Abstract
The combination of poly(ADP-ribose)polymerase (PARP) inhibitors and alkylating agents is currently being investigated in cancer therapy clinical trials. However, the DNA lesions producing the synergistic cell killing effect in tumors is not fully understood. Treatment of human and mouse fibroblasts with the monofunctional DNA methylating agent methyl methanesulfonate (MMS) in the presence of a PARP inhibitor has been shown to trigger a cell cycle checkpoint response. Among other changes, this DNA damage response to combination treatment includes activation of ATM/Chk2 and phosphorylation of histone H2A.X. These changes are consistent with DNA double-strand break (DSB) formation during the response, but the measurement of DSBs has not been addressed. Such DSB evaluation is important in understanding this DNA damage response because events other than DSB formation are known to lead to ATM/Chk2 activation and H2A.X phosphorylation. Here, we examined the structural integrity of genomic DNA after the combined treatment of cells with MMS and a PARP inhibitor, i.e., exposure to a sub-lethal dose of MMS in the presence of the PARP inhibitor 4-amino-1,8-napthalimide (4-AN). We used pulsed field gel electrophoresis (PFGE) for measurement of DSBs in both human and mouse embryonic fibroblasts, and flow cytometry to follow the phosphorylated form of H2A.X (γ-H2A.X). The results indicate that DSBs are formed with the combination treatment, but not following treatment with either agent alone. Our data also show that formation of γ-H2A.X correlates with PARP-1-expressing cells in S-phase of the cell cycle. The observations support the model that persistence of PARP-1 at base excision repair intermediates, as cells move into S-phase, leads to DSBs and the attendant checkpoint responses.
Keywords: PARP-1, PARP inhibitor, Methyl methanesulfonate, Cell cycle, Double-strand breaks, γ-H2A.X
1. Introduction
Genomic DNA is constantly subjected to a plethora of damaging events resulting in potentially toxic DNA lesions. Well known cytotoxic events include oxidative modifications of DNA bases, alkylation of DNA bases, spontaneous base loss and strand breakage, and irradiation-induced chemical changes. If not repaired, DNA lesions of all types can adversely affect the ability of DNA to execute its cellular functions. Moreover, un-repaired base excision repair (BER) intermediates can lead to DSB formation as a function of replication fork arrest [1-3]. A DSB is considered a highly detrimental form of DNA damage, and cells maintain elaborate mechanisms for both detecting and repairing this type of lesion. When a DSB is detected, normal cells typically signal a replication and cell division arrest, thus allowing more time for DSB repair. The replication and cell cycle arrests occur through a signaling cascade that includes activation of the ATM and Chk2 kinases and downstream effectors [4].
BER is the primary defense mechanism against simple DNA base lesion damage and single-strand breaks in mammalian cells. Incomplete processing of intermediates during BER can result in accumulation of toxic AP sites and single-strand breaks [1,2]. After initiation of BER, the abundant PARP-1 protein is considered one of the first BER factors to bind to the single-strand break intermediate [5-8]. Following binding, PARP-1 becomes activated and undergoes auto-poly(ADP-ribosyl)ation. This activation of PARP-1 facilitates BER, since treatment of cells with PARP inhibitor blocks BER [9-12] and strand break intermediates accumulate [13,14], especially when there is a deficiency in a key BER enzyme/cofactor such as XRCC1 or DNA polymerase β (Pol β) [13,14]. In addition to PARP-1, PARP-2 has PAR synthesis activity and may function as a back up in the absence of PARP-1 [15,16]. Wild-type human and mouse fibroblasts in culture do not exhibit sensitivity when treated with a low dose MMS or with the PARP inhibitor 4-AN alone [17-19]. However, treatment with the combination of MMS and 4-AN (M+4) causes a profound decrease in cell survival [14,18,20]. This is accompanied by activation of an S-phase cell cycle checkpoint [18,19] and eventual cell death by apoptosis [20]. Investigation into the extreme cytotoxic effect revealed features suggesting the formation of DSBs: M+4 treatment resulted in rapid phosphorylation of ATM and the ATM signaling partner Chk2 [17]; and M+4 treated cells exhibited phosphorylation of histone H2A.X. Although these features are consistent with DSB formation, it is known that phosphorylation of H2A.X and activation of ATM and Chk2 are not restricted to the DSB response [21-23]. To further examine the cytotoxic effect of M+4 treatment, we used pulsed field gel electrophoresis (PFGE) of genomic DNA to directly quantify formation of DSBs. The results are discussed in the context of roles for PARP activity in DNA repair and DNA damage responses and of models explaining the cytotoxicity of PARP inhibition. Since inhibited PARP-1 protein is proposed to be involved in the checkpoint response, we also evaluated DSB formation in PARP-1 −/− cells.
2. Materials and methods
2.1. Cell lines and maintenance
The human fibroblasts studied were AT cells (SV40 transformed cells obtained from an Ataxia Telangiectasia patient) complemented with an ATM construct as described [24]. These cells were obtained from the Corriell Cell Repository, Catalog ID, GM16667. Cells were grown in Dulbeco’s Modified Eagle’s Medium (DMEM) that contained glutamine (Invitrogen) and 10% fetal bovine serum (FBS) (HyClone) and was supplemented with hygromycin (100 μg/ml) to maintain complementation, at 37°C in a 10% CO2 chamber. SV40-transformed NBS1 fibroblasts were derived from primary cells obtained from a patient with Nijmegen Breakage Syndrome, and were then NBS1-complemented with one extra human chromosome 8 [25]. These cells were obtained and used with the permission of Dr. M.Z. Zdzienicka and were grown in DMEM containing glutamine and 10% FBS, at 37°C in a 10% CO2 chamber. Wild type MEFs were generated from a clone obtained from the SV40-transformed mouse embryonic cell line, M16tsA as previously reported [26]. They were grown in DMEM containing GlutaMAX I (Invitrogen), 10% FBS and hygromycin (80 μg/ml). MEFs were grown in 10% CO2 incubators at 34°C. Wild type mouse embryonic fibroblasts (MEFs) were generated from a clone obtained from the SV40-transformed MEF cell line, M16tsA as previously reported [26]. They were grown in DMEM containing GlutaMAX I (Invitrogen), 10% FBS and hygromycin (80 μg/ml). MEFs were grown in 10% CO2 incubators at 34°C. The spontaneously immortalized PARP-1 wild-type MEFs and the isogenic PARP-1 null cells were obtained from Dr. Josianne Ménissier-de Murcia [27]. They were maintained in DMEM containing glutamine (Invitrogen) and 10% FBS and were grown in 10% CO2 at 37°C.
2.2. Genotoxic treatment
For PFGE studies, either 7 × 105 or 1 × 106 cells were grown in 10-cm dishes. Cells were mock-treated or treated with 0.1 mM MMS (low-dose for human fibroblasts) or 0.25 mM MMS (low-dose for MEFs) and/or 10 μM 4-AN. In all experiments, the combined M+ 4 treatment was for 1 h in culture medium. Cells were then washed with Hanks’ balanced salt solution (HBSS) (HyClone) and then cultured in fresh cell-specific culture medium with or without 10 μM 4-AN. For 4-AN alone and M+4 studies, cells were continuously exposed to 4-AN for the entire period of the experiment. Mock-treated cells were washed, and the medium was replaced in the same fashion as with the drug treated cells.
2.3. PFGE
After treatment, cells were washed twice using HBSS on ice and trypsinized for 30 sec at room temperature. Cells were harvested in 5 ml of the cell-specific medium noted above. Cell pellets were obtained using centrifugation at 6°C at 1500 rpm for 5 min. Cell pellets were re-suspended in an equal volume of cell re-suspension buffer (Bio-Rad, CHEF mammalian DNA plug kit) and 2% clean-cut agarose (Bio-Rad kit). The resulting cell plugs were equilibrated in Proteinase K buffer (Bio-Rad kit) on ice for at least 1 h prior to addition of Proteinase K (Ambion) to a final concentration of 1 mg/ml. Proteinase K digestion was performed at 16°C for 16 to 24 h. Plugs were washed three times for 1 h each at 4 °C, using 1× washing buffer (Bio-Rad). Plugs were equilibrated for 1 h in 0.5× TBE running buffer prior to loading onto the 1% PFGE-grade agarose (Bio-Rad) gel. Electrophoresis was conducted on a CHEF-mapper XA PFGE system. Electrophoresis conditions were 120° using 10-90 second switch times at 6 V/cm for 24 h. To ensure that our electrophoresis conditions were optimal and that we were capturing all of the DSBs entering the gel, a 60-240 second switch time at 4 V/cm for 18 h also was also used (data not shown). The electrophoresis conditions were different for the ionizing radiation (IR) experiments presented in Fig. 1A and C; in this case, the conditions were 120° with a 50-5000 second switch time at 1.5 V/cm for 66 h.
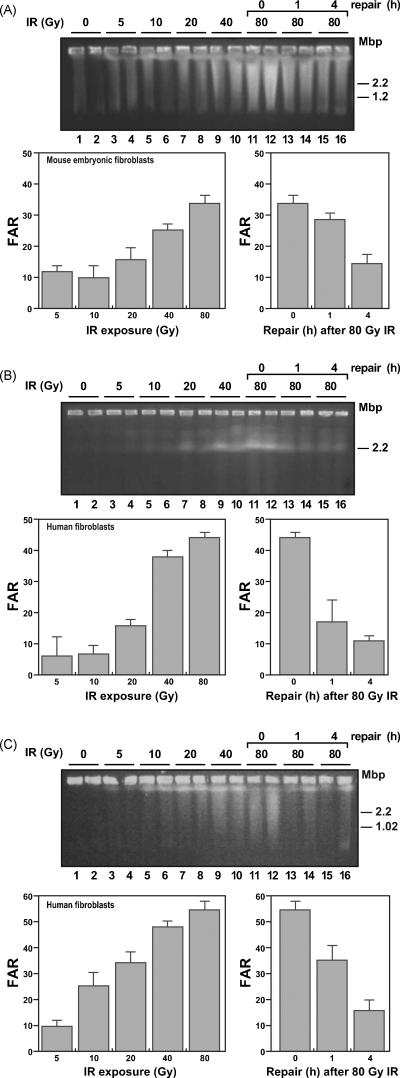
Fig. 1.
Detection of DSBs using PFGE in model cellular systems.
(A) Top, photograph of a representative agarose gel prepared using agarose plugs from MEFs treated with increasing amounts of IR; plugs were electrophoresed in a 1% agarose gel as described in Section 2. DNA was stained using Sybr Gold. Bottom, quantification of the DNA signal in panel above. The percentage of total DNA that entered the gel, or fraction of activity released x100 (FAR), was plotted. The disappearance of the 80 Gy-induced FAR signal over a 4 h period is shown separately in the plot on the right. (B) Top, representative agarose gel obtained using agarose plugs prepared from GM 166667 human fibroblasts, IR treated as in panel A. Bottom, quantification of the FAR signal from panel B. The graph on the right shows the rate of disappearance of the signal. (C) Top, representative agarose gel obtained using agarose plugs prepared from NBS1 human fibroblasts Itreated with IR as in panel A. Bottom, quantification of the FAR signals as described above. Error bars for the panels were determined from two separate samples for each experimental condition.
2.4. Calculation of fraction of activity released (FAR)
CHEF-grade agarose gels were stained using Sybr Gold (Invitrogen) according the manufacturer’s instructions. The signal in each lane was separated into three zones. The first zone corresponded to the well at the top of the gel, the second and third zones corresponded to regions within the gel where DNA signals were observed immediately below the well and elsewhere in the gel, respectively. The total signal was taken as the sum of all three zones. The fraction of DNA entering the gel was calculated using the sum of zones two and three divided by the total signal for all three zones (i.e., the FAR). The FAR values reported were then multiplied by 100, and the numbers plotted in the figures represent the values after subtracting the background (mock-treated). Gels also were Southern blotted to verify the sensitivity of the Sybr Gold staining method. This was accomplished by transferring the DNA from the gel onto a Hybond (GE) nylon membrane and then probing the transferred DNA with HindIII-digested genomic DNA (human or mouse) radiolabeled with [α-32P]dCMP. Similar results were obtained by this method as with the Sybr Gold staining method described above.
2.5. Flow cytometry and γ-H2A.X staining
Cells were seeded in 10-cm dishes at a density of 2 × 106. Cells were cultured and mock-treated, or treated with MMS, 4-AN or M+4, as described above. After treatment and culture, cells were harvested by trypsinization, collected by centrifugation and washed with phosphate buffered saline (PBS). The washed cell pellet was prepared using the H2A.X Phosphorylation Assay Kit for flow cytometry (Millipore) following the manufacturer’s instructions except for minor variations. Briefly, cells were resuspended in 500 μl of 1X Fixation solution for 20 min on ice, washed with PBS and resuspended in 500 μl of Permeabilization solution for 30 min on ice. 50 μl of this mixture was added to a tube containing 5 μl of γ-H2A.X-FITC antibody, 0.5% Tween 20, 1% BSA and 5 μg of RNase A (Sigma). Cells were placed on ice and periodically mixed with gentle agitation. Cells were collected by centrifugation and resuspended in 5 μg/ml propidium iodide (PI) (Sigma-Aldrich) and stored in the dark for 30 min. Samples were read on a FACS flow cytometer (BD Biosciences) and analyzed using Cell Quest software (BD Biosciences).
3. Results
3.1. Measurement of DSBs by PFGE
To make use of PFGE for measurement of DSBs (reviewed in [28]), we first characterized wild-type mouse and human fibroblasts for DSB formation after exposure to IR (Fig. 1). In this assay for DSBs, chromosome-size DNA and replicating DNA molecules do not enter the gel, whereas DNA fragments 10 Mbp or less enter the gel [28] and are detected by Sybr Gold staining. We were able to readily detect DSBs over an increasing range of IR exposure from 5 to 80 Gy (Fig. 1). The amount of DNA entering the gel was quantified at each dose, and the background obtained from untreated cells was subtracted. A dose-dependent response was observed with all cell types used (Fig. 1), and at the highest dose of IR used (80 Gy), 35 to 55% of the cellular DNA (Fig. 1, top panels, lanes 11 and 12) entered the gel. The background observed in this set of experiments was negligible compared with the DSBs observed with increasing IR. However, in the M+4 experiments to be described below where the level of DSBs was lower, the background was a significant consideration.
Repair of DSBs was assessed over a 4 h period after exposure to 80 Gy IR. A time-dependent decrease in the amount of DNA entering the gel was observed, reflecting DSB repair (Fig. 1, top panels, lanes 11-16). In each cell type, a significant decrease in signal was observed by 4 h. Results for accumulation and repair of DSBs are presented graphically under each gel panel. These results with PFGE assay and IR exposure in human and mouse fibroblasts are consistent with earlier findings [29-34].
3.2. DSB formation as a function of the combination of low-dose alkylating agent and PARP inhibition
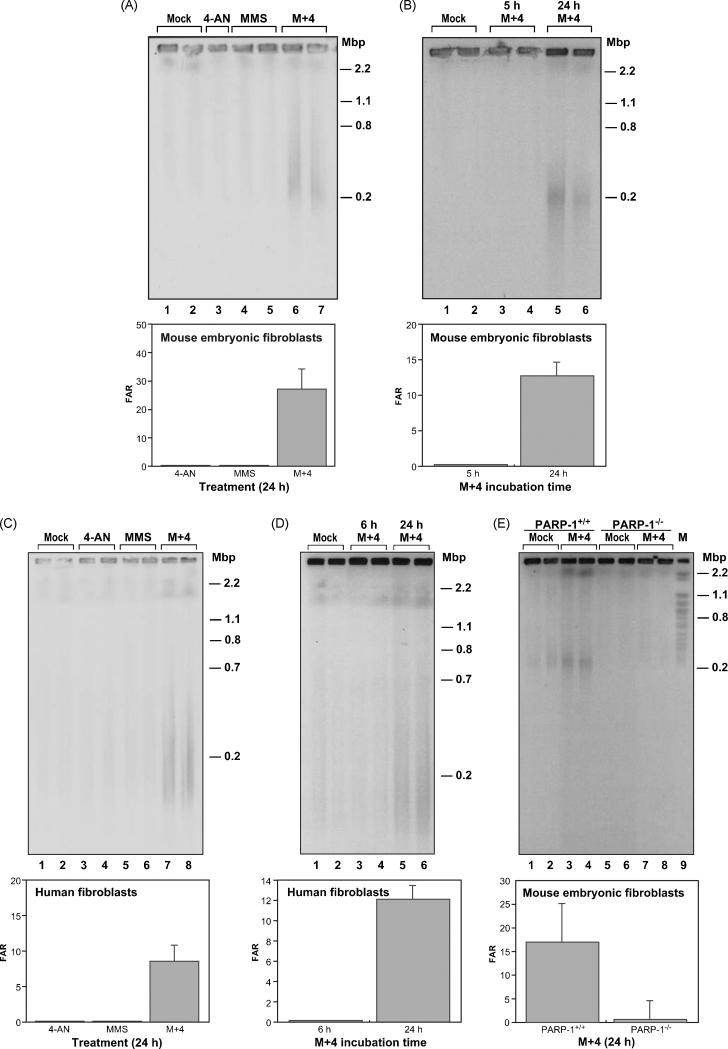
Human and mouse fibroblasts treated with low-dose MMS or 4-AN as single agents exhibited DSBs that were equivalent to the background level (Fig. 2A and C). However, the M+4 combination treatment for 24 h consistently produced DSB levels that were higher than the background or the single agent samples (Fig. 2A, lanes 6 and 7 and Fig. 2C, lanes 7 and 8). A summary of the results with human and mouse fibroblasts is shown in Table 1. With M+4 treatment, DSBs were detected in both cell types at a level higher than background. DSB measurements expressed as FAR value in cells treated with M+4 ranged from ~17 to 23 (Table 1). With the M+4 combination in both mouse and human cells, DSBs were found at a higher level at 24 h, than at the earlier times studied, 5-6 h (Fig. 2B and D, compare lanes 3 and 4 with 5 and 6). In further experiments, to ascertain the effect of a higher level of alkylating agent-induced DNA damage, the MMS dose was increased by 10-fold in the M+4 combination treatment. There was an increase in DSB formation under this condition (data not shown). Finally, in light of the use of PARP inhibitors in mono-therapy for BRCA1/2-deficient tumors, it is noteworthy that we did not observe DSB formation after treatment of wild-type MEFs with 4-AN alone. These cells are wild-type for HR and BER, and this circumstance probably accounts for the difference in our results and those obtained by others with BRCA-deficient cancer cells [35,36].
Fig. 2.
Treatment with M+4 results in an increased FAR signal.
(A) Top panel, representative agarose gel stained with Sybr Gold to visualize the DNA obtained from MEFs mock-treated (lanes 1 and 2), or treated with 10 μM 4-AN (lane 3), 0.25 mM MMS for 1 h (lanes 4 and 5) or M+4 (lanes 6 and 7) as described in Section 2. The total duration of treatment was 24 h. (B) Top panel, representative agarose gel stained with Sybr Gold from MEFs mock-treated (lanes 1 and 2), treated with M+4 for 5 h (lanes 3 and 4), or for 24 h (lanes 5 and 6). (C) Top panel, representative agarose gel stained with Sybr Gold to visualize the DNA obtained from human fibroblasts mock-treated (lanes 1 and 2), or treated with 4-AN (lanes 3 and 4), MMS (lanes 5 and 6) or M+4 (lanes 7 and 8). The duration of treatment was 24 h. (D) Top panel, representative agarose gel stained with Sybr gold from human cells that were mock-treated (lanes 1 and 2), treated with M+4 for 6 h (lanes 3 and 4), or treated with M+4 for 24 h (lanes 5 and 6). (E) Top panel, representative agarose gel stained with Sybr gold from isogenic PARP-1+/+ (lanes 1 - 4) or PARP-1−/− MEFs (lanes 5 - 8) that were mock-treated (lanes 1, 2, 5 and 6) or treated with M+4 (lanes 3, 4, 8 and 9). In each of the bottom panels, A-E, are shown quantification of the DNA signals from the agarose gels above. Error bars represent the difference error from the mean of two samples for each condition after the background was subtracted. For each gel, size is indicated on the right in mega base pairs (Mbp).
Table 1.
Summary of PFGE FAR values obtained with wild-type cells that were either mock-treated or treated with M+4
| Cell type | Mock- treated (Number of Exp.) |
Mock- treated FAR (Average) . |
Std . Err. |
M+4 (Number of Exp.) |
M+4 FAR (Average) |
Std Err. |
Net FAR values |
Diff. Err. |
|---|---|---|---|---|---|---|---|---|
| MEFs | 6 | 34.3 | 5.0 | 6 | 57.4 | 4.0 | 23.1 | 6.4 |
| Human fibroblasts |
4 | 32.9 | 5.0 | 8 | 50.3 | 4.2 | 17.4 | 6.5 |
3.3 DSB formation as a function of PARP-1 expression
The results described so far are consistent with a model where activity-inhibited PARP-1 protein is involved in the production of DSBs (reviewed in [37]). To explore this possibility, we asked if DSBs could be observed in the absence of PARP-1. PARP-1 −/− cells were evaluated for DSB formation after combination M+4 treatment using the PFGE assay. We observed a low, but above background, level of DBSs in PARP-1 −/− cells treated with M+4 (Fig. 2E). The isogenic PARP-1 wild-type cells examined in parallel exhibited a higher level of DSBs upon M+4 treatment, as expected. These observations indicated that DSBs are formed as a consequence of M+4 treatment in the absence of PARP-1, but that the level of DSBs produced was extremely low and near the detection limit of our PFGE assay (Table 2). The results illustrate the importance of PARP-1 in the DSB formation phenotype of the M+4 treated cells. It is noteworthy that MMS-treated PARP-1 null cells continue to exhibit hypersensitivity to PARP inhibition, but the extent of the hypersensitivity is considerably less than that seen with the wild-type cells [19].
Table 2.
Summary of PFGE FAR values obtained with PARP-1 −/− cells that were either mock-treated or treated with M+4
| Cell type | Ex p |
Mock-treated FAR (Average). |
Std. Err. |
M+4 FAR (Average) |
Std Err. |
Net FAR values |
Diff. Err. |
|---|---|---|---|---|---|---|---|
| Wild-type | 1 | 27.9 | 0.6 | 45.1 | 2.8 | 17.2 | 2.9 |
| 2 | 40 | 11.2 | 57.1 | 1.8 | 17.1 | 8.1 | |
| 3 | 58.3 | 1.9 | 69.1 | 1.1 | 10.8 | 2.2 | |
| PARP-1 −/− | 1 | 33.4 | 2.6 | 30.7 | 2.5 | 0 | 3.6 |
| 2 | 37.6 | 1.7 | 38.2 | 3.8 | 0.7 | 4.1 | |
| 3 | 56.2 | 2.6 | 56.6 | 2.1 | 0.4 | 3.3 |
3.4. DSB and γ-H2A.X levels as a function of time after M+4 treatment
It is known that an S-phase checkpoint is activated in response to M+4 treatment [14,17,18] and that this effect requires cells to be in S-phase. To further investigate the relationship between M+4 treatment, S-phase and DSBs, we used unsynchronized human cells and evaluated various time points for the presence of γ-H2A.X. Flow cytometry analysis was employed to simultaneously monitor DNA content by PI staining.
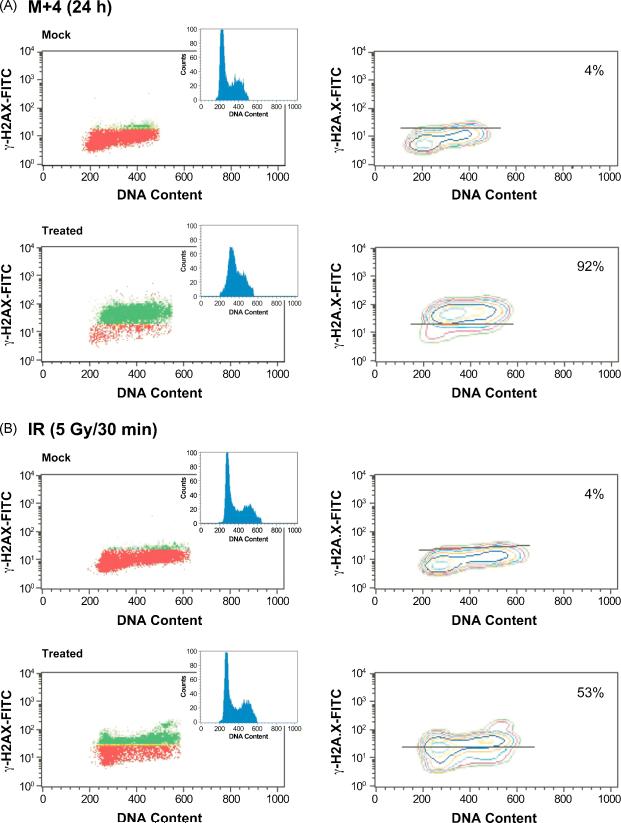
The majority (92%) of M+4-treated cells were positive for γ-H2A.X staining and corresponded to cells accumulated in S-phase (see DNA content histogram inset of Fig. 3A, bottom panel). These results supported the interpretation that following M+4 treatment, the appearance of DSBs was S-phase-dependent. Since the majority of M+4 treated cells were in S-phase by 24 h [17-19], we also evaluated the γ-H2A.X signal in M+4 treated cells at 5 h, a time when only 60% of the cells were found in S-phase. Similar to the results at 24 h, the γ-H2A.X signal was confined to cells in S-phase (data not shown). We verified that the γ-H2A.X signal was specific to M+4 treatment by following untreated cells and cells treated with MMS or 4-AN alone (Fig. 3A, top panel). Very few (4%) of these cells were γ–H2A.X positive. Finally, in contrast to the results obtained with M+4 treatment, cells treated with 5 Gy IR displayed immediate γ-H2A.X formation in all phases of the cell cycle (Fig. 3B). Together, these observations indicate that the measured γ-H2A.X signal was S-phase-dependent after M+4 treatment.
Fig. 3.
S-phase dependent γ-H2A.X signal in M+4-treated human fibroblasts. (A) Top panel, flow cytometry analysis using mock-treated cells stained with a FITC-conjugated antibody to γ-H2A.X as described in Section 2. DNA containing a signal for γ-H2A.X above background levels (red) is shown in green in the plots on the left. DNA content was measured using PI staining. Bottom panels illustrate γ-H2A.X and cell cycle analysis 24 h after exposure to M+4. (B) Flow cytometry analysis as described in (A), but cells were mock-treated (top) or treated with 5 Gy of IR and assessed 30 min after exposure (bottom). The horizontal line drawn on the γ-H2A.X contour plot in the graphs on the right distinguishes background (below the line) from positive signal (above the line). Percentages shown in the panels on the right indicate the percentage of total DNA positive for γ-H2A.X staining.
4. Discussion
Previous results [14,17,38] indicated that cell cycle checkpoints are triggered in cultured mammalian cells when they are treated with MMS in the presence of the PARP inhibitor 4-AN. In the current study, we used two approaches, PFGE and staining for γ-H2A.X, to evaluate whether DSB formation accompanies this response. DSB formation was observed by both approaches when cells were subjected to the combination of MMS-induced BER and PARP inhibition with 4-AN. Treatment with either MMS or 4-AN alone did not lead to an increase in DSBs. Yet, the amount of DSBs detected by PFGE appeared to be lower compared with γ-H2A.X staining (compare Figs. 2 and 3). An explanation for this difference may be that γ-H2A.X staining is more sensitive, involving amplification of signal stemming from a small number of DSBs. In fact, the H2A.X phosphorylation is known to radiate outward several kilo base pairs in each direction from the site of a DSB [23].
In support of our hypothesis that DSBs formation occurred during S-phase, flow cytometric analysis showed that the M+4-induced γ-H2A.X signal occurred during S-phase (Fig. 3A), and DSBs measured by PFGE were predominantly observed 24 h after treatment (Fig. 2B and D), at a time when cells had accumulated in S-phase. In contrast, with IR treated cells, DSBs measured by PFGE (Fig. 1) and γ-H2A.X signals (Fig. 3B) were observed in all phases of the cell cycle. The results also showed that DSBs were observed in untreated cells (Figs. 2 and 3). The origin of this background level of DSBs was not investigated, but such DSBs could have been due to “general” replication fork stalling or intrinsic background occurring during the experimental procedure. Others also have observed background levels of DBSs in untreated mammalian cells [30,39-42].
A working model to explain DSB formation in response to M+4 treatment calls for retention of inhibited PARP-1 at BER intermediates secondary to MMS-induced lesions. The PARP-1 bound to DNA eventually acts as a barrier to the replicative machinery during S-phase, leading to DBS formation after fork collapse and/or replication run-off. Additionally, the absence of poly(ADP-ribose) synthesis may hinder important protein-protein interactions that are required for efficient repair of strand breaks [43].
The present study indicated that the M+4-induced DSBs occur predominantly in S-phase, consistent with involvement of DNA replication in DSB formation and with proposed models explaining the effects of PARP inhibition [37]. Our observation that PARP-1 −/− cells treated with M+4 exhibit a reduced level of DSB formation is consistent with the proposed model and with PARP-2 serving in a back up role in the absence of PARP-1. We previously found that PARP-1 null cells retained a modest degree of enhanced cytotoxicity in response to M+4 treatment, consistent with a proposed role for PARP-2 as a backup for PARP-1 deficiency [19]. PARP-1 activity appears to be critical in the base lesion repair process. This is consistent with recent findings on cell-based repair of base lesions where inhibition of PARP resulted in decreased repair capacity [10] and with findings in PARP-1 null chicken DT40 cells that strand break repair intermediates accumulated after MMS treatment [44]. PARP-1 has been shown to be involved in the replication fork restart process, where it is thought to assist in recruitment of the Mre11/Rad50/NBS1 (MRN complex). The role of PARP-1 in the recruitment of the MRN complex was dependent on PARP-1 activity [43], thus, when PARP-1 was unable to auto-poly(ADP-ribosyl)ate, the MRN complex was not recruited, resulting in a failure of fork restart. Additionally, the importance of PARP-1 in the context of chromatin remodeling has been reported [45,46]. These recent results showed that a chromatin-remodeling complex, ALC1, had decreased binding to DNA damage response (DDR) proteins when PARP activity was inhibited. Ahel et al. also demonstrated that recruitment of ALC1 to damage sites was reduced when PARP activity was inhibited [45].
In summary, DSBs are produced in response to treatment of human and mouse fibroblasts with the combination of low-dose MMS and the PARP inhibitor 4-AN. PARP inhibition in combination with DNA damaging agents is in clinical evaluation for cancer treatment, and the results described here may contribute to the understanding of the molecular consequences of such an approach [47,48].
Acknowledgements
We thank Jennifer Zeng for technical assistance, Michael Carrozza for discussion and critical reading of this manuscript and Bonnie Mesmer for careful editing. We thank Carl Bortner and the Flow Cytometry Center for assistance. We are also grateful to the Michael Resnick laboratory for use of their Bio-Rad CHEF-mapper XA PFGE system, as well as for technical assistance from Wenjian Ma and Jim Westmoreland. This research was supported by Research Project Number Z01-ES050159 in the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Conflict of interest The authors declare there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ma W, Panduri V, Sterling JF, Van Houten B, Gordenin DA, Resnick MA. The transition of closely opposed lesions to double-strand breaks during long-patch base excision repair is prevented by the coordinated action of DNA polymerase δ and Rad27/Fen1. Mol. Cell. Biol. 2009;29:1212–1221. doi: 10.1128/MCB.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- [5].El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J. Biol. Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- [7].Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- [8].Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Creissen D, Shall S. Regulation of DNA ligase activity by poly(ADP-ribose) Nature. 1982;296:271–272. doi: 10.1038/296271a0. [DOI] [PubMed] [Google Scholar]
- [10].Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase β and PARP activities in base excision repair in living cells. DNA Repair (Amst.) 2009;8:1290–1299. doi: 10.1016/j.dnarep.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Satoh MS, Poirier GG, Lindahl T. NAD(+)-dependent repair of damaged DNA by human cell extracts. J. Biol. Chem. 1993;268:5480–5487. [PubMed] [Google Scholar]
- [12].Satoh MS, Poirier GG, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- [13].Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase β in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst.) 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- [16].Sallmann FR, Vodenicharov MD, Wang ZQ, Poirier GG. Characterization of sPARP-1. An alternative product of PARP-1 gene with poly(ADP-ribose) polymerase activity independent of DNA strand breaks. J. Biol. Chem. 2000;275:15504–15511. doi: 10.1074/jbc.275.20.15504. [DOI] [PubMed] [Google Scholar]
- [17].Carrozza MJ, Stefanick DF, Horton JK, Kedar PS, Wilson SH. PARP inhibition during alkylation-induced genotoxic stress signals a cell cycle checkpoint response mediated by ATM. DNA Repair (Amst.) 2009;8:1264–1272. doi: 10.1016/j.dnarep.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Horton JK, Stefanick DF, Kedar PS, Wilson SH. ATR signaling mediates an S-phase checkpoint after inhibition of poly(ADP-ribose) polymerase activity. DNA Repair (Amst.) 2007;6:742–750. doi: 10.1016/j.dnarep.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest after DNA methylating agent exposure. J. Biol. Chem. 2005;280:15773–15785. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- [20].Horton JK, Stefanick DF, Wilson SH. Involvement of poly(ADP-ribose) polymerase activity in regulating Chk1-dependent apoptotic cell death. DNA Repair (Amst.) 2005;4:1111–1120. doi: 10.1016/j.dnarep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- [21].Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- [22].Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum. Mol. Genet. 2004;13:2937–2945. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- [23].Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. αH2AX and cancer. Nature Reviews: Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
- [25].Kraakman-van der Zwet M, Overkamp WJ, Friedl AA, Klein B, Verhaegh GW, Jaspers NG, Midro AT, Eckardt-Schupp F, Lohman PH, Zdzienicka MZ. Immortalization and characterization of Nijmegen Breakage syndrome fibroblasts. Mutat. Res. 1999;434:17–27. doi: 10.1016/s0921-8777(99)00009-9. [DOI] [PubMed] [Google Scholar]
- [26].Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- [27].Trucco C, Oliver FJ, de Murcia G, Ménissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Herschleb J, Ananiev G, Schwartz DC. Pulsed-field gel electrophoresis. Nat. Protoc. 2007;2:677–684. doi: 10.1038/nprot.2007.94. [DOI] [PubMed] [Google Scholar]
- [29].Ager DD, Dewey WC. Calibration of pulsed field gel electrophoresis for measurement of DNA double-strand breaks. Int. J. Radiat. Biol. 1990;58:249–259. doi: 10.1080/09553009014551601. [DOI] [PubMed] [Google Scholar]
- [30].DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ, Jr., Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- [31].Iliakis GE, Metzger L, Denko N, Stamato TD. Detection of DNA double-strand breaks in synchronous cultures of CHO cells by means of asymmetric field inversion gel electrophoresis. Int. J. Radiat. Biol. 1991;59:321–341. doi: 10.1080/09553009114550311. [DOI] [PubMed] [Google Scholar]
- [32].Joshi N, Grant SG. DNA double-strand break damage and repair assessed by pulsed-field gel electrophoresis. Methods Mol. Biol. 2005;291:121–129. doi: 10.1385/1-59259-840-4:121. [DOI] [PubMed] [Google Scholar]
- [33].Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- [34].Stamato TD, Denko N. Asymmetric field inversion gel electrophoresis: a new method for detecting DNA double-strand breaks in mammalian cells. Radiat. Res. 1990;121:196–205. [PubMed] [Google Scholar]
- [35].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- [36].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- [37].Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kedar PS, Stefanick DF, Horton JK, Wilson SH. Interaction between PARP-1 and ATR in mouse fibroblasts is blocked by PARP inhibition. DNA Repair (Amst.) 2008;7:1787–1798. doi: 10.1016/j.dnarep.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gradzka I, Iwanenko T. A non-radioactive, PFGE-based assay for low levels of DNA double-strand breaks in mammalian cells. DNA Repair (Amst.) 2005;4:1129–1139. doi: 10.1016/j.dnarep.2005.06.001. [DOI] [PubMed] [Google Scholar]
- [40].Rydberg B. Radiation-induced heat-labile sites that convert into DNA double-strand breaks. Radiat. Res. 2000;153:805–812. doi: 10.1667/0033-7587(2000)153[0805:rihlst]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [41].Hoglund E, Blomquist E, Carlsson J, Stenerlow B. DNA damage induced by radiation of different linear energy transfer: initial fragmentation. Int. J. Radiat. Biol. 2000;76:539–547. doi: 10.1080/095530000138556. [DOI] [PubMed] [Google Scholar]
- [42].Cedervall B, Wong R, Albright N, Dynlacht J, Lambin P, Dewey WC. Methods for the quantification of DNA double-strand breaks determined from the distribution of DNA fragment sizes measured by pulsed-field gel electrophoresis. Radiat. Res. 1995;143:8–16. [PubMed] [Google Scholar]
- [43].Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009 doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pachkowski BF, Tano K, Afonin V, Elder RH, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in PARP-1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP-1-proficient cells exposed to MMS. Mutat. Res. 2009;671:93–99. doi: 10.1016/j.mrfmmm.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol. Med. 2005;11:456–463. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- [48].Plummer ER, Calvert H. Targeting poly(ADP-ribose) polymerase: a two-armed strategy for cancer therapy. Clin. Cancer. Res. 2007;13:6252–6256. doi: 10.1158/1078-0432.CCR-07-0617. [DOI] [PubMed] [Google Scholar]