOP gene expression in isolation, we generated a series of transgenic fly lines and analyzed reporter gene expression within the embryo. While each individual element in a single copy was incapable of driving strong gene expression (data not shown), transgenic reporter lines containing multiple copies of RhoB (RhoBB-lacZ), RhoA (RhoAAA-lacZ), and RhoD sequences (RhoDD-lacZ) yielded specific expression patterns (Figure 1). Like RhoBAD-lacZ, both the RhoAAA-lacZ and RhoDD-lacZ reporters activate SOP gene expression within the body segments (Figure 1D and 1G). In contrast, RhoBB-lacZ is only expressed in a limited number of cells in the embryonic head (arrow in Figure 1J) and analysis of RhoBAD-lacZ revealed a similar head expression pattern (arrow in Figure 1A). While it is currently unclear if these head cells express the endogenous rho gene, we did detect elevated levels of a marker (phospho-ERK staining) consistent with activated EGF signaling in nearby cells (data not shown).
Figure 1. RhoBAD contains separable elements that drive reporter expression in sensory cells.
A-C. Diagram of the RhoBAD enhancer highlighting the relative position of the conserved RhoA, RhoB, and RhoD elements at top. Lateral views of RhoBAD-lacZ stage 11 (A) and stage 16 (B) embryos immuno-stained for β-gal (green) and a PNS-specific neuronal marker (mAb22C10, red) as noted. A lateral view close-up of four abdominal segments shows the β-gal pattern within the lch5 organ (C). The attachment (a), cap (c), scolopale (s), neuron (n), and ligament (l) cells of the C1 lineage are noted. D-F. Diagram of the RhoAAA enhancer at top. Lateral views of RhoAAA-lacZ stage 11 (D) and stage 16 (E) embryos immuno-stained for β-gal (green) and mAb22C10 (red) as noted. A lateral view close-up of four abdominal segments shows the β-gal pattern within the lch5 organ (F). Note, RhoAAA-lacZ is expressed in all the neurons and scolopale cells of the lch5 organ as well as some extra cap and ligament cells. G-I. Diagram of the RhoDD enhancer at top. Lateral views of RhoDD-lacZ stage 11 (G) and stage 16 (H) embryos immuno-stained for β-gal (green) and mAb22C10 (red) as noted. A lateral view close-up of four abdominal segments shows the β-gal pattern within the lch5 organ (I). J-K. Diagram of the RhoBB enhancer at top. Lateral views of RhoBB-lacZ stage 11 (J) and stage 16 (K) embryos immuno-stained for β-gal (green) and mAb22C10 (red) as noted.
Since RhoA and RhoD are each sufficient to drive gene expression in C1 SOP cells, we focused our studies on these two transgenes. Importantly, we found two differences in the ability of RhoAAA and RhoDD to drive gene expression in SOP cells compared to RhoBAD. First, while RhoAAA-lacZ behaves like RhoBAD-lacZ and drives higher expression in abdominal than thoracic C1 SOP cells (Figure 1D), RhoDD-lacZ drives similar low levels of β-gal in both the thoracic and abdominal C1 SOP cells (Figure 1G). Second, we found that RhoAAA-lacZ consistently labels the neurons and scolopale cells of the entire lch5 organ in older embryos (Figure 1F), whereas RhoBAD-lacZ (Figure 1C) and RhoDD-lacZ (Figure 1I) are predominantly restricted to only the C1 SOP cell lineage. Altogether, these findings support the following model for RhoBAD activity: RhoA contributes to abdominal ch organ expression, RhoD contributes to general C1 ch organ expression in all body segments, and RhoB contributes to gene expression within a cluster of head cells.
Hox-dependent and Hox-independent regulation of rho activity in SOP cells
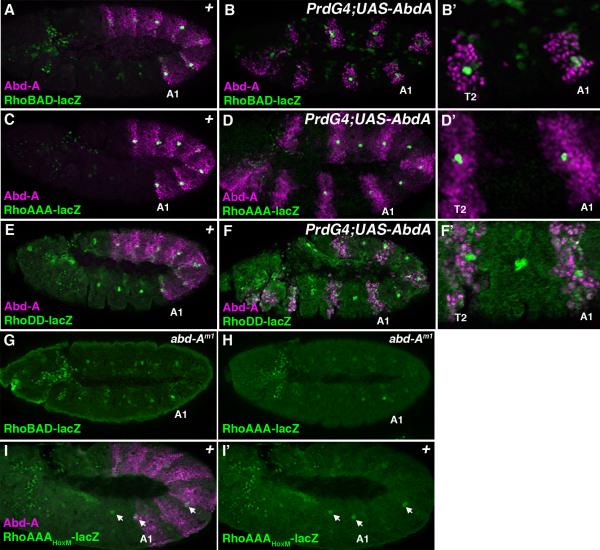
Analysis of the RhoBAD, RhoAAA, and RhoDD reporter lines shows that each is expressed within C1 SOP cells, but only RhoBAD and RhoAAA are expressed at higher levels in the abdominal segments that express Abd-A (Figure 2A, 2C, and 2E). These data suggest that the Abd-A Hox input is mediated through the RhoA element. We used the Gal4-UAS system to test this hypothesis by driving Abd-A expression in every other segment with the PrdG4 driver. As shown in Figure 2, both RhoBAD-lacZ (Figure 2B) and RhoAAA-lacZ (Figure 2D) activity are enhanced when Abd-A is expressed in thoracic segments. In contrast, the RhoDD-lacZ reporter is not affected by ectopic Abd-A within either the thorax or the abdomen (Figure 2F). In addition, we analyzed abd-A mutant embryos and found that the enhanced abdominal activity seen with RhoBAD-lacZ and RhoAAA-lacZ was compromised with only sporadic segments showing significant β-gal, which could be detected in both thoracic and abdominal segments (Figures 2G and 2H). Finally, we found that a point mutation that was previously shown to disrupt Abd-A binding to RhoA (HoxM) and RhoBAD activity in vivo (Li-Kroeger et al., 2008), results in a similar failure of the RhoAAA reporter to be up-regulated in abdominal SOP cells (Figure 2I). Importantly, Brodu et al. described a similar pattern of expression for the endogenous rho gene, suggesting that the initial rho activation is Hox-independent whereas its up-regulation in abdominal SOP cells is dependent upon abd-A (Brodu et al., 2002). Thus, the RhoBAD and RhoAAA enhancer elements behave similarly to the endogenous rho gene, and the abdomen-specific up-regulation of these rho enhancers is dependent upon Abd-A binding RhoA.
Figure 2. Hox dependent and Hox-independent regulation of Rho enhancer activity.
Lateral views of stage 11 Drosophila embryos immunostained for β-gal (green) and Abd-A (purple). The first abdominal segment (A1) of each embryo is noted. A. A RhoBAD-lacZ embryo reveals higher β-gal levels in abdominal than thoracic SOP cells. B-B’. A PrdG4;UAS-Abd-A;RhoBAD-lacZ embryo reveals the induction of thoracic β-gal to abdominal levels within the T2 thoracic segment expressing Abd-A protein. Close-up view of the T2, T3, and A1 segments is shown at right. C. A RhoAAA-lacZ embryo reveals high β-gal levels in the abdominal C1 SOP cells. D-D’. A PrdG4;UAS-Abd-A;RhoAAA-lacZ embryo reveals the induction of thoracic β-gal to abdominal levels within the T2 thoracic segment expressing Abd-A protein. Close-up view of the T2, T3, and A1 segments is shown at right. E. A RhoDD-lacZ embryo reveals low equal β-gal levels in both the thoracic and abdominal C1 SOP cells. F-F’. A PrdG4;UAS-Abd-A;RhoDD-lacZ embryo reveals that ectopic Abd-A has no affect on β-gal levels within either the thorax or abdomen. Close-up view of the T2, T3, and A1 segments is shown at right. G. A abd-Am1;RhoBAD-lacZ embryo reveals low β-gal levels in both abdominal and thoracic segments. H. A abd-Am1;RhoAAA-lacZ embryo reveals low β-gal levels in both abdominal and thoracic segments. I. A RhoAAAHoxM-lacZ embryo reveals sporadic low β-gal levels (arrows) in both abdominal and thoracic segments.
Atonal and Senseless expression precede rho enhancer activity in ch organ SOPs
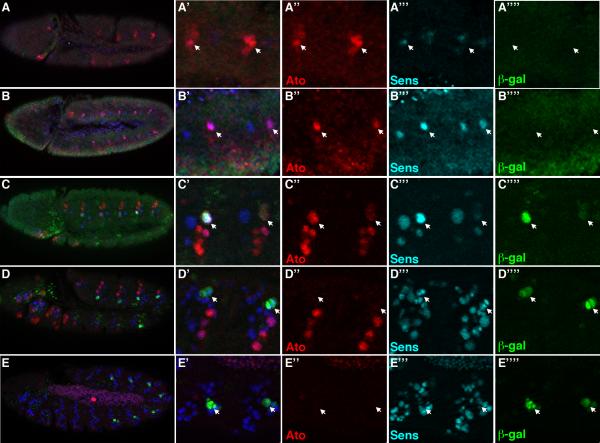
The Abd-A input into rho and the rho enhancers reveals how EGF signaling and oenocyte specification occurs within abdominal segments. However, both rho and the rho enhancers are also weakly expressed within C1 SOP cells in the absence of abd-A, suggesting a general ch organ input contributes to rho expression. To better understand how rho expression is initiated in C1 SOP cells, we first characterized the relative timing of Rho enhancer activity during SOP development using two markers of ch organ fate: Ato and Sens. As shown in Figure 3, co-stains for Ato, Sens, and β-gal proteins on different staged RhoAAA-lacZ embryos revealed the following: 1) Ato is weakly expressed in a cluster of cells, a subset of which weakly express Sens and none of which yet express RhoAAA-lacZ (Figure 3A). 2) Ato and Sens are detected in individual SOP cells prior to RhoAAA-lacZ activity (Figure 3B). 3) Ato, Sens, and β-gal are co-expressed in SOP cells of slightly older embryos (Figure 3C). 4) Ato is extinguished whereas Sens and β-gal are detected after SOP cell division (Figure 3D). 5) Sens decreases after subsequent cell divisions and is extinguished in mature cells of the ch organ (Figure 3E, data not shown). 6) No significant difference in either Ato or Sens levels is observed between thoracic and abdominal C1 SOP cells (Supplemental Figure 2). Since β-gal protein has a long half-life, we also performed in situ hybridizations on RhoAAA-lacZ and RhoBAD-lacZ embryos and observed similar results except that lacZ mRNA is only detected into the two-cell stage of the C1 lineage (Supplemental Figure 2). This result is similar to previous reports for rho mRNA within the C1 lineage (Lage et al., 1997). Thus, Ato and Sens are co-expressed prior to and at the time of Rho reporter activation, but Ato is extinguished while Sens persists into later stages of the SOP cell lineage.
Figure 3. Time course of Ato, Sens, and RhoAAA-lacZ activity in the Drosophila embryo.
Lateral views of RhoAAA-lacZ Drosophila embryos immunostained for Ato (red), β-gal (green), and Sens (blue). Close-up of two abdominal segments are shown at right with arrows depicting the C1 SOP cell or its lineage. A-A’’’’. Stage 9 embryo reveals broad Ato and weak Sens expression precedes RhoAAA-lacZ activity. B-B’’’’. Stage 10 embryo reveals Ato and Sens expression precedes RhoAAA-lacZ activity. C-C’’’’. Early stage 11 embryo reveals co-expression of Ato, Sens and β-gal in C1 SOP cells. Note that only one of the C1 SOP cells at right is in focal plane. D-D’’’’. Mid-stage 11 embryo reveals Ato expression disappears prior to C1 SOP cell division, whereas Sens and β-gal are detected in both progeny. E-E’’’’. Stage 12 embryo reveals Sens expression begins to fade within the C1 SOP lineage.
atonal and senseless are required for normal RhoBAD activity and oenocyte specification
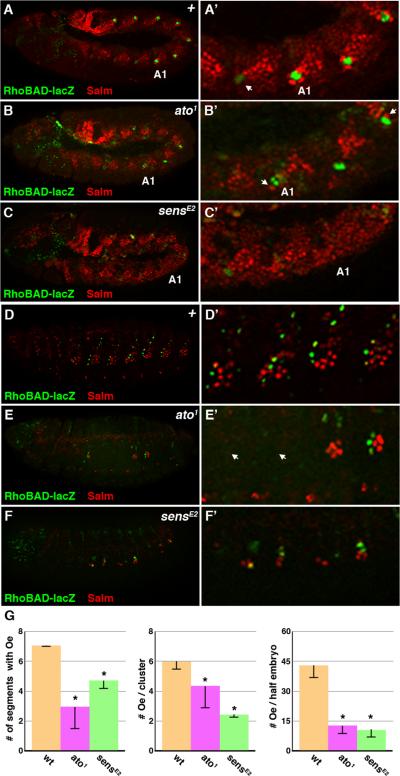
While we previously showed that Sens directly represses RhoBAD activity, Sens has also been shown to function as a co-activator in the presence of Ato (Acar et al., 2006; Li-Kroeger et al., 2008). Moreover, the expression of both Ato and Sens precedes Rho enhancer activity in C1 SOP cells, indicating they may participate in rho activation. Hence, we analyzed RhoBAD-lacZ expression in both ato1 and sensE2 mutant embryos. In ato1 embryos, we found that RhoBAD-lacZ activity is lost in some, but not all, segments and those segments that express β-gal have less compared to control segments (Figure 4B, see methods for details). Importantly, the C1 SOP cells that do form in ato1 embryos express Sens (Supplemental Figure 3). These findings are consistent with reports that, unlike other ch organ SOP cells, the C1 SOP cell occasionally forms in the absence of ato through redundancy with additional bHLH genes (Jarman et al., 1993). Similar to ato1 embryos, RhoBAD-lacZ activity is greatly diminished in sensE2 mutants (Figure 4C). However, the decreased β-gal levels may be due to the indirect loss of Ato expression in sens mutant embryos as re-supplying Ato enhances RhoBAD-lacZ activity, although not to wild type levels ((Li-Kroeger et al., 2008) and data not shown). Nevertheless, our data reveal that the normal RhoBAD-lacZ expression pattern and levels are dependent upon ato and sens function within SOP cells.
Figure 4. Atonal regulates RhoBAD activity.
A-C. Lateral views of stage 11 RhoBAD-lacZ Drosophila embryos immunostained for β-gal (green) and Salm (red). Close-ups of one thoracic and three abdominal segments (A1 labeled) are shown at right. Note that wild type embryos (A, A’) contain abdominal C1 SOP cells (marked by β-gal) surrounded by whorls of oenocytes (high Salm). The thoracic C1 SOP cells show weak β-gal without adjacent whorls of Salm-positive cells (arrow). In contrast, both ato1 (B, B’) and sensE2 (C, C’) mutant embryos have fewer abdominal segments that express β-gal, and those segments that do are surrounded by fewer oenocytes (arrow) than wild type embryos. D-F. Lateral views of RhoBAD-lacZ stage 16 embryos immunostained for β-gal (green) and Salm (red). Close up views of four abdominal segments is shown at right. Wild type embryos (D, D’) have RhoBAD-lacZ activity in the abdominal C1 SOP lineage with clusters of Salm-positive oenoyctes in close proximity, whereas both ato1 (E, E’) and sensE2 (F, F’) mutant embryos show a loss of both β-gal and Salm-positive cells in many abdominal segments. Close-up views show segments completely lacking β-gal and oenocytes (arrow), whereas other segments express weak β-gal and have few oenocytes. G. Quantification of oenocyte (Oe) numbers in wild type, ato1 and sensE2 embryos. * denotes p-value < 0.001.
In contrast to our findings that some segments of ato1 embryos have RhoBAD-lacZ activity, a previous study found that rho expression was lost in all segments of ato mutant embryos (Lage et al., 1997). However, two additional studies reported that oenocytes, which are dependent upon rho expression and EGF signaling for their specification, can form in a subset of ato mutant segments (Elstob et al., 2001; Rusten et al., 2001). To reconcile these findings, we analyzed RhoBAD-lacZ;ato1 embryos for the formation of oenocytes. As shown in Figure 4B, we found few cells expressing high levels of an oenocyte marker (Salm) in close proximity to the RhoBAD-positive (β-gal) cells in ato1 embryos in comparison to wild type controls (Figure 4A). As oenocytes rapidly delaminate into the embryo, we also analyzed late stage embryos (Stage 15-16) where their numbers are easier to quantify. As shown in Figure 4D, control embryos have clusters of approximately six oenocytes (5.8 +/- 0.2 per cluster) in each of seven abdominal segments (7.0 +/- 0.0 segments). In contrast, many segments of ato1 mutant embryos completely lack oenocytes with only 2.8 +/- 1.3 of the abdominal segments containing this cell type (Figure 4E). Moreover, when oenocytes do develop in ato1 segments fewer form than in control segments (4.4 +/- 1.9 per cluster, p-value < 0.001), and they are almost always associated with a C1 SOP cell that has RhoBAD-lacZ activity (Figure 4E). Thus, the total number of oenocytes that form is significantly less per embryo (Figure 4G, 12.5 +/- 4.5 in ato1 embryos versus 40.5 +/- 2.8 in control embryos, p-value < 0.001). We observed similar results in sens mutants as well with a significant decrease in the number of segments containing oenocytes (4.7 +/- 0.5), the number of oeoncytes per segment (2.3 +/- 0.1), and the total number of oenocytes per half embryo (10.6 +/- 4.6) (Figure 4F, 4G). Taken together, these data indicate impaired oenocyte recruitment by ato and sens mutant SOP cells, a finding that correlates well with the diminished, but not absent, RhoBAD-lacZ activity in these embryos.
Ato stimulates Rho enhancer expression through both the RhoA and RhoD elements
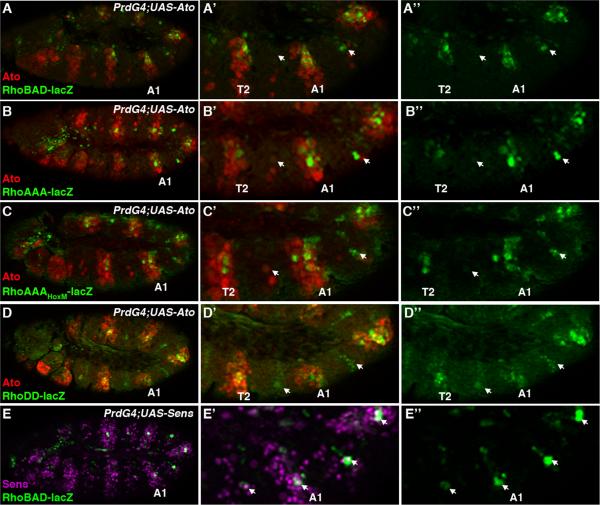
To further test the idea that ato and sens regulate Rho enhancer activity, we used PrdG4 to ectopically express each factor in every other segment of the Drosophila embryo. As shown in Figure 5A, PrdG4;UAS-Ato embryos show enhanced RhoBAD-lacZ activity in both the thoracic and abdominal segments that express Ato. To determine which regions of RhoBAD are responsive to ectopic Ato, we analyzed PrdG4;UAS-Ato embryos for RhoAAA-lacZ and RhoDD-lacZ activity. As shown in Figure 5B, RhoAAA-lacZ is frequently stimulated in more cells of the abdomen and is weakly responsive to Ato in the thorax. This finding suggests that while Ato expression within the Abd-A expression domain causes more cells to activate RhoAAA, Ato weakly stimulates RhoAAA expression independent of the abdominal Hox factor. Consistent with this idea, Ato activates a RhoA reporter that lacks a functional Hox binding site (PrdG4;AAAHoxM-lacZ;UAS-Ato) to similar levels within both the thorax and abdomen (Figure 5C). Lastly, we also found that RhoDD-lacZ is activated by Ato at equivalent levels in both the thorax and abdomen (Figure 5D). These data suggest that Ato can positively regulate RhoBAD activity through both the RhoA and RhoD elements. In contrast, PrdG4;UAS-Sens embryos show no significant change in RhoBAD-lacZ activity (Figure 5E). This data was surprising as Sens directly binds and represses RhoBAD (Li-Kroeger et al., 2008). However, we noted that even though PrdG4 enhanced the number of cells expressing Sens, it did not dramatically increase Sens levels within the C1 SOP cells that express RhoBAD-lacZ (compare PrdG4 “on” versus PrdG4 “off” segments). Thus, while both ato and sens are required for normal RhoBAD activity, only ato is sufficient to trigger rho enhancer activation in additional cells.
Figure 5. Ato stimulates RhoBAD activity through the RhoA and RhoD elements.
A-E. Lateral views of stage 11 Drosophila embryos immunostained for β-gal (green), Ato (red), and/or Sens (purple) as indicated. Close-ups of thoracic and abdominal segments (A1 labeled) are shown at right. PrdG4 is expressed in every other segment including the first abdominal segment (A1) and the T2 thoracic segment. The PrdG4-negative segments that do not drive UAS gene expression are noted by arrows in A-D. PrdG4;UAS-Ato embryos reveal that ectopic Ato induces RhoBAD-lacZ (A), RhoAAA-lacZ (B), RhoAAAHoxM-lacZ (C), and RhoDD-lacZ (D) activity. PrdG4;UAS-Sens embryo shows that ectopic Sens does not significantly alter RhoBAD activity (E). Note, however, that while many more cells express Sens, Sens levels in the C1 lineage (arrows) are not dramatically different between the PrdG4-on versus PrdG4-off segments.
Ato indirectly regulates RhoBAD by inhibiting Senseless binding to RhoA
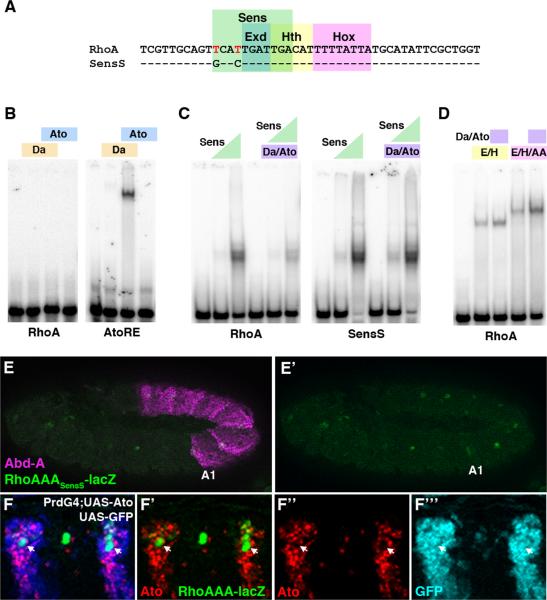
While our genetic studies reveal that Ato may be a direct activator of RhoBAD-lacZ activity, RhoBAD does not contain a good match for the E-Box sequence bound by Ato and its partner protein Daughterless (Da). Moreover, purified Ato and Da proteins bind a control site (E1 of AtoRE, zur Lage et al., 2004) but not RhoBAD (data not shown), RhoA (Figure 6B), or RhoD (data not shown). These data suggest Ato does not regulate RhoBAD activity by direct DNA binding. However, RhoA does contain a Sens binding site (Figure 6A) and Sens is known to physically interact with Ato (Acar et al., 2006). Hence, we performed gel shift analysis using all three proteins and found that while no higher order complexes are observed, Sens binding was consistently reduced by 50% in the presence of Ato and Da (Figure 6C). In contrast, Ato and Da do not inhibit the binding of three other regulators of RhoA as an Abd-A Hox transcription factor complex containing Extradenticle (Exd) and Homothorax (Hth) binds well in their presence (Figure 6D). Intriguingly, we previously reported that this Abd-A Hox complex regulates RhoBAD-lacZ activity by interfering with Sens-mediated repression through direct competition for overlapping binding sites (Li-Kroeger et al., 2008). Thus, we propose that both Ato and Abd-A contribute to RhoBAD activity by antagonizing Sens binding to RhoA through distinct mechanisms: Ato by sequestering Sens protein and Abd-A by competing with Sens for RhoA.
Figure 6. Ato decreases the ability of Senseless to bind RhoA.
A. Sequence of wild type RhoA element highlighting the presence of the Sens, Exd, Hth, and Hox binding sites. The red nucleotides denote a mis-match to the consensus Sens binding site. The SensS sequence is shown. B. Gel shift assays showing that Ato and Da do not bind RhoA. The Ato and Da proteins are able to bind a positive control probe (AtoRE). C. Gel shift assays showing that Ato and Da significantly decrease the ability of Sens to bind RhoA but not the optimized SensS probe. D. Gel shift assays showing that Ato and Da have no significant effect on Exd/Hth or Exd/Hth/Abd-A binding to RhoA. E-E’. Lateral view of a stage 11 RhoAAASensS-lacZ embryo immunostained for β-gal (green) and Abd-A (purple) revealing that strengthening of the Sens site in all three copies of RhoA results in decreased β-gal activity in the abdomen. F-F’’’. Lateral view of three abdominal PrdG4;UAS-Ato;UAS-GFP segments (stage 11) immunostained for β-gal (green), GFP (blue), and Ato (red). Note that while GFP is detected in the β-gal positive C1 cells, Ato expression is low (arrows). However, Ato protein is detected in many other cells of the PrdG4-on stripe.
The complex relationship between ato and sens makes genetically testing this model in vivo difficult, since the removal of either factor compromises the expression of the other and severely disrupts the development of ch organ SOP cells (Jafar-Nejad et al., 2003; Jafar-Nejad and Bellen, 2004; Nolo et al., 2000). To circumvent these problems, we instead generated transgenic reporter lines (RhoBADSensS-lacZ and RhoAAASensS-lacZ) containing a higher affinity Sens binding site in place of the endogenous low affinity DNA binding sequence. This higher affinity site changes two nucleotide mismatches from the Sens consensus site and enhances Sens binding for RhoA approximately 20-fold without affecting the formation of the Exd/Hth/Abd-A Hox complex on this probe (SensS in Figure 6A and Li-Kroeger et al., 2008). When tested in vivo, we found that RhoAAASensS-lacZ, like RhoBADSensS-lacZ, results in decreased β-gal expression in abdominal SOP cells (Figure 6E and Li-Kroeger et al., 2008). These findings suggest that Sens binding on SensS takes precedence over Abd-A and Ato inhibition resulting in gene repression. Consistent with this result, we found that Ato/Da does not significantly decrease Sens binding for the high affinity RhoA-SensS sequence (Figure 6C). Thus, Ato's ability to interfere with Sens DNA binding is site-specific and dependent upon the relative affinity of Sens DNA binding.
Together, these data suggest that Ato and Sens are co-expressed at the time RhoBAD is activated in C1 SOP cells and that Ato diminishes Sens binding to RhoA, thereby interfering with Sens-mediated repression of RhoBAD. A prediction of this model is that prolonging Ato expression within the C1 SOP cell lineage should interfere with Sens-mediated repression of RhoBAD and result in higher β-gal. Indeed, we observe an increase in Rho enhancer-dependent β-gal levels within Ato mis-expressing segments of the thorax, but these levels do not consistently reach abdominal levels (Figure 5). However, a recent paper by Chang et al. found that the Achaete and Scute proneural proteins are post-transcriptionally degraded prior to SOP cell division even when expressed by a Gal4 driver (Chang et al., 2008). To determine if the Ato protein is also rapidly down-regulated in the SOP cell lineage, we co-expressed Ato and GFP (PrdG4;UAS-Ato;UAS-GFP) and found that Ato protein is cleared while GFP is detected within cells of the C1 SOP lineage (Figure 6F). Neighboring cells, however, express both Ato and GFP protein, suggesting that only SOP cells readily turn-over proneural proteins. This finding, coupled with our expression analysis showing the continued detection of Sens protein after SOP cell division (Figure 3), demonstrates that cells of the ch organ lineage experience Ato and Sens co-expression as well as Sens alone during their development.
DISCUSSION
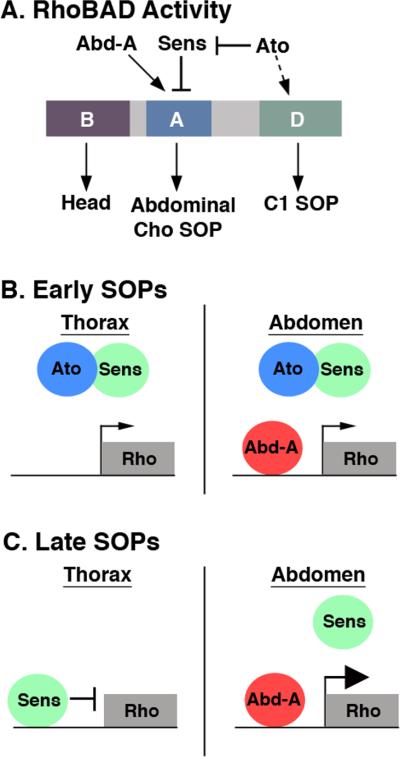
In this study, we found that the Atonal proneural factor is required for both normal rho enhancer function and the proper specification of abdominal oenocytes. In addition, we determined that two distinct regions of the RhoBAD enhancer contribute to gene activity within the C1 SOP cells. The RhoA element preferentially drives gene expression within abdominal SOP cells, whereas RhoD drives weaker gene expression within the C1 SOP cells of both the thoracic and abdominal segments (Figure 7A). Using a combination of genetic and biochemical analyses, we found that the Ato, Sens, and Abd-A inputs contribute to proper rho enhancer activity. In particular, we show that RhoA, but not RhoD, is directly responsive to the Abd-A Hox factor. In addition, we found that Ato indirectly stimulates RhoBAD activity through both the RhoA and RhoD elements. Although we currently do not understand how Ato stimulates RhoD, we found that Ato limits the DNA binding activity of the Sens repressor protein to RhoA. Coupled with other recent findings on proneural gene function, our results have two major implications: 1) We describe a model for how Ato and Sens inputs are integrated to differentially regulate gene expression during SOP cell lineage development, and 2) We discuss how a proneural input (Ato) and a Hox factor (Abd-A) cooperate to regulate Rho enhancer activity, at least in part, by limiting Sens-mediated repression.
Figure 7. Model of RhoBAD enhancer function in SOP cells.
A. Schematic of RhoBAD enhancer highlighting the positive (Ato and Abd-A) and negative (Sens) transcriptional inputs into the conserved RhoA and RhoD elements. The RhoB element drives gene expression in the embryonic head, the RhoA region drives gene expression within the abdominal ch organ (Cho) SOP cells, and the RhoD region drives gene expression within the C1 SOP cells of both the thorax and abdomen. B-C. Schematic of RhoA function in early (B) and late (C) SOP cells. In early SOP cells the thoracic and abdominal SOPs express Ato, which limits Sens-mediated repression of rho resulting in low levels of gene expression within both the thorax and abdomen. In late SOP cells, Ato has been degraded freeing Sens to bind RhoA and repress gene expression within the thorax. Within the abdomen, however, Abd-A continues to limit Sens binding allowing for enhanced rho gene expression and the induction of abdomen-specific oenocyte cells.
Ato and Sens interactions affect gene expression in SOP cells
Sens and the proneural factors are intricately linked during PNS development in Drosophila (Jafar-Nejad et al., 2003; Jafar-Nejad and Bellen, 2004; Nolo et al., 2000). Loss-of-function mutations in proneural genes disrupt sens expression resulting in a decrease in sensory organ formation and sens mutations result in decreased proneural gene expression and widespread sensory organ deficits. While both encode transcription factors required for PNS development, they have opposite effects on gene expression when bound to DNA. Proneural factors bind E-box DNA sequences with Daughterless to activate gene expression, whereas Sens binds a distinct DNA sequence to repress gene expression (Bertrand et al., 2002; Jafar-Nejad et al., 2003; Powell et al., 2004). However, recent data revealed that proneural proteins can convert Sens from a transcriptional repressor to a co-activator. Acar et al. showed that three different proneural factors (Ac, Sc, and Ato) interact with Sens in GST-pulldown and/or co-immunoprecipitation assays (Acar et al., 2006). In addition, cell culture assays showed that Sens stimulates the activation potential of proneural factors bound to E-Box sequences (Acar et al., 2006; Powell et al., 2008; Powell and Jarman, 2008). Thus, Sens is a transcriptional repressor when directly bound to DNA through its zinc finger motifs whereas it is a potent co-activator when recruited to DNA by proneural proteins.
In this study, we provide two pieces of information that add to our understanding of how Sens and proneural factors regulate gene expression. First, we used purified Sens and Ato/Da proteins to show that Ato decreases the ability of Sens to bind the RhoA enhancer element. As RhoA contains a relatively low affinity Sens site, we performed a parallel experiment using a high affinity Sens site (SensS) and found that Ato does not significantly alter Sens binding to an optimized site. This data reveals that Ato's ability to interfere with Sens binding to DNA is site-specific and dependent upon binding affinity. How might Ato interfere with Sens binding to DNA? Acar et al. showed that Ato, Ac, and Sc all directly interact with Sens through the second and third Sens zinc finger motifs (Acar et al., 2006). Since Sens requires these motifs to bind DNA, it is likely that the proneural factors compete with DNA for the same zinc fingers. Thus, we propose the following model: if the binding affinity of Sens to DNA is high, Ato cannot interfere with Sens-mediated repression. However, if the binding affinity of Sens to DNA is low, Ato binds Sens and interferes with its ability to repress gene expression.
Secondly, expression analysis revealed that cells of the C1 SOP lineage differentially express Ato and Sens during their maturation. The initial SOP cell (SOPI) expresses both Ato and Sens during sensory organ specification. However, Ato protein is rapidly extinguished and no longer detectable once the SOP cell divides, whereas Sens persists into the SOPII cells. The rapid loss of Ato, even when it is expressed using a Gal4 driver, is consistent with recent findings that proneural proteins activate an E3 ubiquitin ligase pathway to trigger their own degradation (Chang et al., 2008). Thus, these findings suggest that the early SOP cell expresses both Ato and Sens and that Ato can alter Sens function in two ways: 1) by recruiting Sens to E-Box sequences as a co-activator, and 2) by interfering with Sens's ability to bind low affinity DNA sites (Figure 7B).
Integrating the proneural and Hox pathways to regulate rho in the Drosophila abdomen
Brodu et al. reported that rho is initially weakly expressed in C1 SOP cells in both the thorax and abdomen, and is only up-regulated in the abdominal SOP cells by the Abd-A Hox factor (Brodu et al., 2002). We found that the RhoBAD-lacZ reporter is also expressed in this pattern and propose that Ato is part of an initiator pathway that allows rho expression in early C1 SOP cells. Ato does so in two ways: 1) by inhibiting Sens binding to RhoA through direct protein-protein interactions, and 2) by indirectly stimulating RhoD through an unknown mechanism. In total, these interactions result in the initiation of rho expression in early C1 SOP cells of both thoracic and abdominal segments (Figure 7B). Ato's subsequent degradation releases Sens to bind RhoA and repress gene expression in thoracic SOP cells (Figure 7C). Consistent with this idea, mutations that abolish Sens binding (SensM) result in de-repression of Rho reporters in the thorax (Li-Kroeger et al., 2008). In the abdomen, however, an Abd-A complex out-competes Sens for RhoA to allow continued rho expression, subsequent EGF signaling, and the specification of additional cell types (Figure 7B,C). Thus, Ato cooperates with the Abd-A Hox factor to stimulate EGF signaling by up-regulating rho expression via interfering with Sens-mediated repression.
While our findings provide insight into how rho is up-regulated in abdominal SOP cells, they uncover an interesting question: why is rho activated at all within thoracic SOP cells? Currently, there is no known function for rho activity within the thorax as rho mutant embryos show no phenotypic defect in cells surrounding the thoracic SOPs. As the lack of oenocyte production within the thorax is solely due to insufficient Spi secretion (oenocytes form in the thorax if rho is ectopically expressed, Brodu et al., 2002), these data suggest that Rho levels are too low to trigger enough Spi secretion to affect neighboring cell fate. Consistent with this prediction is that the levels of an activated kinase downstream of EGF signaling (phospho-ERK) are very low in cells neighboring the thoracic C1 SOP cells compared to the abdominal SOP cells. So, why is rho activated within the thorax if it has no functional consequences? One interpretation is that Ato may provide competency for rho expression so that an additional positional factor such as Abd-A can fully stimulate rho and trigger Spi secretion and EGF signaling. In support of this idea, the widespread expression of Abd-A within the thorax activates RhoBAD-lacZ expression only within the C1 SOP cells and oenocytes form only in close proximity to these thoracic SOP cells (Figure 2 and data not shown). Thus, weak rho expression downstream of ato may provide a flexible and responsive system for activating Spi secretion in different body regions.
MATERIALS AND METHODS
Plasmid and transgenic fly generation
A full-length His-Sens-V5 tagged protein was made by cloning the Sens cDNA (kind gift of Hugo Bellen) in-frame with a C-terminal V5 epitope of the pAc5.1 vector (Invitrogen). The entire Sens-V5 cDNA was subsequently cloned into the pET14b His-tagged bacterial protein expression vector (Novagen). Three copies of RhoA (Insert sequence), two copies of RhoB (Insert sequence), or two copies of RhoD (Insert sequence) were multimerized using oligonucleotides and PCR. Restriction enzyme sites (EcoR1 and BamH1) were engineered into the oligonucleotides (details available upon request) and the PCR product was cloned into the hs43-nuc-lacZ P-element vector to generate the RhoAAA-lacZ, RhoBB-lacZ, and RhoDD-lacZ transgenic fly constructs (Gebelein et al., 2004). Specific mutations in all three copies of RhoA were generated by PCR. The RhoA wild type and SensS sequences are shown in Figure 6A. The HoxM sequence changes the highlighted Hox site in Figure 6A from TTTTAT to TTTGCT. All constructs were confirmed by DNA sequencing and transgenic fly lines were established using standard P-element transformation (Rainbow Transgenic Flies).
Fly stocks and embryo staining
Fly stocks used were as follows: yw1118, UAS-GFP, and PrdG4 (Bloomington Stock Center); ato1 and UAS-Ato (gift of Andrew Jarman); sensE2 and UAS-Sens (gift of Hugo Bellen); abdAM1, UAS-AbdA, (gift of Richard Mann); RhoBAD-lacZ and RhoBADHoxM-lacZ (Li-Kroeger et al., 2008); RhoAAA-lacZ, RhoAAAHoxM-lacZ, RhoAAASensS-lacZ, RhoBB-lacZ, and RhoDD-lacZ (this study). Expression of lacZ (anti-β-gal, Abcam, 1:1000), Abd-A (GP4, 1:500) (Jarman et al., 1993; Li-Kroeger et al., 2008), mAb22C10 (DSHB, 1:50), Ato (1:5000) (Jarman et al., 1993), Sens (1:200) (Xie et al., 2007), Pros (1:500), Salm (1:1000) (Xie et al., 2007), and GFP (Rb, A11122 - Molecular Probes, 1:500) were detected by indirect immunofluorescent antibody staining using an apotome-configured Zeiss fluorescent microscope. All flies were raised at 25°C. Embryos were harvested, fixed and immunostained using standard protocols. Comparative β-gal levels of RhoBAD-lacZ activity were determined by imaging 10 different age-matched heterozygous and homozygous mutant embryos using identical settings. Oenocyte identity was confirmed using anti-Salm and counterstaining with anti-Pros to discriminate between oenoyctes (Salm-positive only) and scolopale cells (Salm- and Pros-positive cells, data not shown). All quantifications were done using a minimum of 10 Drosophila embryos (7 abdominal segments per embryo).
Protein purification, EMSAs, and antibody production
The full-length His-tagged Ato and Da proteins (constructs were kind gifts of Andrew Jarman) were purified from BL21 bacteria under denaturing conditions using Ni-chromatography. Proteins were re-folded using step-wise decreases in urea through dialysis as previously described (Gebelein et al., 2002). The His-tagged Abd-A, Exd, and Hth proteins were purified as previously described (Gebelein et al., 2002; Gebelein et al., 2004). Protein concentrations were measured by the Bradford assay and confirmed by SDS PAGE and Coomassie blue analysis. EMSAs were performed using native PAGE as described (Gebelein and Urrutia, 2001). The dried acrylamide gels were exposed to phosphor-screens and densitometry was performed using ImageQuant 5.1 software.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andrew Jarman, Hugo Bellen, Yuh Nung Jan, Richard Mann, the Bloomington Drosophila Stock Center (Indiana University), and the Developmental Studies Hybridoma Bank (University of Iowa) for reagents. We thank Richard Mann, Nadean Brown, and Mike Crickmore for critically reading the manuscript. Special thanks to Masato Nakafuku for many helpful discussions and suggestions regarding this project. This work was supported by an Ohio Cancer Research Associate grant and an NIH grant (R01GM079428) to B.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development. 2006;133:1979–1989. doi: 10.1242/dev.02372. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Bray S. Notch signalling in Drosophila: three ways to use a pathway. Semin Cell Dev Biol. 1998;9:591–597. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- Brodu V, Elstob PR, Gould AP. abdominal A specifies one cell type in Drosophila by regulating one principal target gene. Development. 2002;129:2957–2963. doi: 10.1242/dev.129.12.2957. [DOI] [PubMed] [Google Scholar]
- Chang PJ, Hsiao YL, Tien AC, Li YC, Pi H. Negative-feedback regulation of proneural proteins controls the timing of neural precursor division. Development. 2008;135:3021–3030. doi: 10.1242/dev.021923. [DOI] [PubMed] [Google Scholar]
- Elstob PR, Brodu V, Gould AP. spalt-dependent switching between two cell fates that are induced by the Drosophila EGF receptor. Development. 2001;128:723–732. doi: 10.1242/dev.128.5.723. [DOI] [PubMed] [Google Scholar]
- Gebelein B. The control of EGF signaling and cell fate in the Drosophila abdomen. Fly (Austin) 2008;2:257–258. doi: 10.4161/fly.7084. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BP, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafar-Nejad H, Bellen HJ. Gfi/Pag-3/senseless zinc finger proteins: a unifying theme? Mol Cell Biol. 2004;24:8803–8812. doi: 10.1128/MCB.24.20.8803-8812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Kunisch M, Haenlin M, Campos-Ortega JA. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc Natl Acad Sci U S A. 1994;91:10139–10143. doi: 10.1073/pnas.91.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage P, Jan YN, Jarman AP. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr Biol. 1997;7:166–175. doi: 10.1016/s0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B. Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell. 2008;15:298–308. doi: 10.1016/j.devcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Okabe M, Okano H. Two-step induction of chordotonal organ precursors in Drosophila embryogenesis. Development. 1997;124:1045–1053. doi: 10.1242/dev.124.5.1045. [DOI] [PubMed] [Google Scholar]
- Powell LM, Deaton AM, Wear MA, Jarman AP. Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes Cells. 2008;13:915–929. doi: 10.1111/j.1365-2443.2008.01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Cantera R, Urban J, Technau G, Kafatos FC, Barrio R. Spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development. 2001;128:711–722. doi: 10.1242/dev.128.5.711. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. Regulating the dynamics of EGF receptor signaling in space and time. Development. 2005;132:4017–4027. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- zur Lage P, Jarman AP. Antagonism of EGFR and notch signalling in the reiterative recruitment of Drosophila adult chordotonal sense organ precursors. Development. 1999;126:3149–3157. doi: 10.1242/dev.126.14.3149. [DOI] [PubMed] [Google Scholar]
- zur Lage PI, Powell LM, Prentice DR, McLaughlin P, Jarman AP. EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev Cell. 2004;7:687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.