Abstract
Earlier, we have shown that doxorubicin-loaded liposomes (Doxil) modified with a chimeric phage fusion coat protein specific towards MCF-7 breast cancer cells identified from a phage landscape library demonstrated a significantly enhanced association with target cells and an increased cytotoxicity. Based on some structural similarities between the N-terminus of the phage potein and known fusogenic peptides, we hypothesized that, in addition to the specific targeting, the phage protein may possess endosome-escaping potential and an increased cytotoxicity of drug-loaded phage protein-targeted liposomes may be explained by an advantageous combination of both, cell targeting and endosomal escape of drug-loaded nanocarrier. The use of the fluorescence resonance energy transfer (FRET) technique allowed us to clearly demonstrate the pH-dependent membrane fusion activity of the phage protein. Endosomal escape and cytosolic delivery of phage-liposomes was visualized with fluorescence microscopy. Endosome acidification inhibition by bafilomycin A 1 resulted in decreased cytotoxicity of the phage-Doxil, while the endosome disruption by chloroquine had a negligible effect on efficacy of phage-Doxil, confirming its endosomal escape. Our results demonstrated an endosome-escaping property of the phage protein and provided an insight on mechanism of the enhanced cytotoxicity of phage-Doxil.
Keywords: Drug delivery, Liposome, Cytoplasmic delivery, Endosomal escape, Membrane fusion, Phage display, Landscape phage, Doxil®, Breast cancer
Introduction
Cytoplasmic delivery of drugs and drug-loaded nanocarriers is highly desirable but difficult to achieve. The poor in vivo performance of conventional therapeutics and novel biomedicines, such as proteins, DNA, antisense oligonucletides (ODN), and siRNA, is often associated with the lack of efficient drug carriers to transport drugs to their sites of action in the cytosol or nuclei. An ideal delivery system should circumvent both extracellular and intracellular barriers. Liposome formulations have proved to be versatile pharmaceutical carriers with increasing number of clinical applications1. While surface modification with biocompatible polymers (such as PEG) allows for stability and longevity of liposomes in biological fluids2–4, the attachment of ligands specific for cell surface receptors enables liposomes to access target cells and induces internalization via receptor-mediated endocytosis1, 5, 6. However, upon endocytic uptake, the majority of liposomes and their contents still are transported to lysosomes for subsequent degradation 7, 8. Thus, the promotion of rapid endosomal release is a critical strategy for improved efficiency of liposomal drugs8.
Viruses have naturally evolved an effective strategy to escape from the endosome to the cytosol by exploiting the biological process of endosomal acidification. Influenza virus, for example, bears a fusogenic peptide with a short chain of N-terminal amphiphilic anionic peptide residues (termed as HA2). At neutral pH, the HA2 subunit exists in a non-helical conformation due to charge repulsions from the ionized glutamic acid residues at positions 11 and 15 and the ionized aspartic acid residue at position 19 within HA2. Upon virus entry into the endosomal compartment, however, the HA2 subunit undergoes a structural transition into a stable helical secondary structure as a consequence of the protonations of glutamic and aspartic acids triggered by the endosomal acidic interior. Helix conformation is believed to promote membrane interactions between internalized virus and the endosome of host cells, resulting in cytosolic release of the viral genome9.
Inspired by the mechanism of virus invasion, synthetic peptides mimicking virus fusogenic peptides have been designed. Their capacity for pH-dependent release of payload from liposomes has been explored to facilitate their endosome escape10. Synthetic peptides, such as diINF7, INF6, are hemagglutinin derivatives with an N-terminal domain similar to that of the influenza virus HA2 subunit and have pH-sensitive fusogenic properties. Co-loading diINF7 and the diphtheria toxin A chain (DAT) into liposomes provoked an enhanced cytotoxicity of DAT towards ovarian carcinoma cells as a result of increased cytoplasmic delivery of DAT under the action of pH-driven membrane fusion of diINF711. Additionally, 1000-fold increased transfection efficiency has been observed upon INF6 modification of transfectam/DNA complexes12. Synthetic amphipathic peptide GALA, composed of 30 amino acid residues with a repeated glutamic-ananine-leucine-aninine unit, has shown pH-responsive membrane destabilizing activity and demonstrated cytoplasmic delivery 13–15.
Recently, we have used the phage-display technique to identify a chimeric phage fusion protein specific towards MCF-7 breast cancer cells from a phage landscape library as a targeting moiety for drug-loaded pharmaceutical nanocarriers16. We have found that doxorubicin-loaded liposomes (Doxil®) modified with the hybrid phage fusion coat protein (phage-Doxil) demonstrated a significantly enhanced association with target cells and a remarkably increased cytotoxicity17. However, the mechanisms of phage protein-mediated intracellular delivery of phage-Doxil remain to be elucidated.
We have noticed, however, that the phage fusion coat protein shares some common structural features with the fusogenic components of virus and synthetic amphipathic peptides. The phage fusion protein is composed of 55 amino acid residues: ADMPGTVLPDPAKAAFDSLQASATEYIGYAWAMVVVIVGATIGIKLFKKFTSKAS. The amino-acid sequence from the residues 1 to 26 is the hydrophilic N-terminus, which displays the cancer cell-binding properties. The residues from 27 to 40 represent a highly hydrophobic “membranophilic” segment, which allows the phage coat protein to accommodate readily into the liposomal membrane. At the same time, the phage fusion coat protein, similar to HA2 and GALA, contains acid residues such as aspartic acid and glutamic acid in its N-terminal (position 2, 5, 10, and 17). Therefore, one may speculate that, within the acidic environment of the endosome, the protonation of carboxylic groups of the aspartic acid and glutamic acid residues will allow the phage protein to acquire more hydrophobicity, which could induce the interaction between phage-liposomes and the endosomal membrane, with carboxylic groups in acidic surrounding absorbing protons like a “proton sponge” and resulting in swelling and rupture of the endosomal membrane. Thus, we expect that the phage protein attached to the surface of a drug-loaded pharmaceutical nanocarrier not only mediates the specific recognition and targeting, but also facilitates the cytoplasmic delivery of the nanocarrier and loaded drug via endosomal membrane destabilization. Presumably, the dual function of the phage protein contributes to enhanced cytotoxicity induced by phage liposomes.
In this study, we examined the pH-dependent membrane fusion activity of the phage fusion protein using the fluorescence resonance energy transfer (FRET) technique and the cytosolic delivery of phage-liposomes using fluorescence microscopy. The cytosolic delivery of doxorubicin-loaded phage-liposomes was confirmed by the results of endosome acidification inhibition and use of an endosome disrupting agent.
Experimental Section
Materials and Reagents
Doxil® was purchased from Ben Venue Laboratories Inc. (Bedford, OH). L-α-phosphatidylcholine (egg; EPC); 1,2-dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt; DPPG); 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt; DOTAP); 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)2000] (ammonium salt; PEG2000-PE ); cholesterol (98%); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-DOPE); and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt; Rho-PE) were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). NH4Cl; Bafilomycin A1 and chloroquine diphosphate salt were from Sigma (St Louis, MO); trihydrochloride, trihydrate (Hoechst 33342); 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS; pyranine); p-xylene-bis-pyridinium bromide (DPX), and Alexa Fluor 488 transferrin were from Invitrogen Inc (Eugene, OR); Sodium cholate and BCA protein assay kits were from Pierce (Rockford, IL); Cell Titer Blue assay kit from Promega (Madison, WI); Fluor Mounting Medium was from Trevigen Inc (Gaithersburg, MD). MCF-7 human breast adenocarcinoma (HTB 22™) cells were obtained from the ATCC (Manassas, VA). All cells were grown as recommended by the ATCC at 37°C, 5% CO2.
Preparation of Liposomes
Phage-liposomes were prepared using a post-insert protocol17 and liposomes with the compositions as shown in Table 1. Briefly, plain liposomes were prepared by the hydration of the lipid film followed by a 30 min bath sonication and extrusion through 200 nm polycarbonate membrane. To prepare phage-liposomes, plain liposomes were incubated with phage fusion coat protein at the indicated phage protein-to-lipid weight ratios and with 15 mM final concentration of sodium cholate. After the overnight incubation at 37°C, the crude formulation was dialyzed at 4°C against PBS to remove sodium cholate. To prepare double-labeled liposomes, Rho-PE and NBD-DOPE were added to the liposome composition. The liposomes were dialyzed against PBS to remove non-incorporated markers. Phage-Doxil was prepared by incubating Doxil with the cholate-stabilized phage fusion coat protein at 0.5% protein-to-lipid weight ratio and subsequent dialysis as describe above. Liposomes encapsulating 35 mM HPTS and 50 mM DPX were prepared using a freezing-thawing protocol 18.
Table 1.
Liposomes used in the study
| The use of liposomes | Designation | Lipid composition and % molar ratio | Phage proteins % (w/w) |
|---|---|---|---|
| a Artificial Membrane Fusion | Double-labeled liposomes | EPC: DPPG: CHOL: Rho-PE: NBD-DOPE | 0.0% |
| 47 : 46. 5 : 5 : 0.5 : 0.1. | |||
| Phage -liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE | 1%, 0.5% or 0.05% | |
| 45 : 30 : 20 : 2 : 3 | |||
| Plain liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE | 0.0% | |
| 45 : 30 : 20 : 2 : 3 | |||
| a Intracellular Membrane Fusion | Phage-liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE: Rho-PE: NBD-DOPE | 0.5% |
| 43.5 : 30 : 20 : 2 : 3 : 0.5 : 0.1 | |||
| Plain liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE: Rho-PE: NBD-DOPE | 0.0% | |
| 43.5 : 30 : 20 : 2 : 3 : 0.5 : 0.1 | |||
| a Endocytic Uptake | Phage-liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE: Rho-PE | 0.5% |
| a Endosome Release | 44 : 30 : 20 : 2 : 3 : 1 | ||
| b Cytoplasmic Delivery of HPTS | Plain liposomes | EPC: CHOL: DPPG: DOTAP: PEG-PE: Rho-PE | 0.0% |
| 44 : 30 : 20 : 2 : 3 : 1 | |||
| c Cytotoxicity | Phage-Doxil | HSPC: CHOL: PEG-PE | 0.5% |
| 56.2 : 38.3 : 5.3 | |||
| Doxil | HSPC: CHOL: PEG-PE | 0.0% | |
| 56.2 : 38.3 : 5.3 | |||
Liposomes with PBS, pH 7.4;
Liposomes with 35mM of HPTS and 50mM of DPX;
Liposomes with 2mg/ml of doxorubicin..
Artificial Membrane Fusion
The fusion between the membranes of plain- or phage-liposomes and double-labeled liposomes with the FRET donor NBD-PE and acceptor Rho-PE was followed by monitoring the fluorescence resonance energy transfer between FRET pairs. Briefly, double-labeled liposomes with 0.1mol% NBD-PE and 0.5mol% Rho-PE were added to plain liposomes or phage-liposomes with a varying concentration of phage fusion coat protein in either citrate-PBS buffer (pH 5.3) or PBS buffer (pH 7.4) at a 1:4 molar ratio and a total lipid concentration of 100 µM. Samples were incubated at 37°C, and fluorescence intensities of the samples were measured. An emission spectrum between 500–650 nm was obtained at predetermined time points using the excitation wavelength of 470 nm. Changes in the fluorescence intensity ratio (R-value) of NBD-PE (530 nm) to Rho-PE (585 nm) served as an indicator of the occurrence of the fusion. The net membrane fusion activity mediated by phage protein was indicated by the normalized R value, which is the difference in R values between phage-liposomes and plain liposomes.
Intracellular Membrane Fusion
MCF-7 cells were grown in a 6-well microplate to 70–80% confluence, and then treated with 1µM of double-labeled phage-liposomes or plain liposomes at 4°C for 1h, then incubated at 37°C in the presence or absence of NH4Cl (20 mM) for 1h. Cells were scraped and suspended in 1 ml of PBS buffer, pH 7.4. Fluorescence intensities of the samples were monitored at excitation wavelength of 470 nm, emission wavelength of 530 nm and 585 nm. Changes in the fluorescence intensity ratio (R-value) of NBD-PE (530 nm) to Rho-PE (585 nm) served as an indicator of the fusion. The R-value was normalized to protein concentration of the sample detected by the BCA protein assay.
Effect of Bafilomycin A1 and Chloroquine on Cytotoxicity of Doxil and phage-Doxil
MCF-7 cells were seeded into 96 well microplates at a density of 4×104 cells/well and grown until cells reached 40–50% confluence. For Bafilomycin A1 (BFA) inhibition studies, MCF-7 cells were treated with 0.1 µM BFA for 30 min and then incubated with 51.7 µM of Doxil or phage-Doxil in a serum-free MEM containing 0.1 µM of BFA for 24 h. For chloroquine inhibition, MCF-7 cells were treated with 51.7 µM of Doxil or phage-Doxil for 24 h in a serum-free MEM containing 50 µM of chloroquine. As a control, MCF-7 cells were treated with 51.7 µM of Doxil or phage-Doxil, 0.1 µM of BFA or 50 µM chloroquine in a serum-free MEM for 24 h. After cells were washed 3 times with PBS, pH 7.4, cell viability was evaluated by the Cell Titer Blue assay as described in the manufacturer’s manual. Briefly, cells were incubated with the fresh complete MEM medium (100µl/well) along with the Cell Titer Blue assay reagent (20µl/well) at 37°C for 2h. The fluorescence intensity was measured using the multi-detection microplate reader (Bio-Tek, Winooski, VT) with 525/590 nm excitation/emission wavelengths. The percent of cell viability was calculated by dividing the treated sample value by the value for the untreated cell sample. The percent of cell viability upon the treatment with phage-Doxil with BFA or chloroquine inhibition was normalized to that with BFA or chloroquine treatment alone.
Fluorescence Microscopy
MCF-7 cells were seeded on sterile coverslips in 6-well plates at the density of 2.5 × 105 cells/well and grown to 70–80% confluence. To study the uptake mechanism of phage liposomes, cells were co-incubated with 1 µM of rhodamine-labeled phage-liposomes and 20 µg/ml of Alexa Fluor 488 transferrin in serum-free MEM medium for 30 min at 37°C. After triple washing with PBS at 4°C, the cells were fixed with 2.5% paraformaldehyde. To study the endosomal release of rhodamine-labeled phage- or plain liposomes, cells were incubated with 1 µM of rhodamine-labeled phage-liposomes or plain liposomes in a serum-free MEM for 30 min at 37°C, and washed once with PBS, pH 7.4. The cells were then incubated for an additional 2 h in a serum-free MEM at 37°C. Cell nuclei were counterstained with 5 µg/ml of Hoechst 33342 for 10 min. For determination of the cytosolic delivery of liposome-encapsulated HPTX-DPX, cells were incubated with 1 µM of HPTS-DPX encapsulating phage-liposomes or plain liposomes in a serum-free MEM for 1 h at 37°C, and washed with PBS, pH 7.4. Cells were incubated for additional 16 h in a serum-free MEM at 37°C. After triple washing with PBS, a coverslip was mounted on the glass slide over mounting medium and visualized with a fluorescence microscopy (Zeiss Co. Ltd. Germany) at 100 × magnification with FITC, TxR and DAPI filters. All images were taken with monochromatic CCD cameras, and the data was collected using Openlab software and exported as tagged image files (TIF).
Statistical Analysis
The statistical significance of the results was analyzed using the SPSS (version 16). Differences between experiment groups were compared using ANOVA followed by a Bonferroni post hoc test. The results were considered statistically significant if the p value was less than 0.05.
Results
Endocytic Uptake of Phage-Liposomes
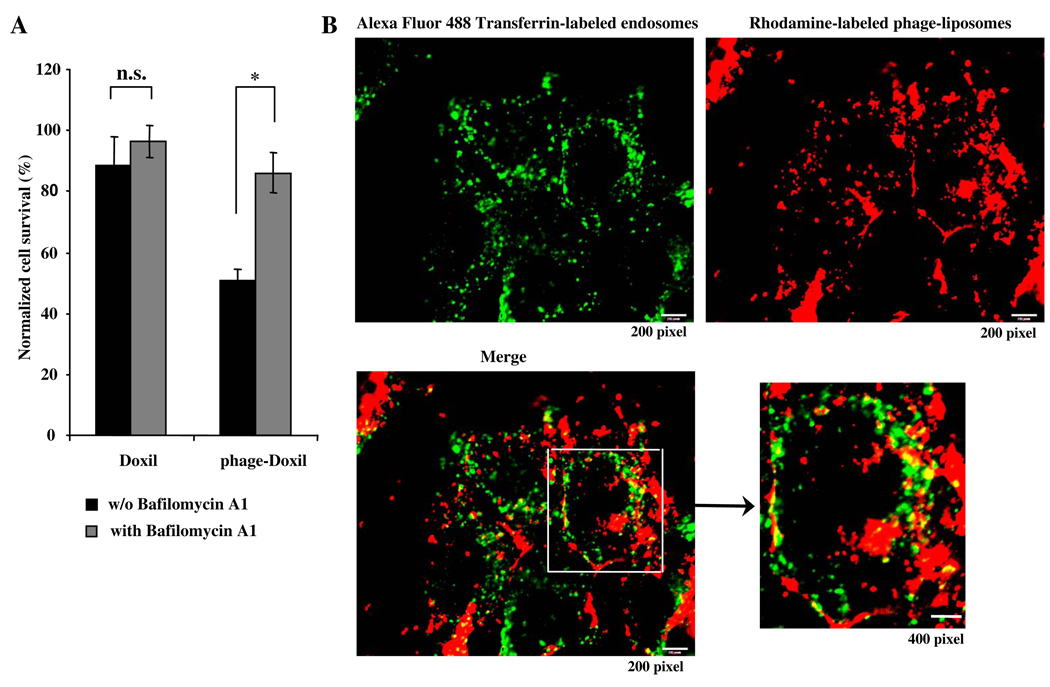
Using the endosomal acidification inhibitor, bafilomycin A1 (BFA), we examined the possible involvement of the endocytic pathway in phage-Doxil-induced cell death. The presence of BFA significantly reduced tumor cell killing by phage-Doxil, suggesting that endosomal acidification is required for phage-Doxil-mediated cell death, and that endocytosis is a pathway for phage-Doxil internalization (Fig 1A). We also found that rhodamine-labeled phage-liposomes were entering transferrin-labeled early endosomes, further confirming the endocytic traffic of phage liposomes (Fig 1B). BFA inhibition of the endosomal acidification lowered the phage-Doxil-induced cell death to a level comparable to that induced by the non-modified Doxil, which showed no significant change with or without BFA inhibition. Taken together, these results imply that endosome entrapment of Doxil may be one of reasons for the lower cell death by Doxil, and suggest that the phage protein may play a role in endosome escape of phage-Doxil, which, in turn, provokes its increased cytotoxicity (Fig 1A).
Figure 1. Endocytic uptake of phage-liposomes.
A) shows the effect of endosome acidification on phage-Doxil-mediated cytotoxicity as revealed by Bafilomycin A1 inhibition.(*p<0.05; n=5, mean ± SEM). B) Fluorescence microscopy shows that rhodamine-labeled phage liposomes (red) co-localized with transferrin-labeled early endosomes (green).
Artificial Membrane Fusion
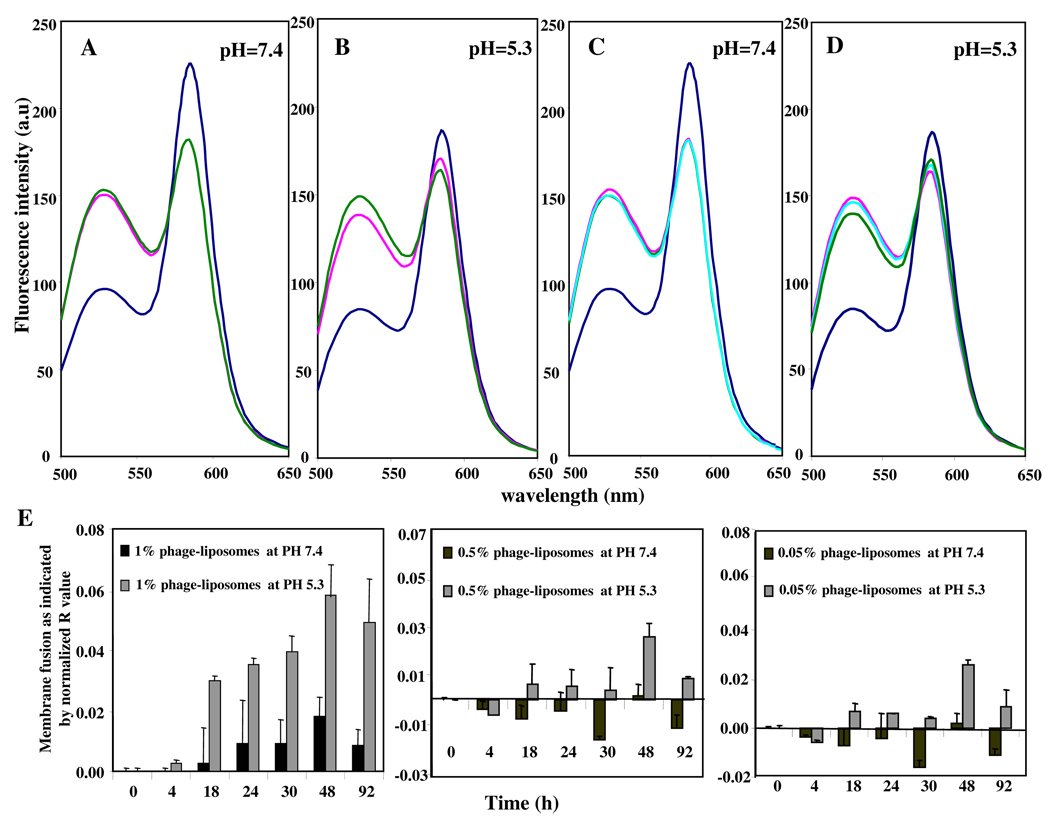
Endosomal escape is usually associated with destabilization of the endosomal membrane under the action of certain fusogenic components incorporated into drug carriers. We have used a FRET lipid mixing assay to examine the potential fusogenic property of the phage protein. When control liposomes double-labeled with FRET pairs (such as NBD-DOPE and Rho-PE) were exitated at 470 nm, the FRET acceptor Rho-PE has quenched the donor NBD-DOPE fluorescence, resulting in FRET. After the addition of either plain liposomes or phage-liposomes, FRET was decreased, indicating membrane fusion between double-labeled liposomes and plain- or phage-liposomes. At neutral pH, there was no difference in membrane fusion induced by plain- and phage-liposomes (Fig 2A). At acidic pH value (5.3), however, the extent of membrane fusion induced by phage-liposomes was significantly larger than that by plain liposomes (Fig 2B), suggesting the phage protein facilitated/provoked the pH-sensitive membrane fusion.
Figure 2. Membrane fusion activity of phage protein detected by FRET.
(A, B) pH-dependent membrane fusion by phage protein.( – double-labeled liposomes only; – double-labeled liposomes + plain liposomes; – double-labeled liposomes + 1% phage-liposomes); (C, D) Effect of dose of phage protein on membrane fusion at acidic pH.(– double-labeled liposomes only; – double labeled liposomes + 1% phage-liposomes; – double-labeled liposomes + 0.5% phage-liposomes; – double-labeled liposomes + 0.05% phage-liposomes). (E) Time course of the pH- and dose-dependent membrane fusion mediated by the phage protein. (n=3; mean ± SD).
Furthermore, the membrane fusion activities of phage-liposomes was increasing with the increase in the concentration of phage fusion coat protein in phage-liposomes, but only in an acidic and not in a neutral medium (compare Figs. 2C and 2D).
After 92 h monitoring of the membrane fusion induced by phage-liposomes with varying concentrations of phage protein (1%, 0.5% and 0.05%), the net membrane fusion activity mediated by phage protein [as indicated by the normalized R value] was significantly higher in acidic pH than in neutral pH, confirming the pH-dependent and dose-dependent membrane fusion properties of the phage fusion coat protein (Fig 2E).
Intracellular Membrane Fusion and Endosomal Escape Potential
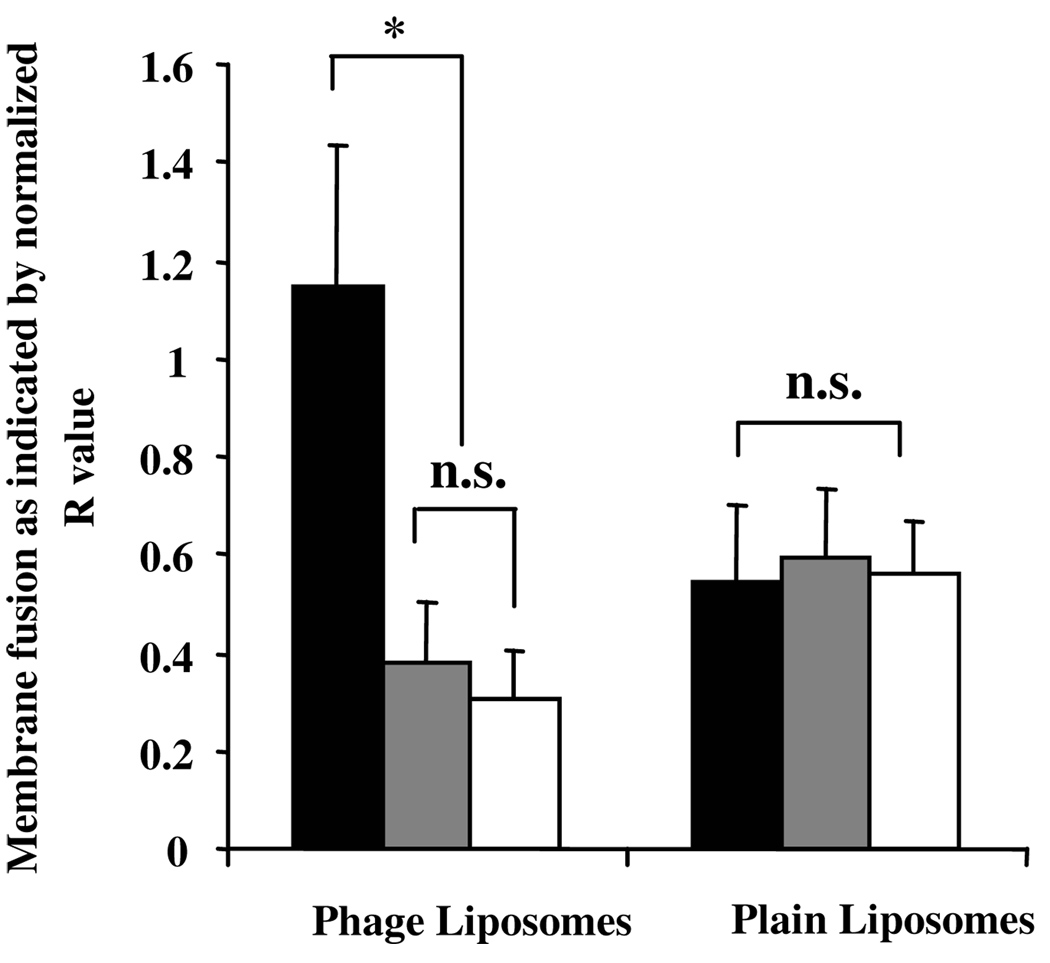
To further assess the ability of the phage protein to destabilize cellular membranes, we treated MCF-7 cells with phage- or control plain-liposomes double-labeled with FRET pairs (e.g. NBD-DOPE and Rho-PE). The 1h treatment at 4°C showed destabilization of the plasma membrane, whereas the 1h treatment at 4°C, followed by the 1h treatment at 37°C, resulted in total membrane fusions (both, plasma and intracellular membrane fusion). Figure 3 showed that while no significant difference in plasma membrane fusion induced by either phage-liposomes or plain liposomes occurred, phage-liposomes induced much more pronounced intracellular membrane fusion than plain liposomes did. Furthermore, the inhibition of the endosomal acidification with NH4Cl blocked the intracellular membrane fusion induced by phage-liposomes, indicating the pH-dependency of intracellular membrane destabilization with phage-protein. Contrary to phage-liposomes, plain liposomes did not appear to induce intracellular membrane fusion. NH4Cl had no effect on their membrane fusion, confirming the specific role of phage protein in the endosomal membrane destabilization and its endosomal escape potential.
Figure 3. Intracellular membrane fusion and endosomal escape potential of phage protein.
The inhibition of the endosomal acidification by NH4Cl blocks phage protein–induced intracellular membrane fusion detected by FRET. Black bar: 1 h binding at 4 °C followed by 1 h internalization at 37 °C; Grey bar: 1 h binding at 4 °C followed by 1 h internalization at 37 °C with NH4Cl ; White bar: 1 h binding at 4 °C . (*p<0.05; n=3, mean ± SD).
Endosomal Escape of Phage-Liposomes by Mediation of the Endosome Acidification
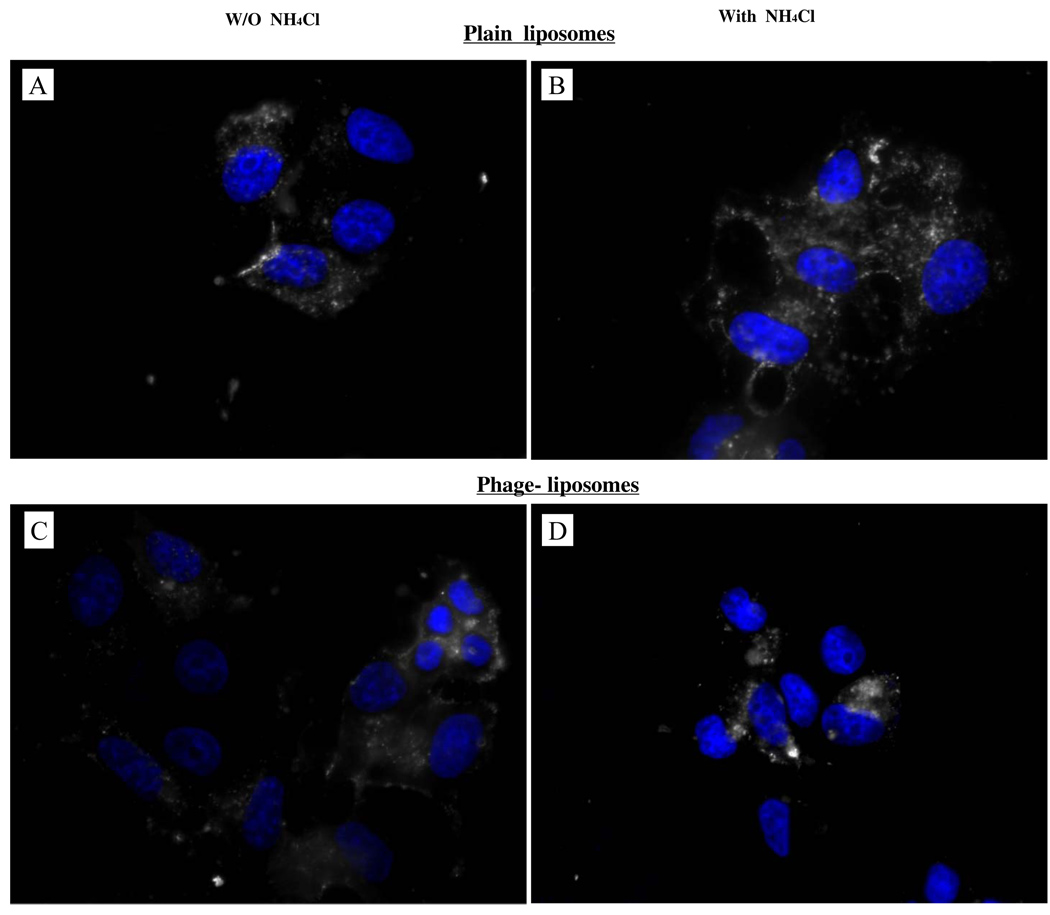
Phage-liposomes and plain liposomes (control) were labeled with rhodamine, and their subcellular localization was visualized with a fluorescence microscopy. In the cells treated with plain liposomes, the fluorescence was visible as a perinuclear punctuate (vacuolar) pattern (Fig. 4A), indicating that plain liposomes were taken up by cells via the endocytic pathway and were ultimately confined within late endosomes or lysosomes. In the case of phage-liposomes, however, a diffuse and relatively weak rhodamine fluorescence was observed throughout the cells (Fig. 4C), suggesting that phage-liposomes were released from endosomes into the cytoplasm. To examine the pH-dependence of the endosomal release by phage-liposomes, again the endosome acidification inhibitor NH4Cl was used. In the presence of 20mM NH4Cl, the diffuse distribution pattern of phage-liposomes returned to the perinuclear punctuate (vacuolar) pattern (Fig. 4D). At the same time, NH4Cl treatment had no effect on the fluorescence distribution pattern of plain liposomes (Fig. 4B). This result clearly confirms the pH-dependent endosomal release of phage-liposomes mediated by the liposome-attached phage protein.
Figure 4. Endosome release of rhodamine-labeled phage-liposomes.
A) Perinuclear punctuate pattern of plain liposomes in the absence of the endosome acidification inhibitor - NH4Cl, suggesting their entrapment into endosomes or lysosomes. B) Punctuate pattern of plain liposomes in the presence of the endosome acidification inhibitor - NH4Cl, C) Diffuse subcellular pattern of phage-liposomes, indicating their subcellular locations in cytosol; D) Perinuclear punctuate pattern, indicating NH4Cl inhibition on endosome release of phage-liposomes.
Endosomal Escape of Phage-Liposomes by the Cytoplasmic Delivery of HPTS
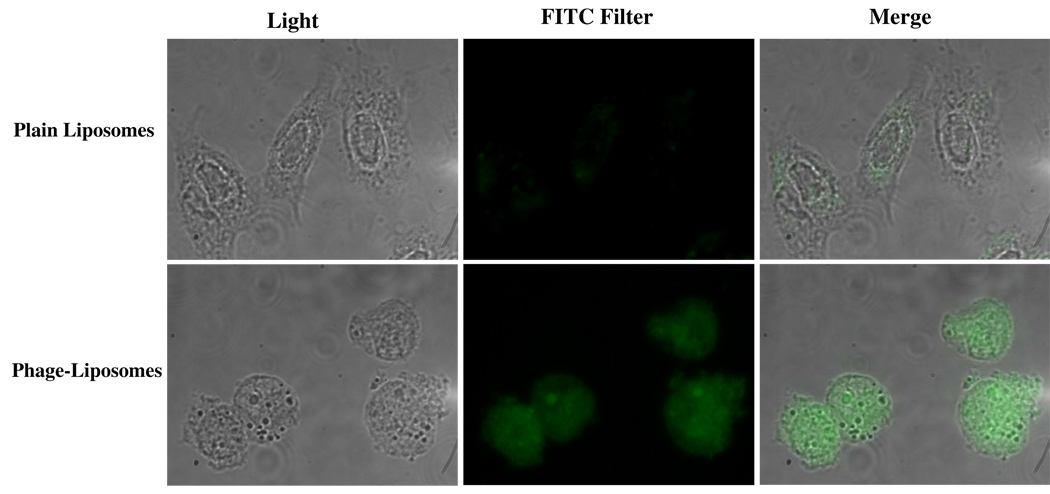
HPTS is a membrane-impermeable and pH-dependent fluorescent dye (at neutral pH, HPTS shows strong fluorescence at 450 nm excitation, while at acidic pH, HPTS fluorescence at 390 nm excitation). Figure 5 showed that with phage-liposomes, strong green fluorescence throughout the cells confirms the cytoplasmic delivery of the HPTS, while much fewer plain liposomes could be discovered in the cytosol (significantly weaker fluorescence).
Figure 5. Cytosolic delivery of HPTS encapsulated by phage-liposomes.
While much less green fluorescence is observed in the case of plain liposomes, phage-liposome treatment shows strong fluorescence emission at the 494nm excitation, indicating that HPTS is released into the neutral cytosol.
Endosomal Escape of Phage-Liposomes by Cytotoxicity
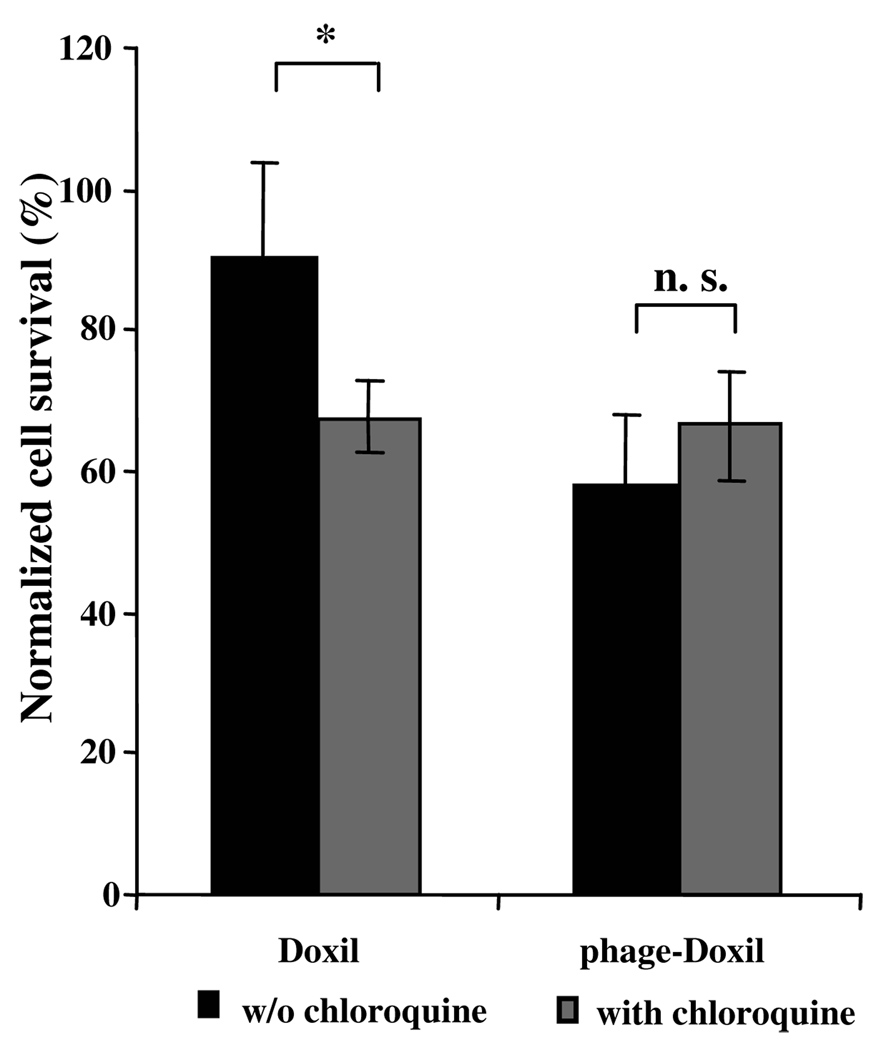
The endosomal escape of phage-liposomes was also confirmed by examining the tumor cell killing activities of Doxil and phage-Doixl in the presence of the chloroquine, which is an endosome-disrupting agent. The chloroquine treatment enhanced Doxil-induced tumor cell death, but had a negligible effect on the phage-Doxil-triggered cell death (Fig. 6), suggesting that endosome disruption by chloroquine facilitates the escape of the endosome-entrapped Doxil into the cytosol thus increasing its cytotoxicity, whereas phage-Doxil escapes from endosomes without the help of chloroquine.
Figure 6. Endosomal escape of phage-Doxil.
Endosome disruption by chloroquine showing enhanced cytotoxicity of Doxil but a negligible effect on phage-Doxil. (*p<0.05; n=6, Mean±SEM).
Discussion
Previously, we constructed and described a phage fusion protein composed of a MCF-7 cell-specific-targeting peptide genetically fused to phage pVIII coat protein17. The incorporation of such a phage protein into doxorubicin-loaded liposomes (Doxil) resulted in a remarkable increase in the killing efficiency of targeted tumor cells17. The enhanced cytotoxicity was shown to be mediated by the increased association of phage-Doxil with target cells17. However, more specifically, the cytotoxicity of phage-Doxil could depend on multiple events including cellular uptake of the drug, its cytoplasmic transport, and nuclear delivery. This, in turn, suggests a possible involvement of other phenomena, such as the endosomal escape of the preparation and nuclear translocation, in the enhanced Doxil cytotoxicity induced by the phage protein.
The structural similarity between the phage protein and anionic fusogenic peptides, such as HA2, INF1-4, INF7, E5, JTS-1 and GALA10, 13–15, suggests a possible endosome-escaping activity of the phage protein. Earlier detailed studies have demonstrated that the pH-sensitive endosome disruption by anionic fusogenic peptides is attributable to carboxylic groups within glutamic acid residues, which protonate in the acidic endosomal environment resulting in transition of the secondary structure of the peptide from a random coil to a stable α-helix and subsequent destabilization of the endosomal membrane10, 14, 19. In the case of the phage protein, its N-terminus contains several residues of aspartic and glutamic acids, which should protonate similarly in acidic conditions and display similar structural and endosome-destabilizing properties. In addition, a poly (aspartamide) derivative has been reported to trigger higher transfection efficiency by facilitating the endosomal escape20. This led us to explore fusogenic activity and endosomal escape of the phage protein.
Initially, we confirmed that phage-liposomes are taken by the target cells via a normal endocytic pathway. Current tools for the identification of uptake mechanisms fall into two categories: the use of inhibitors, which perturb intracellular trafficking, and the use of the markers, which label intracellular organells21. To better understand the uptake pathway of phage-liposomes, we combined both tools in our study. A decreased tumor cell killing by phage-Doxil upon the Bafilomycin A1 treatment served as the first indication that phage-liposomes are internalized into endosomes, and that the cytotoxicity of phage-Doxil depends on endosome acidification. We have also found that rhodamine-labeled phage-liposomes co-localize with positive transferrin-marked endosomes (Fig 1). The punctate subcellular distribution upon the treatment with the inhibitor of the endosome acidification (Fig 4), along with temperature sensitivity and energy dependence of phage-liposome-mediated induction of the intracellular membrane fusion (Fig 3) further confirm the endocytic pathway of cellular uptake of phage-liposomes.
PH-responsive membrane fusion and destabilization is believed to underlay the mechanism employed by envelop viruses as well as by synthetic anionic peptides to escape from the endosomes10, 15, 19. In the present study, we used a well-established method, FRET, to detect the membrane fusion. With artificial membranes, phage protein triggered the membrane fusion in an acidic environment, and protein mediated-membrane fusion was enhanced with increasing protein concentration, which confirms that phage protein contributes to the pH-sensitive membrane fusion. In cell experiments, it was further shown that phage protein triggers the endosomal membrane fusion. A temperature decrease to 4°C and an endosome acidification inhibition by NH4Cl both significantly decreased the membrane fusion, suggesting energy- and pH-dependency of membrane fusion mediated by the phage protein. These results not only clearly demonstrate the endocytic uptake of phage-liposomes, but also suggest the endosome-escaping potential of the phage protein.
When visualizing the endosomal escape of phage-liposomes with the fluorescence microscopy, we found different intracellular distribution patterns for phage-liposomes and non-modified plain liposomes, which reflect their different subcellular localization. In the case of plain liposomes, the observed perinuclear punctuate localization indicates their entrapment within endosomes and/or lysosomes. In the case of phage-liposomes, their predominantly diffuse distribution pattern suggests the escape of phage-liposome from endosomes into the cytosol. This diffuse pattern of endosomal escape agrees well with the previous reports on the photo-triggered endosomal release of 10kDa dextran and polyplex 22and cytoplasmic delivery of calcein-loaded liposomes23. Furthermore, the inhibition of the endosome acidification with NH4Cl results visually observed entrapment of phage-liposomes within perinuclear vesicles, but does not change the punctuate distribution pattern of plain liposomes, further indicating the pH-sensitive membrane fusion property of phage protein, which facilitates the endosomal escape of phage-liposomes. This conclusion is still further supported by the intracellular behavior of phage-liposomes co-loaded with HPTS and its quenching counterpart, DPX, when de-quenching and cytoplasmic fluorescence of the HPTS is observed, which is the result of the endosomal escape of phage-liposomes and release of their contents into the cytoplasm. This result is in good agreement with the data on the cytoplasmic delivery of HPTS by pH-sensitive liposomes composed of fusogenic lipid DOPE and CHEMS24.
The cytosolic delivery of phage-Doxil was also confirmed by the analysis of the effect of chloroquine on its cytotoxicity. Chloroquine is known to disrupt the endosome integrity by swelling and bursting the endosome25, and to inhibit endosome delivery to lysosomes26. Thus, it has been proven to serve as a helper of DNA transfection presumably by facilitating the endosomal release and cytosolic delivery of DNA25. As expected, we found that chloroquine enhanced the efficacy of Doxil, bringing its effect to the level similar to that achieved with phage-Doxil; however, chloroquine has a negligible effect on the efficacy of phage-Doxil. This, along with results of Bafilomycin A1 inhibition, suggests that phage protein, like chloroquine, stimulates the endosomal release of phage-Doxil and promotes drug access to the cytosol.
Certainly, it would be interesting to find out if the MCF-7-specific phage protein can facilitate the endosomal escape in other cells. However, a very high specificity of the selected phage towards target cells compared to non-target ones makes this task very difficult if not impossible. Considering cell binding is a necessary upstream event of endosomal escape, we have not evaluated endosomal escape property of the MCF-7 specific phage protein on other cell lines in the study. Earlier, we have found17 that free DMPGTVLP phage binds with MCF-7 cells 12-to-26 times better than to non-target cells, including HepG2 cells (human hepatocellular carcinoma cells), MCF-10A cells (non-tumorigenic human epithelial cells) and WI-38 cells (normal human lung fibroblasts). The same remains true for phage protein-decorated pharmaceutical nanocarriers. According FACS analysis data17, MCF-7-specific-phage-liposomes bind to almost 50 % of target MCF-7 cells, but only to 2% of non-target C166-GFP cells (mouse yolk sac endothelial cells) in a co-culture system consisting of MCF-7 and C166-GFP cells. The binding of MCF-7-specific phage-liposomes with the co-culture of non-target cells NIH3T3 (mouse fibroblasts) and C166-GFP was also negligible. Such low level of binding of MCF-7-specific phage protein with other cells does not allow for any meaningful estimate of its endosomal escape potential in cells others than target MCF-7 cells. However, we have to assume the ability of phage protein to induce the endosomal membrane fusion in various non-target cells, since the fusogenic property seems not to be cell type-dependent, but only pH-dependent. This assumption is confirmed by the fact that both, MCF-7-specific phage-Doxil 17 and MCF-7-specific phage-decorated polymeric micelles built of polyethylene glycol-phosphatidyl ethanolamine conjugate and loaded with paclitaxel (unpublished data) demonstrated a higher cytotoxicity towards non-target cells (C166 or NIH3T3) compared to controls (e.g. plain Doxil, paclitaxel -loaded plain micelles, non-target, doxorubicin-loaded phage liposomes modified with a unrelated phage protein or non-target, paclitaxel-loaded phage micelles modified with a unrelated phage protein) at a higher drug concentration, suggesting that the increase in the non-target cell death may be a result of the increase endosomal escape mediated by the MCF-7-specific phage protein even after a nonspecific uptake of phage-Doxil and phage-micelles loaded with paclitaxel by non-target cell lines.
Overall, our results demonstrate that phage protein bear not only membrane anchoring and cell-specific targeting activities, but also a pH-dependent fusogenic property required for endosomal escape. Accordingly, the substantial increase in the efficiency of target cancer cell killing by phage-Doxil compared to non-modified Doxil is a result of the favorable combination of several different activities of the phage protein. A drug carrier equipped with the phage protein should achieve a “one stone, two birds” effect with respect to tumor cell-specific targeting and endosome escaping properties.
Acknowledgements
This work was supported by the NIH grant# 1 R01 CA125063-01 and the Animal Health and Disease Research grant 2006-9, College of Veterinary Medicine Auburn University to Valery A Petrenko.
References
- 1.Torchilin VP. Recent Advances with Liposomes as Pharmaceutical Carriers. Nat Rev Drug Discov. 2005;4(2):145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 2.Klibanov AL, Maruyama K, Torchilin VP, Huang L. Amphipathic Polyethyleneglycols Effectively Prolong the Circulation Time of Liposomes. FEBS Lett. 1990;268(1):235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- 3.Blume G, Cevc G. Molecular Mechanism of the Lipid Vesicle Longevity in Vivo. Biochim Biophys Acta. 1993;1146(2):157–168. doi: 10.1016/0005-2736(93)90351-y. [DOI] [PubMed] [Google Scholar]
- 4.Torchilin VP, Omelyanenko VG, Papisov MI, Bogdanov AA, Trubetskoy VS, Herron JN, Gentry CA. Poly(ethylene glycol) on the Liposome Surface: on the Mechanism of Polymer-Coated Liposome Longevity. Biochim Biophys Acta. 1994;1195(1):11–20. doi: 10.1016/0005-2736(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 5.Park JW, Kirpotin DB, Hong K, Shalaby R, Shao Y, Nielsen UB, Marks JD, Papahadjopoulos D, Benz CC. Tumor Targeting Using Anti-Her2 Immunoliposomes. J Control Release. 2001;74(1–3):95–113. doi: 10.1016/s0168-3659(01)00315-7. [DOI] [PubMed] [Google Scholar]
- 6.Torchilin VP. Targeted Pharmaceutical Nanocarriers for Cancer Therapy and Imaging. AAPS J. 2007;9(2):E128–E147. doi: 10.1208/aapsj0902015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dokka S, Rojanasakul Y. Novel Non-Endocytic Delivery of Antisense Oligonucleotides. Adv Drug Deliv Rev. 2000;44(1):35–49. doi: 10.1016/s0169-409x(00)00082-x. [DOI] [PubMed] [Google Scholar]
- 8.Vasir JK, Labhasetwar V. Biodegradable Nanoparticles for Cytosolic Delivery of Therapeutics. Adv Drug Deliv Rev. 2007;59(8):718–728. doi: 10.1016/j.addr.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lear JD, DeGrado WF. Membrane Binding and Conformational Properties of Peptides Representing the NH2 Terminus of Influenza HA-2. J Biol Chem. 1987;262(14):6500–6505. [PubMed] [Google Scholar]
- 10.Martin ME, Rice KG. Peptide-Guided Gene Delivery. AAPS J. 2007;9(1):E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mastrobattista E, Koning GA, van Bloois L, Filipe AC, Jiskoot W, Storm G. Functional Characterization of an Endosome-Disruptive Peptide and Its Application In Cytosolic Delivery of Immunoliposome-Entrapped Proteins. J Biol Chem. 2002;277(30):27135–27143. doi: 10.1074/jbc.M200429200. [DOI] [PubMed] [Google Scholar]
- 12.Kichler A, Mechtler K, Behr JP, Wagner E. Influence of Membrane-Active Peptides on Lipospermine/DNA Complex Mediated Gene Transfer. Bioconjug Chem. 1997;8(2):213–221. doi: 10.1021/bc970009z. [DOI] [PubMed] [Google Scholar]
- 13.Subbarao NK, Parente RA, Szoka FC, Jr, Nadasdi L, Pongracz K. pH-Dependent Bilayer Destabilization by an Amphipathic Peptide. Biochemistry. 1987;26(11):2964–2972. doi: 10.1021/bi00385a002. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Nicol F, Szoka FC., Jr GALA: a Designed Synthetic pH-Responsive Amphipathic Peptide with Applications in Drug and Gene Delivery. Adv Drug Deliv Rev. 2004;56(7):967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 15.Wagner E. Application of Membrane-Active Peptides for Nonviral Gene Delivery. Adv Drug Deliv Rev. 1999;38(3):279–289. doi: 10.1016/s0169-409x(99)00033-2. [DOI] [PubMed] [Google Scholar]
- 16.Jayanna PK, Torchilin VP, Petrenko VA. Liposomes Targeted by Fusion Phage Proteins. Nanomedicine. 2009;5:83–89. doi: 10.1016/j.nano.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, D'Souza GGM, Bedi D, Fagbohum OA, Potturi P, Papahadjopoulos-Sternberg B, Petrenko VA, Torchilin VP. Enhanced Binding and Killing of Target Tumor Cells by Drug-Loaded Liposomes Modified with Tumor-Specific Phage Fusion Coat Protein. Future Medicine: Nanomedicine. 2010 doi: 10.2217/nnm.10.30. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchilin VP, Weissig V. Liposomes. 2th ed. New York: Oxford University Press Inc.; 2003. p. 163. [Google Scholar]
- 19.Cho YW, Kim JD, Park K. Polycation Gene Delivery Systems: Escape from Endosomes to Cytosol. J Pharm Pharmacol. 2003;55(6):721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 20.Kanayama N, Fukushima S, Nishiyama N, Itaka K, Jang WD, Miyata K, Yamasaki Y, Chung UI, Kataoka K. A PEG-Based Biocompatible Block Catiomer with High Buffering Capacity for the Construction of Polyplex Micelles Showing Efficient Gene Transfer Toward Primary Cells. ChemMedChem. 2006;1(4):439–444. doi: 10.1002/cmdc.200600008. [DOI] [PubMed] [Google Scholar]
- 21.Watson P, Jones AT, Stephens DJ. Intracellular Trafficking Pathways and Drug Delivery: Fluorescence Imaging of Living and Fixed Cells. Adv Drug Deliv Rev. 2005;57(1):43–61. doi: 10.1016/j.addr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 22.de Bruin KG, Fella C, Ogris M, Wagner E, Ruthardt N, Brauchle C. Dynamics of Photoinduced Endosomal Release of Polyplexes. J Control Release. 2008;130(2):175–182. doi: 10.1016/j.jconrel.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Chu CJ, Dijkstra J, Lai MZ, Hong K, Szoka FC. Efficiency of Cytoplasmic Delivery by PH-Sensitive Liposomes to Cells in Culture. Pharm Res. 1990;7(8):824–834. doi: 10.1023/a:1015908831507. [DOI] [PubMed] [Google Scholar]
- 24.Morilla MJ, Montanari J, Frank F, Malchiodi E, Corral R, Petray P, Romero EL. Etanidazole in PH-Sensitive Liposomes: Design, Characterization and in Vitro/in Vivo Anti- Trypanosoma Cruzi activity. J Control Release. 2005;103(3):599–607. doi: 10.1016/j.jconrel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Erbacher P, Roche AC, Monsigny M, Midoux P. Putative Role of Chloroquine in Gene Transfer into A Human Hepatoma Cell Line by DNA/Lactosylated Polylysine Complexes. Exp Cell Res. 1996;225(1):186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- 26.Mellman I, Fuchs R, Helenius A. Acidification of the Endocytic and Exocytic Pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]