Abstract
Activation of prophenoloxidase and synthesis of antimicrobial peptides (AMPs) are two important innate immune mechanisms in insects. In the current study, we investigated immune responses activated by three major bacterial components, lipopolysaccharide (LPS) (including rough mutants of LPS), lipoteichoic acid (LTA), and peptidoglycan (PG), in the larvae of a lepidopteran insect, Manduca sexta. We found that two DAP (Diaminopimelic acid)-type PGs from Escherichia coli and Bacillus subtilis were much more potent than LPS and LTA from the respective bacteria as well as a Lysine-type PG in activation of prophenoloxidase in M. sexta larval plasma in vitro. Transcription levels of AMP genes, such as Attacin, Lebocin and Moricin genes, in the hemocytes and fat body of larvae were significantly induced by smooth LPS (TLR4grade) and rough mutants of LPS (TLRgrade™), synthetic lipid A, LTA, and PG. LPS from E. coli and LTA from B. subtilis activated AMP expression to significantly higher levels than PGs from the respective bacterial strains, and smooth LPS were more potent than lipid A and rough mutants of LPS in activation of AMP expression. Our results demonstrated for the first time that LTA can activate AMP expression, and different moieties of LPS may synergistically activate AMP expression in M. sexta.
Keywords: Lipopolysaccharide, Lipoteichoic acid, Peptidoglycan, Phenoloxidase, Antimicrobial peptide, Manduca sexta
1. Introduction
The innate immune system is important to both vertebrates (Mogensen, 2009) and invertebrates (Iwanaga and Lee, 2005), and it relies on a repertoire of germline-encoded pattern recognition receptors (PRRs) to recognize specific pathogen-associated molecular patterns (PAMPs) to initiate cellular and humoral immune responses (Akira et al., 2006; Janeway and Medzhitov, 2002; Kumar et al., 2009). PAMPs are unique molecular patterns found only in pathogens but not in host cells, such as bacterial lipopolysaccharide (LPS), peptidoglycan (PG), lipoteichoic acid (LTA), flagellin, unmethylated CpG DNA, and viral dsRNA (Janeway and Medzhitov, 2002; Mogensen, 2009). PRRs include Toll-like receptors (TLRs) (Armant and Fenton, 2002), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) (Kawai and Akira, 2009), peptidoglycan recognition proteins (PGRPs) (Royet and Dziarski, 2007), and C-type lectin like proteins (Hollmig et al., 2009).
The innate immune system of invertebrates share similarities with that of vertebrates, and it also has distinct mechanisms (Kanost et al., 2004; Kimbrell and Beutler, 2001; Kurata et al., 2006; Lemaitre and Hoffmann, 2007). In insects, cellular immune responses include hemocyte-mediated phagocytosis, nodule formation, encapsulation and melanization, whereas activation of the prophenoloxidase (proPO) system and synthesis of antimicrobial peptides (AMPs) are two important humoral defense mechanisms (Cerenius et al., 2008; Hancock and Sahl, 2006; Imler and Bulet, 2005; Kanost et al., 2004; Lavine and Strand, 2002; Lemaitre and Hoffmann, 2007; Marmaras and Lampropoulou, 2009). ProPO activation can be triggered by bacterial components after their recognition by specific PRRs, leading to activation of a serine proteinase cascade that results in activation of proPO-activating proteinases (PAPs) (Cerenius et al., 2008). Active PAPs then convert proPO to functionally active phenoloxidase (PO) in the presence of essential co-factors, serine proteinase homologs (SPHs) (Jiang et al., 2003; Yu et al., 2003), and PO is the key enzyme in the melanization reaction. For example, LPS and PG can stimulate proPO activation after binding to C-type lectins and PGRPs, respectively (Takehana et al., 2002; Yu et al., 1999; Yu and Kanost, 2000). LPS from Gram-negative bacteria, LTA from Gram-positive bacteria, and PG from both Gram-negative and Gram-positive bacteria are three major bacterial components. However, it is not clear which bacterial component (LPS, LTA or PG) is more potent in activation of proPO.
Expression of AMPs is regulated by signal transduction pathways. In Drosophila melanogaster, AMP expression is regulated by the Toll and immune deficiency (IMD) pathways (Lemaitre and Hoffmann, 2007), and the two pathways are mainly activated by Lys (Lysine)-type and DAP (Diaminopimelic acid)-type PGs, respectively (Choe et al., 2005; Michel et al., 2001). Most Gram-positive bacteria contain the Lys-type PG, while Gram-negative bacteria and Gram-positive Bacilli species contain the DAP-type PG (Schleifer and Kandler, 1972). Interestingly, the DAP in Bacilli species is mostly amidated-DAP (more than 90%) but the DAP in Gram-negative bacteria is a meso-DAP (Leulier et al., 2003; Schleifer and Kandler, 1972). LPS and LTA are also potent elicitors in activation of immune-related genes in several insect species, including Tribolium castaneum (Altincicek et al., 2008), D. melanogaster (Jin et al., 2008), Tenebrio molitor (Haine et al., 2008), Bombus ignitus (Choi et al., 2008), Bombyx mori (Ha Lee et al., 2007), and Thermobia domestica (Altincicek and Vilcinskas, 2007). Although it was reported that crude LPS can induce immune responses in D. melanogaster (Imler et al., 2000), highly purified LPS does not activate the Toll or IMD pathway in adult flies (Kaneko et al., 2004; Leulier et al., 2003). Crude LPS are often contaminated with trace amounts of PG, which may cause activation of immune responses (Ha Lee et al., 2007; Imler et al., 2000). Recently, it was reported that highly purified LPS (TLRgrade™) can activate immune responses in the fat body of B. mori (a lepidopteran insect), but the activation level is lower than that by crude LPS or PG (Tanaka et al., 2009). Thus, it is necessary to confirm activation of AMPs by LPS in another lepidopteran insect. Insects encode a variety of AMPs. Some AMPs, such as Cecropin and Attacin, are commonly found in insect species, but some other AMPs, like Moricin, Lebocin and Gloverin, have been identified only in lepidopteran species but not in dipteran species (Axen et al., 1997; Cheng et al., 2006; Huang et al., 2009; Yamakawa and Tanaka, 1999). Little is known about how these lepidoptera-specific AMPs are activated and regulated.
Smooth LPS is composed of three moieties: O-specific antigen (O-polysaccharide), carbohydrate core, and lipid A (Raetz and Whitfield, 2002). It has been demonstrated in mammals that lipid A moiety of LPS can stimulate the TLR4 signaling pathway (Meng et al., 2010). However, it is not known whether the O-specific antigen and core moieties of LPS play a role in the TLR4 signaling pathway or not. Recently, LTA has been identified as a ligand for Draper in phagocytosis of Staphylococcus aureus by Drosophila hemocytes (Hashimoto et al., 2009). However, no study thus far has been reported about activation of AMPs by LTA in model insects like D. melanogaster and B. mori or in other insects. Here we report AMP expression activated by smooth LPS (TLR4grade), LTA and PG from the respective bacterial strains, and by synthetic lipid A and rough mutants (Ra and Re) of LPS (TLRgrade™), in the hemocytes and fat body of larvae from the tobacco hornworm, M. sexta. We also compared the ability of LPS, LTA and PG in proPO activation using M. sexta larval plasma. Our results showed that DAP-type PGs (from Escherichia coli and Bacillus subtilis) are major elicitors of proPO activation in M. sexta larval plasma compared to LPS and LTA from the respective bacterial strains and a Lys-type PG from S. aureus. Smooth LPS, two rough mutants (Ra and Re) of LPS, synthetic lipid A, LTA and PG all significantly induced expression of AMP genes to various levels in the hemocytes and fat body of M. sexta larvae compared to the controls (injection with saline or DMSO). However, the activation levels of AMPs by LPS-K12 (from E. coli K12) and LTA-BS (from B. subtilis) were significantly higher than those by PG-K12 and PG-BS from the respective bacterial strains (equal amounts of bacterial components), respectively, suggesting that LPS-K12 and LTA-BS can indeed activate AMP expression in M. sexta larvae. But PG-SA (from S. aureus) was more potent than LTA-SA in activation of AMP expression. We also observed that lipid A and two rough mutants (Ra and Re) of LPS activated expression of AMP genes to significantly lower levels than smooth LPS did. Our results demonstrated for the first time that LTA can activate AMP expression, and different moieties of LPS may have synergistic effects on activation of AMPs in M. sexta.
2. Material and methods
2.1. Insects and bacterial cell wall components
M. sexta eggs were kindly provided by Professor Michael Kanost, Department of Biochemistry at Kansas State University. Larvae were reared on artificial diet at 25°C (Dunn and Drake, 1983), and 5th instar larvae were used for all experiments. Ultrapure LPS (TLR4grade, only activates the TLR4 pathway) from E. coli strain 0111:B4 (LPS-0111) and strain K12 (LPS-K12) and PG from the same E. coli strains (PG-0111 and PG-K12), LTA and PG from Staphylococcus aureus (LTA-SA, PG-SA) and Bacillus subtilis (LTA-BS, PG-BS) were purchased from InvivoGen (San Diego, California, USA). Synthetic lipid A was purchased from Peptide Institute, Inc (Minoh-shi, Osaka, JAPAN). Rough mutants of LPS from E. coli EH100 (LPS-Ra, TLRgrade™) and E. coli R515 (LPS-Re, TLR4grade™) were purchased from Alexis Biochemicals (Plymouth Meeting, PA, USA).
2.2 In vitro proPO activation
Individual M. sexta larval hemolymph samples were collected, hemocytes were removed by centrifugation and plasma samples were screened for proPO activation as described previously (Jiang et al., 2001). Plasma samples with low PO activity when incubated at room temperature alone but high PO activity when incubated with Micrococcus luteus were used for proPO activation assays. Plasma sample #15 was selected and used for the following experiments. Aliquots (2 μL each) of plasma were incubated alone or with 1 μg of LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA or PG-SA in a total of 10 μL Tris-Ca2+ buffer (100 mM Tris-HCl, 100 mM NaCl, 1 mM CaCl2, pH 7.5) in wells of a 96-well microtiter plate for 30 min at room temperature, then L-dopamine substrate solution (2 mM L-dopamine in 50 mM phosphate buffer, pH 6.5) (190 μL) was added. Absorbance at 470nm was measured every 30 seconds for a total of 30 readings with a microtiter plate reader (PowerWave XS, Bio-Tek Instrument, Inc). One unit of PO activity is defined as an increase of absorbance (ΔA470) by 0.001 per minute. Data from four replicates of each sample were analyzed. Experiments were repeated 3 times and similar results were obtained. To determine whether LPS-K12 and LTA-BS can activate proPO after prolonged incubation, aliquots of plasma were incubated with LPS-K12 and LTA-BS for a total of two hours. PO activity was measured every 30 min and data were collected as described above.
2.3 Bacterial inhibition zone assay
The inhibition zone assay was performed according to a published protocol (Hultmark, 1998). A single colony of E. coli DH5α or B. subtilis was grown overnight in Luria-Bertani (LB) broth at 37°C. Overnight bacterial cultures (0.1mL) were then inoculated into fresh LB broth (5 mL) and bacteria were grown to late log-phase. Log-phase bacterial cultures were diluted to 2 × 108 cfu/mL, and 1μL bacterial culture (about 200,000 cfu) was added into 5 mL LB containing 0.8% agarose (kept in a 45°C heating block), mixed well and spreaded immediately on a petri dish. After the medium was solidified, 2-mm wells were punched in the agarose, four wells per plate. Hemolymph was collected from naïve larvae or larvae injected with bacterial components (10 μg per larva) at 24 h post-injection (at least four larvae per group), and hemocytes were removed by centrifugation. Plasma samples were collected (from at least 4 larvae) and 4 μL plasma samples were added into each well. The plates were incubated at 37°C overnight, and the diameters of inhibition zones were measured (by subtracting 2-mm from the punched well) and recorded.
2.4 In vivo immune stimulation in M. sexta larvae by bacterial components
Fifth instar M. sexta larvae were anesthetized on ice for 20 min. Each larva was injected with saline (control) or with 20 μg of Lipid A, LPS-Ra, LPS-Re, LPS-0111, LPS-K12, PG-0111, PG-K12, PG-SA, PG-BS, LTA-SA or LTA-BS (four larvae per group). Hemolymph was collected 24 h post-injection and mixed with equal volume of anti-coagulant (AC) saline (4 mM NaCl, 40 mM KCl, 8 mM EDTA, 9.5 mM citric acid-monohydrate, 27 mM sodium citrate, 5% sucrose, 0.1% polyvinylpyrollidone, 1.7 mM PIPES) supplemented with propylthiouracil. Hemocytes were collected after centrifugation at 3000g for 10 min at 4°C and immediately resuspended in 1 mL TRI reagent (Sigma Aldrich) for total RNA extraction. Fat body was also collected from these larvae and washed three times in AC saline for total RNA extraction. For reverse transcription, total RNA (2 μg) was treated with RQ1 RNase-free DNase I (Promega) at 37°C for 30 min to remove contaminated genomic DNA, and DNase I was inactivated by heating to 75°C for 20 min. Reverse transcription was performed using OligodT primer (Promega) and ImProm-II reverse transcriptase (Promega) following the manufacturer’s instructions. RNase H (NEB) was added to remove hybrid RNA, and cDNA was diluted 10-fold with distilled water. Real-time PCR was performed with SYBR Premix (Takara) on a 7500 system (Applied Biosystems) using diluted cDNAs as templates and primers listed in Table 1. Data from three replicates of each sample were analyzed with SDS software (ABI) using a comparative method (2−ΔΔCt). Samples from saline-injected larvae were used as the controls.
Table 1.
Primers used in real-time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Ribosomal protein S3 (Rps3) | 5′-GGAGCTGTACGCTGAGAAAG-3′ | 5′-TTAATTCCGAGCACTCCTTG-3′ |
| Attacin-2 | 5′-TTCGTCGTCCTGGTCTGTCT-3′ | 5′-TTGGAGAGGGAGAAGCCCAT-3′ |
| Lebocin | 5′-ACGTGCGTAGTGTGAACGAG-3′ | 5′-CGCAGATTATGAGTTACGACGA-3′ |
| Moricin | 5′-ATAAAGCAGGCTGGCAAGG-3′ | 5′-AGAAGATTCCGAAGGGAGAAC-3′ |
| Lysozyme | 5′-GTGTGCCTCGTGGAGAATG-3′ | 5′-ATGCCTTGGTGATGTCGTC-3′ |
For stimulation by different forms of LPS, larvae were injected with DMSO, lipid A (dissolved in DMSO), LPS-Ra, LPS-Re, LPS-0111 and LPS-K12 (20 μg per larva) (four larvae per group), hemocytes were collected at 24h post-injection for real-time PCR analysis as described above. All the above experiments were repeated from three independently pooled samples (biological repeats, n=3), similar results were obtained, and figures in this article represent a typical set of data.
2.5 In vivo dose-dependent immune stimulation in M. sexta larvae by LPS, PG, and LTA
For dose-dependent stimulation assay, each larva was injected with 0.02, 0.2 or 2 μg of LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA, or PG-SA (four larvae per group). Hemocytes were collected at 6h post-injection for preparation of RNAs and analysis of AMP gene expression in these hemocytes was performed by real-time PCR the same as described above. These experiments were repeated three times.
2.6 In vitro immune stimulation in the hemocytes and fat body by bacterial components
Hemolymph from six 5th instar M. sexta naïve larvae was collected into an equal volume of AC saline, and hemocytes (5×106) were seeded in wells of 6-well plates. After adhesion, hemocytes were washed three times with the Grace’s medium containing 1× penicillin and streptomycin (HyClone) to remove plasma, then 2 mL of the Grace’s medium were added into each well. Larval fat body was also collected from naïve larvae (at least 4 larvae) into AC saline, and dispensed into wells of 6-well plates. Fat body was washed three times with the Grace’s medium, and 2 mL of the Grace’s medium were added to each well. LPS-K12, LTA-BS, PG-K12 or PG-BS (20 μg per well) was added to each well, and hemocytes and fat body were incubated for 24 h at 25°C. Saline (20 μL of 0.85% NaCl) was added in the control wells. Total RNA was prepared from these hemocytes and fat body, and gene expression was analyzed by real-time PCR as described above. These experiments were repeated three times.
2.7 Data analysis
Figures were made with GraphPad Prism software with one representative set of data. The significance of difference was determined by an unpaired t-test or by ANOVA followed by a Tukey’s multiple comparison test with the same software (GraphPad, San Diego, CA).
3. Results
3.1 Prophenoloxidase is mainly activated by DAP-type PGs
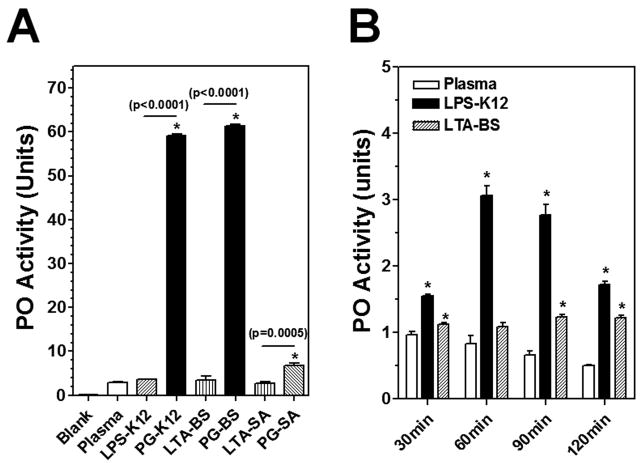
Activation of prophenoloxidase (proPO) is an important immune mechanism in arthropods against large parasites, and the proPO activation cascade can be triggered by bacterial components when specific recognition proteins are present (Takehana et al., 2002; Yu et al., 1999; Yu and Kanost, 2000). We used an in vitro proPO activation assay to test which bacterial components are able to activate proPO in M. sexta larval plasma. When the naïve plasma samples were incubated at room temperature for 30 min, very low PO activity was observed in the plasma sample alone and in the plasma samples incubated with LPS-K12 (from E. coli K12), LTA-BS (from B. subtilis) and LTA-SA (from S. aureus), indicating that proPO was not activated by LPS or LTA under these conditions (Fig. 1A). However, significantly higher PO activity was detected in the naïve plasma samples incubated with three PGs (PG-K12, PG-BS and PG-SA) compared to the plasma alone, and the two DAP-type PGs (PG-K12 and PG-BS) activated PO activity (~60 units) to a significantly higher level than the Lys-type PG (PG-SA) did (~6 units) (Fig. 1A).
Fig. 1. In vitro proPO activation in M. sexta larval plasma by LPS, LTA and PG.
(A) Aliquots of naïve plasma were incubated alone, or with LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA or PG-SA in wells of a 96-well microtiter plate for 30 min at room temperature, and PO activity was determined as described in the Materials and Methods. Data from four replicates of each sample were analyzed. (B) Aliquots of naïve plasma were incubated at room temperature alone, or with LPS-K12 or LTA-BS for 30, 60, 90 or 120 min, and PO activity was determined. Each assay was performed in four replicates. The bars represent the mean of four individual measurements ± S.E.M. The significance of difference was determined by an unpaired t-test using the GraphPad InStat software. Asterisks (*) indicate significant difference compared to the naïve plasma alone group. Significance between two groups was indicated by a horizontal line and p value was shown above the line.
It was reported that synthetic lipid A at high concentrations can activate proPO in the silkworm larval plasma (Kaneko et al., 2004). To further determine whether LPS-K12 and LTA-BS can activate proPO in M. sexta plasma, naïve plasma samples were incubated with LPS-K12 and LTA-BS for a longer period of time (a total of 2 h). Compared to the naïve plasma alone, addition of LPS-K12 and LTA-BS increased PO activity in the plasma samples after prolonged incubation (except for LTA-BS at 60 min) (Fig. 1B). But the amplitude of PO activity (1.2–3.0 units) activated by LPS and LTA after prolonged incubation was lower than that (~ 6 units) activated by the Lys-type PG-SA, and was significantly lower than the PO activity (~60 units) activated by the DAP-type PG-K12 and PG-BS (Fig. 1B). These results suggest that M. sexta proPO is activated rapidly and strongly by DAP-type PG-K12 and PG-BS. LPS-K12 and LTA-BS may activate proPO with prolonged activation time, but they are much less potent than PGs in activation of proPO.
3.2 Antibacterial activity in M. sexta larvae is activated by bacterial components
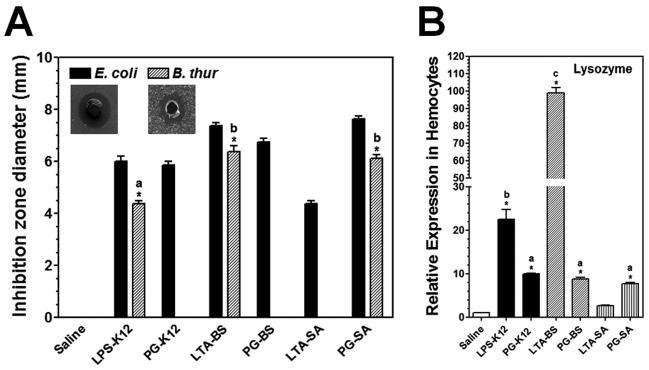
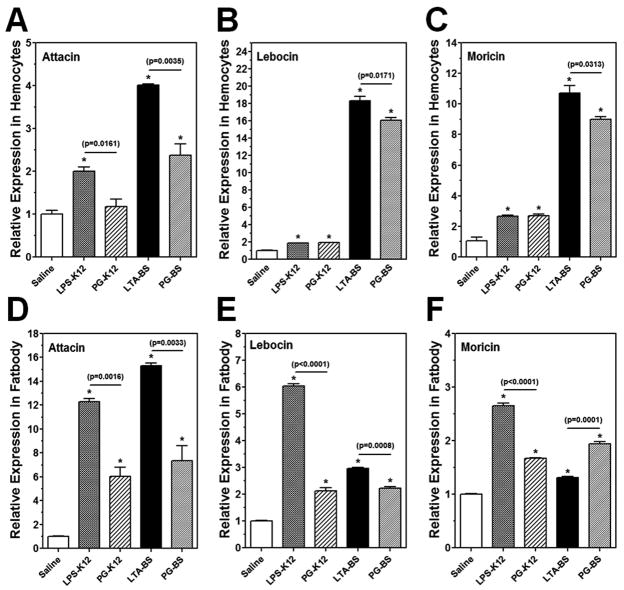
To determine whether LPS, LTA and PG can activate AMP expression in M. sexta larvae, antibacterial activity of plasma from M. sexta naïve larvae injected with bacterial components was measured by a bacterial inhibition zone assay. Plasma from saline-injected larvae did not show antibacterial activity against either Gram-negative E. coli or Gram-positive B. subtilis since no inhibition zones were formed (Fig. 2A). However, plasma samples from larvae injected with LPS-K12, LTA-BS and PG-SA showed high antibacterial activity against both E. coli and B. subtilis, while plasma samples from PG-K12 and PG-BS (DAP-type PGs) as well as LTA-SA injected larvae showed antibacterial activity only to E. coli but not to B. subtilis.
Fig. 2. Induced antibacterial activity in the plasma and induction of lysozyme in M. sexta larvae by bacterial components.
M. sexta 5th instar naïve larvae were injected with LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA, PG-SA or with saline (control), and hemolymph was collected at 24h post-injection. Plasma (cell-free hemolymph) samples were collected for determination of antibacterial activity by an inhibition zone assay (A), while hemocytes were collected for total RNA preparation for real-time PCR analysis of lysozyme expression (B). The bars represent the mean of four (A) or three (B) individual measurements ± S.E.M. The inset in (A) showed typical inhibition zones against E. coli and B. subtilis. Identical letters are not significant difference between groups (p>0.05), while different letters indicate significant difference between groups (p<0.05) determined by ANOVA followed by a Tukey’s multiple comparison test.
Lysozyme has been identified in M. sexta (Mulnix and Dunn, 1994), and it is active against Gram-positive bacteria. To test whether increase in the activity against Gram-positive B. subtilis in the plasma is due to activation of lysozyme in M. sexta larvae by bacterial components, a real-time PCR was performed. Three PGs (PG-SA, PG-BS and PG-K12) activated lysozyme expression in the hemocytes to a similar low level, LPS-K12 increased lysozyme expression to a significantly higher level than the three PGs did (p<0.05), LTA-BS activated lysozyme expression to the highest level among the six bacterial components tested, and LTA-SA almost did not activate lysozyme expression (Fig. 2B). However, the plasma samples of larvae injected with PG-SA and LTA-BS showed similar high activity against B. subtilis, which is signigicantly higher than the activity in the LPS-K12 activated plasma (p<0.05), and no activity against B. subtilis was detected in the plasma samples activated by PG-BS, PG-K12 and LTA-SA (Fig. 2A). These results suggest that increase in lysozyme expression may contribute very little to the activity against B. subtilis in the plasma.
3.3 Bacterial components activate antimicrobial peptide genes in M. sexta larvae
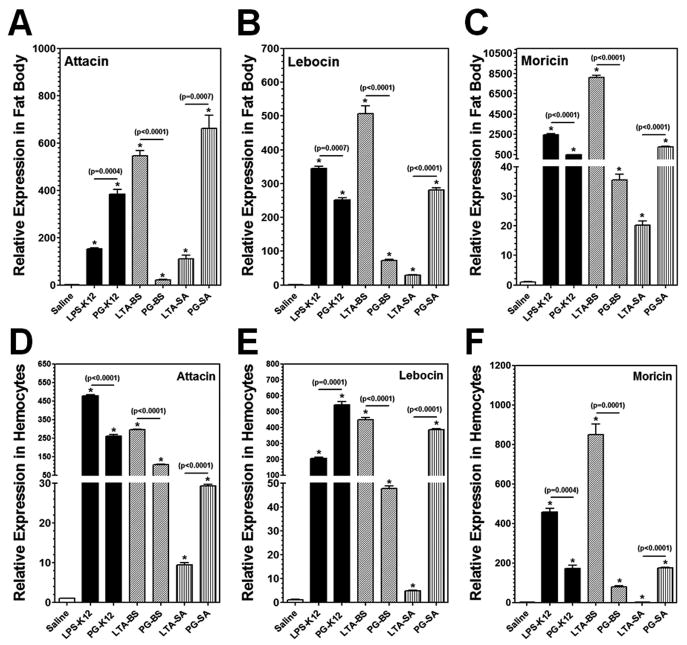
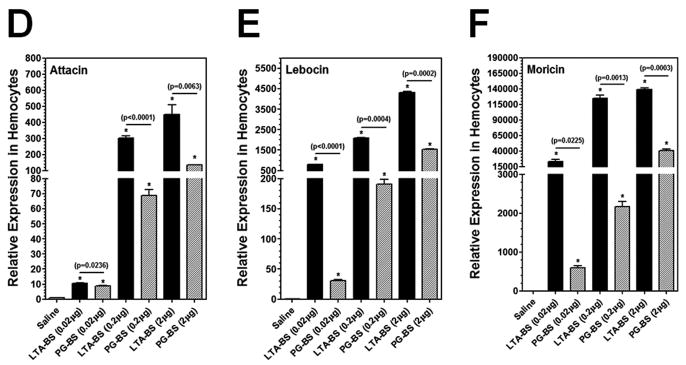
In insects, antimicrobial peptides (AMPs) are small peptides synthesized and secreted mainly by fat body and hemocytes. Expression of AMPs in D. melanogaster is induced primarily by PGs and regulated by the Toll and IMD pathways (Lemaitre and Hoffmann, 2007), but LPS cannot activate immune responses in adult flies (Kaneko et al., 2004; Leulier et al., 2003). Highly purified (TLRgrade™) LPS can activate immune responses in the fat body of B. mori to a lower level than that by crude LPS or PG (Tanaka et al., 2009). So far, it is not known whether LTA can activate AMP expression in insects. Thus, we first tested induction of AMP genes in hemocytes and fat body of M. sexta larvae by LPS, LTA and PG. The real-time PCR results showed that expression of Attacin, Lebocin and Moricin in both hemocytes and fat body of M. sexta larvae was significantly induced by all the bacterial components tested at 24h post-injection, including smooth LPS, LTA and both DAP-type and Lys-type PGs, although the activation levels varied with different bacterial components (Fig. 3). It is noteworthy that for LTA/PG from Gram-positive B. subtilis, LTA-BS activated the AMP genes to a significantly higher level than PG-BS did, but for LTA/PG from Gram-positive S. aureus, the activation levels of AMP genes were significantly lower by LTA-SA than by PG-SA (Fig. 3). For LPS/PG from E. coli strain K-12, LPS-K12 (TLR4grade) activated AMP genes expression to a comparable high level as PG-K12 or to a higher level than PG-K12 did (Fig. 3).
Fig. 3. In vivo activation of AMPs in hemocytes and fat body of M. sexta larvae by LPS, LTA and PG.
M. sexta 5th instar naïve larvae were injected with saline, or with LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA or PG-SA (20 μg per larva), and hemocytes and fat body were collected at 24h post-injection. Expression of Attacin, Lebocin and Moricin mRNAs in the fat body (A–C) and hemocytes (D–F) was analyzed by Real-time PCR. The bars represent the mean of three individual measurements ± S.E.M. Asterisks (*) indicate significant difference compared to the saline-injection group. Significance between two groups was indicated by a horizontal line and p value was shown above the line.
3.4 LPS-K12 and LTA-BS are potent elicitors in activation of antimicrobial peptide genes
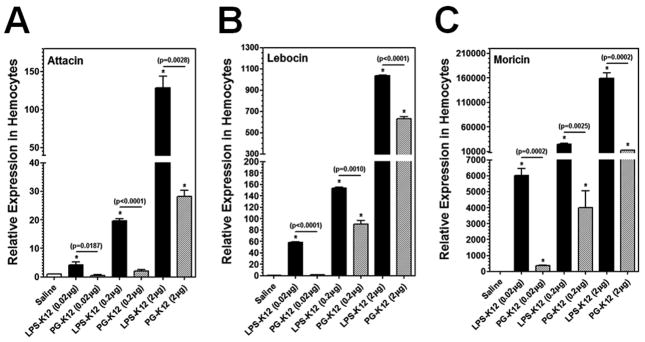
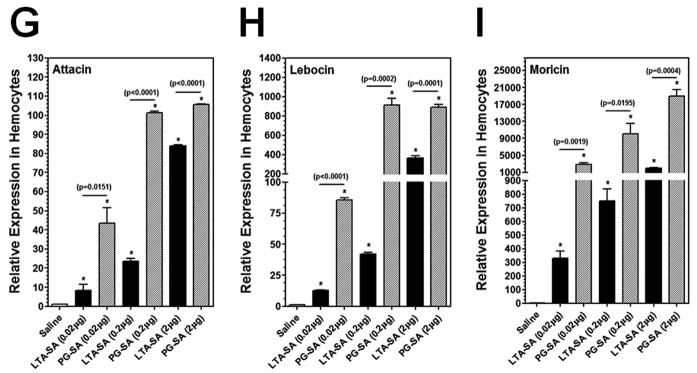
To confirm that LPS-K12 and LTA-BS can indeed activate expression of AMP genes in M. sexta larvae, a dose-dependent activation assay was performed using less but equal amounts of LPS-K12, PG-K12, LTA-BS, PG-BS, LTA-SA and PG-SA, and short activation time (6h post-injection) (Fig. 4). Real-time PCR results showed that expression of Attacin, Lebocin and Moricin genes in the hemocytes at 6h post-injection was significantly higher in the larvae injected with 0.02 μg of LPS-K12, LTA-BS, PG-BS, LTA-SA and PG-SA than in the control larvae (injected with saline) (Fig. 4), but the same amount (0.02 μg) of PG-K12 almost did not activate expression of Attacin and Lebocin (Fig. 4A and B) compared to the control groups. The activation levels of AMP genes were significantly higher in the larvae injected with increasing amounts of LPS, LTA and PG (Fig. 4), indicating a dose-dependent activation of AMP genes by these bacterial components. More importantly, LPS-K12 and LTA-BS activated expressions of AMP genes to significantly higher levels than PG-K12 and PG-BS did, respectively, at all three doses (Fig. 4A–F), but LTA-SA activated AMP expression to a significantly lower level than PG-SA did at all three concentrations (Fig. 4G–I). These results suggest that LPS-K12 and LTA-BS indeed can stimulate AMP gene expression in M. sexta larvae.
Fig. 4. Dose-dependent activation of AMPs in hemocytes of M. sexta larvae by LPS, LTA and PG.
M. sexta 5th instar naïve larvae were injected with saline (control) or different amounts of LPS-K12 and PG-K12 (from E. coli K12) (A–C), LTA-BS and PG-BS (from B. subtilis) (D–F), or LTA-SA and PG-SA (from S. aureus) (G–I), and hemocytes were collected at 6h post-injection. Expression of Attacin, Lebocin and Moricin was analyzed by Real-time PCR as described in Fig. 3.
3.5 Lipid A, core carbohydrate and O-specific antigen moieties of LPS may synergistically activate AMP expression
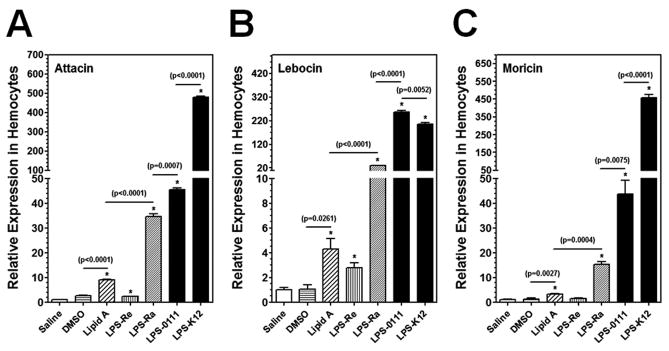
Smooth LPS is composed of the O-polysaccharide chain (O-antigen), the core (outer and inner core) polysaccharide, and lipid A moieties (Raetz and Whitfield, 2002). Each moiety of LPS may induce immune responses differently (Huber et al., 2006). We compared the induction of AMP genes in M. sexta larvae by lipid A, two rough mutants (Ra and Re) of LPS and two smooth LPS from two E. coli strains. LPS-Re is comprised of lipid A and 2-keto-3-deoxyoctonate (KDO), while LPS-Ra lacks only the O-antigen moiety. Injection of lipid A increased expressions of Attacin, Lebocin and Moricin in hemocytes to a low but significantly higher level compared to the DMSO (solvent for Lipid A) control group, and LPS-Re also activated expression of Attacin and Lebocin to a low but significantly higher level than the saline-injection group (Fig. 5). LPS-Ra and two smooth LPS (LPS-0111 and LPS-K12) all activated expression of the three AMP genes in the hemocytes to significantly higher levels than LPS-Re and lipid A did (Fig. 5). The activation level of AMP genes by LPS-K12 and LPS-0111 (smooth LPS) was significantly higher than that by LPS-Ra (Fig. 5). Together, these results suggest that lipid A, the core polysaccharide, and the O-antigen moieties may all induce AMP expression, and the O-antigen moiety may be more important in stimulating immune responses in M. sexta.
Fig. 5. Activation of AMPs in hemocytes of M. sexta larvae by lipid A and different forms of LPS.
M. sexta 5th instar naïve larvae were injected with saline, DMSO, lipid A, LPS-Re, LPS-Ra, LPS-0111 or LPS-K12 (20 μg per larva), and hemocytes were collected at 24h post-injection. Expression of Attacin, Lebocin and Moricin was analyzed by Real-time PCR as described in Fig. 3.
3.6 Bacterial components activate antimicrobial peptide genes in fat body and hemocytes in vitro
Highly purified LPS can activate AMP expression in the fat body of B. mori, but does not activate AMP genes in a cell line derived from the B. mori fat body (Tanaka et al., 2009), suggesting that some plasma factors may play an important role in LPS recognition. To test whether plasma factors/proteins are required for stimulation of AMP expression by bacterial components in M. sexta, hemocytes and fat body were collected from naïve larvae separately and then cultured in vitro in the presence of LPS-K12, PG-K12, LTA-BS and PG-BS. Expression of AMP genes was determined by real-time PCR. We found that all four bacterial components could induce expression of Attacin, Lebocin and Moricin in both hemocytes (Fig. 6A–C) (except PG-K12 for Attacin in Fig. 6A) and fat body (Fig. 6D–F) in vitro to various low levels, but in vitro activation of AMPs (Fig. 6) was several orders lower than in vivo activation (Fig. 3).
Fig. 6. In vitro activation of AMPs in hemocytes and fat body of M. sexta larvae by LPS, LTA and PG.
Hemocytes and fat body from M. sexta 5th instar naïve larvae were collected and cultured in the Grace’s medium in the presence of LPS-K12, PG-K12, LTA-BS, PG-BS or saline (control) at 25°C for 24h. Expression of Attacin, Lebocin and Moricin was analyzed by Real-time PCR as described in Fig. 3.
4. Discussion
Insects have effective innate immune responses, including activation of prophenoloxidase (proPO) and synthesis of AMPs. The proPO activation system is a complicated process that involves a cascade of serine proteinases and other hemolymph proteins (Cerenius et al., 2008; Jiang et al., 2003; Yu et al., 2003). ProPO activation can be triggered by different microbial components after their binding to specific recognition proteins. We compared the ability of LPS, LTA and PG in activation of proPO in M. sexta plasma. Surprisingly, PG-K12 (from E. coli K12) and PG-BS (from B. subtilis), two DAP-type PGs which were less potent than LPS-K12 and LTA-BS, respectively, in activation of AMP expression (Figs. 3 and 4), were more potent elicitors in proPO activation (Fig. 1A). PG-SA (from S. aureus), a Lys-type PG, also activated PO (~6 units), but to a significantly lower level than PG-K12 and PG-BS did (~60 units) (Fig. 1B). LPS-K12 and LTA-BS activated PO to a low level even after prolonged incubation (Fig. 1B). These results suggest that PG is a stronger elicitor in proPO activation compared to LPS and LTA from the respective bacterial strains, and DAP-type PG is more potent than Lys-type PG in proPO activation.
In D. melanogaster, bacteria-activated expression of AMPs is mainly induced by PGs and is regulated by the Toll and IMD pathways (Kaneko et al., 2004; Michel et al., 2001). However, highly purified LPS, a major component of Gram-negative bacteria, cannot activate the Toll or IMD pathway in adult flies (Kaneko et al., 2004; Leulier et al., 2003), although LPS weakly activates AMP production in the fat body of B. mori (Tanaka et al., 2009). In addition, there has been no report about the activation of AMPs in insects by LTA, another major component of Gram-positive bacteria. In this study, we used smooth and rough mutants of LPS (TLR4grade, TLR4grade™ and TLRgrade™), LTA and PG to activate AMP expression in the tobacco hornworm, M. sexta. Our results showed that LPS, LTA and PG all were able to stimulate expression of AMPs in hemocytes and fat body of M. sexta larvae (Figs. 3–5), and activation of AMPs by these bacterial components was dose-dependent (Fig. 4).
LPS and LTA preparations may be contaminated with small amounts of PGs, which are potent elicitors in activation of the Toll and IMD pathways in Drosophila (Kaneko et al., 2004; Leulier et al., 2003). The bacterial components used in this study are high quality preparations. LPS-K12 and LPS-0111 (smooth LPS) are TLR4grade (only activates TLR4), and LPS-Ra and LPS-Re (rough mutants of LPS) are TLRgrade™ and TLR4grade™ that have been tested in mammalian cells. The LPS used in activation of immune responses in the fat body of B. mori [36] was TLRgrade™ from the same company (Alexis Biochemicals). LTA-BS and LTA-SA are also high quality samples with only 12.5 and 125 EU (endotoxin units)/mg of LTA-BS and LTA-SA, respectively. The endotoxin contamination in the three PGs was also very low (endotoxin-free for PG-SA, 0.25 and 125 EU/mg for PG-BS and PG-K12, respectively). Even so, we still cannot rule out the trace amounts of PGs in LPS and LTA samples. In order to confirm that activation of AMPs by LPS and LTA is indeed by the respective bacterial components but not by any contaminated PGs, activation levels of AMPs by LPS and LTA were compared with those by PGs from the respective bacterial strains at three different doses (Fig. 4). We clearly showed that LPS-K12 (from E. coli K-12) and LTA-BS (from B. subtilis) were more potent than PG-K12 and PG-BS, respectively, in activation of AMP expression in hemocytes at three different doses tested (Fig. 4A–F), indicating that LPS-K12 and LTA-BS can activate AMP expressions in M. sexta larvae. To our knowledge, this is the first report about activation of AMPs in insects by LTA-BS. But we also observed that LTA-SA (from S. aureus) activated AMP expression to a significantly lower level than PG-SA did (Fig. 4G–I). Interestingly, LTA-SA is the least potent elicitor among the six bacterial components tested in activation of the AMP genes and lysozyme in M. sexta (Figs. 2B and 3, except for Attacin activation in fat body, Fig. 3A).
It has been demonstrated that LPS cannot activate the IMD or Toll pathway in D. melanogaster (Kaneko et al., 2004; Leulier et al., 2003). Our results clearly showed that LPS-K12 could activate AMP expression in M. sexta larvae (Figs. 3–5), a result consistent with that of B. mori (Tanaka et al., 2009). Since B. mori and M. sexta are lepidopteran insects, while D. melanogaster is a dipteran insect, one possibility is that lepidopteran and dipteran insects may use different PRRs for recognition of bacterial components. For example, in D. melanogaster, there are 13 PGRP genes encoding about 20 functional PGRP proteins (Charroux et al., 2009) that can serve as PRRs to recognize PGs from both Gram-negative and Gram-positive bacteria (Royet, 2004; Royet and Dziarski, 2007). In M. sexta and B. mori, C-type lectins and other plasma proteins that can bind LPS and LTA may function as PRRs for recognition of LPS and LTA (Lee et al., 1996; Ohta et al., 2006; Watanabe et al., 2006; Xu et al., 1995; Yu et al., 2005; Yu and Kanost, 2000; Yu et al., 2006).
Smooth LPS is composed of the O-antigen, outer core, inner core, and lipid A moieties, and rough mutants of LPS (R form LPS) lack the O-antigen moiety and parts of the outer and/or inner cores (Huber et al., 2006). It is not clear whether different moieties of LPS can activate AMP expression. We showed that two smooth LPS (LPS-K12 and LPS-0111), two rough mutants of LPS (LPS-Re and LPS-Ra) and lipid A could all activated AMP expression in hemocytes of M. sexta larvae, however, smooth LPS (LPS-K12 and LPS-0111) stimulated AMP expression to a significantly higher level than rough LPS (LPS-Re and LPS-Ra) and lipid A did (Fig. 5). These results suggest that multiple moieties of LPS may be recognized by different PRRs simultaneously, leading to a synergistic effect in stimulation of AMP expression in insects, a phenomenon that has not been reported in mammals in which lipid A is the major moiety in stimulating immune reactions (Meng et al., 2010; Raetz and Whitfield, 2002). It is also possible that the O-antigen moiety of LPS may interfere with binding of the core moiety and/or lipid A moiety to their receptors if the receptor for the O-antigen is not present at the same time. Thus, not all smooth LPS can activate AMP expression to a higher level than rough mutants of LPS.
Activation of AMPs in in vitro cell culture of M. sexta larval hemocytes and fat body in the presence of LPS, LTA and PG was also observed (Fig. 6). However, in vitro activation level was several orders lower compared to in vivo activation (Figs. 3 and 4). These results suggest that some plasma factors/proteins may be required for recognition of LPS, LTA and PG. These plasma factors/proteins may facilitate binding of bacterial components to receptors or serve as co-receptors in the receptor complexes to activate AMP genes. In the absence of plasma factors/proteins in the in vitro cell culture, very little bacterial components bound to hemocytes and fat body, resulting in low activation level of AMPs. Plasma proteins with such functions may include C-type lectins, PGRPs and beta-glucan recognition/Gram negative bacteria binding proteins, which have the ability to bind LPS, LTA and PG (Ao et al., 2008a; Jomori and Natori, 1991; Lee et al., 2000; Metheniti et al., 2003; Xu et al., 1995; Yu and Kanost, 2002; Yu et al., 2006).
In M. sexta, five AMP genes (Attacin, Cecropin, Moricin, Lebocin and Gloverin) and a lysozyme have been identified (Gorman et al., 2004; Mulnix and Dunn, 1994). The five AMPs have activity mainly against Gram-negative bacteria, while lysozyme is active against Gram-positive bacteria. We measured antibacterial activity in the plasma of larvae injected with bacterial components against Gram-negative E. coli and Gram-positive B. subtilis. High activity against E. coli was detected in the plasma samples of the larvae activated by all six bacterial components tested, but activity against B. subtilis was detected only in the larvae activated by LPS-K12, LTA-BS and PG-SA but not in the larvae injected with PG-K12, PG-BS and LTA-SA (Fig. 2A). Real-time PCR showed that the three PGs (PG-SA, PG-BS and PG-K12) activated lysozyme expression to a similarly low level, LPS-K12 and LTA-BS activated lysozyme expression to significantly higher levels than the three PGs did, and LTA-SA did not activate lysozyme expression (Fig. 2B). However, the activity against B. subtilis was significantly higher in the larvae activated by PG-SA than by LPS-K12, and no activity against B. subtilis was detected in the larvae activated by PG-K12, PG-BS and LTA-SA. These results indicate that activation of lysozyme has trivial effect on the activity against B. subtilis in the plasma. PG-SA is a Lys-type PG that can activate the Toll pathway, while PG-K12 and PG-BS are DAP-type PGs, which can activate the IMD pathway. It is likely that some yet unidentified M. sexta AMPs with activity against Gram-positive bacteria are activated through the Toll pathway, resulting in the activity against B. subtilis. One such candidate AMP gene may be defensin, as defensin is active against Gram-positive bacteria, and it has been identified in B. mori (Kaneko et al., 2008). Thus, activity against E. coli in M. sexta larvae activated by the six bacterial components may result from activation of AMPs via the Toll and/or IMD pathways, but the activity against B. subtilis in the larvae activated by PG-SA, LPS-K12 and LTA-BS may be due to activation of the Toll pathway. A Toll receptor has been identified in hemocytes of M. sexta larvae (Ao et al., 2008b). This M. sexta Toll receptor could be up-regulated to a significantly higher level by PG-SA, LPS-K12 and LTA-BS than by LTA-SA, PG-K12 and PG-BS, respectively (data not shown), a result consistent with that of the antibacterial activity assay (Fig. 2A), further supporting the hypothesis that PG-SA, LPS-K12 and LTA-BS may activate the Toll pathway, while PG-K12, PG-BS and LTA-SA may activate the IMD pathway. The difference in activation of AMP genes and signaling pathways by LTA-BS and LTA-SA may lie in different structures of LTA and teichoic acid in B. subtilis and S. aureus (Greenberg et al., 1996; Morath et al., 2002), as well as different recognition proteins/receptors in insects for LTA-BS and LTA-SA. Future work is to investigate how signaling pathways are activated by LPS and LTA in M. sexta.
Acknowledgments
This work was supported by National Institutes of Health Grant GM066356.
Abbreviations
- AC
anti-coagulant
- AMP
antimicrobial peptide
- IMD
immune deficiency
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- PG
peptidoglycan
- PGRP
peptidoglycan recognition protein
- PO
phenoloxidase
- proPO
prophenoloxidase
- TLR
Toll-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Altincicek B, Knorr E, Vilcinskas A. Beetle immunity: Identification of immune-inducible genes from the model insect Tribolium castaneum. Dev Comp Immunol. 2008;32:585–95. doi: 10.1016/j.dci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Altincicek B, Vilcinskas A. Identification of immune-related genes from an apterygote insect, the firebrat Thermobia domestica. Insect Biochem Mol Biol. 2007;37:726–31. doi: 10.1016/j.ibmb.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Ao JQ, Ling E, Rao XJ, Yu XQ. A novel ML protein from Manduca sexta may function as a key accessory protein for lipopolysaccharide signaling. Mol Immunol. 2008a;45:2772–81. doi: 10.1016/j.molimm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ao JQ, Ling E, Yu XQ. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol Immunol. 2008b;45:543–52. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Armant MA, Fenton MJ. Toll-like receptors: a family of pattern-recognition receptors in mammals. Genome Biol. 2002;3:3011.1–6. doi: 10.1186/gb-2002-3-8-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axen A, Carlsson A, Engstrom A, Bennich H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur J Biochem. 1997;247:614–9. doi: 10.1111/j.1432-1033.1997.00614.x. [DOI] [PubMed] [Google Scholar]
- 8.Cerenius L, Lee BL, Soderhall K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–71. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009;11:631–6. doi: 10.1016/j.micinf.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, et al. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87:356–65. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci USA. 2005;102:1122–6. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YS, Choo YM, Lee KS, Yoon HJ, Kim I, Je YH, et al. Cloning and expression profiling of four antibacterial peptide genes from the bumblebee Bombus ignitus. Comp Biochem Physiol B: Biochem Mol Biol. 2008;150:141–6. doi: 10.1016/j.cbpb.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J Invertebr Pathol. 1983;41:77–85. [Google Scholar]
- 14.Gorman MJ, Kankanala P, Kanost MR. Bacterial challenge stimulates innate immune responses in extra-embryonic tissues of tobacco hornworm eggs. Insect Mol Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg JW, Fischer W, Joiner KA. Influence of lipoteichoic acid structure on recognition by the macrophage scavenger receptor. Infect Immun. 1996;64:3318–25. doi: 10.1128/iai.64.8.3318-3325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha Lee J, Hee Lee I, Noda H, Mita K, Taniai K. Verification of elicitor efficacy of lipopolysaccharides and peptidoglycans on antibacterial peptide gene expression in Bombyx mori. Insect Biochem Mol Biol. 2007;37:1338–47. doi: 10.1016/j.ibmb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Haine ER, Pollitt LC, Moret Y, Siva-Jothy MT, Rolff J. Temporal patterns in immune responses to a range of microbial insults (Tenebrio molitor) J Insect Physiol. 2008;54:1090–7. doi: 10.1016/j.jinsphys.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, et al. Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J Immunol. 2009;183:7451–60. doi: 10.4049/jimmunol.0901032. [DOI] [PubMed] [Google Scholar]
- 20.Hollmig ST, Ariizumi K, Cruz PD., Jr Recognition of non-self-polysaccharides by C-type lectin receptors dectin-1 and dectin-2. Glycobiology. 2009;19:568–75. doi: 10.1093/glycob/cwp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, Cheng T, Xu P, Cheng D, Fang T, Xia Q. A genome-wide survey for host response of silkworm, Bombyx mori during pathogen Bacillus bombyseptieus infection. PLoS ONE. 2009;4:e8098. doi: 10.1371/journal.pone.0008098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber M, Kalis C, Keck S, Jiang Z, Georgel P, Du X, et al. R-form LPS, the master key to the activation of TLR4/MD-2-positive cells. Eur J Immunol. 2006;36:701–11. doi: 10.1002/eji.200535593. [DOI] [PubMed] [Google Scholar]
- 23.Hultmark D. Quantification of Antimicrobial Activity, Using the Inhibition-Zone Assay. In: Wiesner A, Dunphy GB, et al., editors. Techniques in Insect Immunology. SOS Publications; New Jersey: 1998. pp. 103–7. [Google Scholar]
- 24.Imler JL, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy. 2005;86:1–21. doi: 10.1159/000086648. [DOI] [PubMed] [Google Scholar]
- 25.Imler JL, Tauszig S, Jouanguy E, Forestier C, Hoffmann JA. LPS-induced immune response in Drosophila. J Endotoxin Res. 2000;6:459–62. [PubMed] [Google Scholar]
- 26.Iwanaga S, Lee BL. Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol. 2005;38:128–50. doi: 10.5483/bmbrep.2005.38.2.128. [DOI] [PubMed] [Google Scholar]
- 27.Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H, Wang Y, Kanost MR. Proteolytic activation of prophenoloxidase in an insect Manduca sexta. Adv Exp Med Biol. 2001;484:313–7. doi: 10.1007/978-1-4615-1291-2_31. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–60. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 30.Jin LH, Choi JK, Cho HS, Shim J, Kim YJ. Microarray analysis of the gene expression profiles of SL2 cells stimulated by LPS/PGN and curdlan. Mol Cells. 2008;25:553–8. [PubMed] [Google Scholar]
- 31.Jomori T, Natori S. Molecular cloning of cDNA for lipopolysaccharide-binding protein from the hemolymph of the American cockroach, Periplaneta americana. Similarity of the protein with animal lectins and its acute phase expression. J Biol Chem. 1991;266:13318–23. [PubMed] [Google Scholar]
- 32.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–49. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko Y, Tanaka H, Ishibashi J, Iwasaki T, Yamakawa M. Gene expression of a novel defensin antimicrobial peptide in the silkworm, Bombyx mori. Biosci Biotechnol Biochem. 2008;72:2353–61. doi: 10.1271/bbb.80263. [DOI] [PubMed] [Google Scholar]
- 34.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 35.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–37. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimbrell DA, Beutler B. The evolution and genetics of innate immunity. Nat Rev Genet. 2001;2:256–67. doi: 10.1038/35066006. [DOI] [PubMed] [Google Scholar]
- 37.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- 38.Kurata S, Ariki S, Kawabata S. Recognition of pathogens and activation of immune responses in Drosophila and horseshoe crab innate immunity. Immunobiology. 2006;211:237–49. doi: 10.1016/j.imbio.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 40.Lee SY, Wang R, Söderhäll K. A Lipopolysaccharide- and β-1,3-Glucan-binding Protein from Hemocytes of the Freshwater Crayfish Pacifastacus leniusculus. J Biol Chem. 2000;275:1337–43. doi: 10.1074/jbc.275.2.1337. [DOI] [PubMed] [Google Scholar]
- 41.Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 1996;93:7888–93. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemaitre B, Hoffmann J. The Host Defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 43.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–84. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 44.Marmaras VJ, Lampropoulou M. Regulators and signalling in insect haemocyte immunity. Cell Signal. 2009;21:186–95. doi: 10.1016/j.cellsig.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Meng J, Lien E, Golenbock DT. MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J Biol Chem. 2010;285:8695–702. doi: 10.1074/jbc.M109.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metheniti A, Giannakas N, Katsoulas HL, Soldatos AN, Tsakas S, Lambropoulou M. Evidence for a LPS-binding protein in medfly hemocyte surface: Mediation in LPS internalization but not in LPS signaling. Arch Insect Biochem Physiol. 2003;54:25–36. doi: 10.1002/arch.10096. [DOI] [PubMed] [Google Scholar]
- 47.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–9. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 48.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–73. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morath S, Geyer A, Spreitzer I, Hermann C, Hartung T. Structural decomposition and heterogeneity of commercial lipoteichoic Acid preparations. Infect Immun. 2002;70:938–44. doi: 10.1128/iai.70.2.938-944.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulnix AB, Dunn PE. Structure and induction of a lysozyme gene from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 1994;24:271–81. doi: 10.1016/0965-1748(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 51.Ohta M, Watanabe A, Mikami T, Nakajima Y, Kitami M, Tabunoki H, et al. Mechanism by which Bombyx mori hemocytes recognize microorganisms: direct and indirect recognition systems for PAMPs. Dev Comp Immunol. 2006;30:867–77. doi: 10.1016/j.dci.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Royet J. Drosophila melanogaster innate immunity: an emerging role for peptidoglycan recognition proteins in bacteria detection. Cell Mol Life Sci. 2004;61:537–46. doi: 10.1007/s00018-003-3243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–77. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 55.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–77. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. 2002;99:13705–10. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka H, Sagisaka A, Fujita K, Kaneko Y, Imanishi S, Yamakawa M. Lipopolysaccharide elicits expression of immune-related genes in the silkworm, Bombyx mori. Insect Mol Biol. 2009;18:71–5. doi: 10.1111/j.1365-2583.2009.00851.x. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe A, Miyazawa S, Kitami M, Tabunoki H, Ueda K, Sato R. Characterization of a novel C-type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: mechanism of wide-range microorganism recognition and role in immunity. J Immunol. 2006;177:4594–604. doi: 10.4049/jimmunol.177.7.4594. [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Nishijima M, Kono Y, Taniai K, Kato Y, Kadono-Okuda K, et al. Identification of a hemocyte membrane protein of the silkworm, Bombyx mori, which specifically binds to bacterial lipopolysaccharide Insect Biochem. Mol Biol. 1995;25:921–8. [Google Scholar]
- 60.Yamakawa M, Tanaka H. Immune proteins and their gene expression in the silkworm, Bombyx mori. Dev Comp Immunol. 1999;23:281–9. doi: 10.1016/s0145-305x(99)00011-7. [DOI] [PubMed] [Google Scholar]
- 61.Yu XQ, Tracy ME, Ling E, Scholz FR, Trenczek T. A novel C-type immulectin-3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem Mol Biol. 2005;35:285–95. doi: 10.1016/j.ibmb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 62.Yu XQ, Gan H, Kanost MR. Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem Mol Biol. 1999;29:585–97. doi: 10.1016/s0965-1748(99)00036-3. [DOI] [PubMed] [Google Scholar]
- 63.Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- 64.Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J Biol Chem. 2000;275:37373–81. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- 65.Yu XQ, Kanost MR. Binding of hemolin to bacterial lipopolysaccharide and lipoteichoic acid. An immunoglobulin superfamily member from insects as a pattern-recognition receptor. Eur J Biochem. 2002;269:1827–34. doi: 10.1046/j.1432-1033.2002.02830.x. [DOI] [PubMed] [Google Scholar]
- 66.Yu XQ, Ling E, Tracy ME, Zhu Y. Immulectin-4 from the tobacco hornworm Manduca sexta binds to lipopolysaccharide and lipoteichoic acid. Insect Mol Biol. 2006;15:119–28. doi: 10.1111/j.1365-2583.2006.00618.x. [DOI] [PubMed] [Google Scholar]