Abstract
The first committed step in the classical biosynthetic route to menaquinone (vitamin K2) is a Stetter-like conjugate addition of α-ketoglutarate with isochorismate. This reaction is catalyzed by the thiamine diphosphate and metal-ion-dependent 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase (MenD). The medium-resolution (2.35 Å) crystal structure of Bacillus subtilis MenD with cofactor and Mn2+ has been determined. Based on structure–sequence comparisons and modeling, a two-stage mechanism that is primarily driven by the chemical properties of the cofactor is proposed. Hypotheses for the molecular determinants of substrate recognition were formulated. Five basic residues (Arg32, Arg106, Arg409, Arg428, and Lys299) are postulated to interact with carboxylate and hydroxyl groups to align substrates for catalysis in combination with a cluster of non-polar residues (Ile489, Phe490, and Leu493) on one side of the active site. The powerful combination of site-directed mutagenesis, where each of the eight residues is replaced by alanine, and steady-state kinetic measurements has been exploited to address these hypotheses. Arg409 plays a significant role in binding both substrates while Arg428 contributes mainly to binding of α-ketoglutarate. Arg32 and in particular Arg106 are critical for recognition of isochorismate. Mutagenesis of Phe490 and Ile489 has the most profound influence on catalytic efficiency, indicating that these two residues are important for binding of isochorismate and for stabilizing the cofactor position. These data allow for a detailed description of the structure–reactivity relationship that governs MenD function and refinement of the model for the catalytic intermediate that supports the Stetter-like conjugate addition.
Abbreviations: CoA, coenzyme A; PDB, Protein Data Bank; SAD, single-wavelength anomalous diffraction; SEPHCHC, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate; SeMet, selenomethionine; ThDP, thiamine diphosphate; PEG, polyethylene glycol
Keywords: crystal structure, enzyme mechanism, menaquinone biosynthesis, thiamine diphosphate cofactor
Introduction
Menaquinone, or vitamin K2, is a lipid-soluble molecule with distinct functions according to the type of organism in which it is found. Its chemical properties are exploited in the electron transport (respiratory) chain in Gram-positive aerobes and facultative Gram-negative bacteria under anaerobic conditions.1 Gram-negative bacteria use menaquinone only under anaerobic conditions, using ubiquinone under aerobic conditions.2 In mammals, which acquire menaquinone from intestinal microflora and diet, it is an important cofactor for glutamate γ-carboxylation, an essential posttranslational modification of proteins involved in blood coagulation, bone metabolism, and vascular biology.3
Menaquinone biosynthesis has been most studied in Escherichia coli.1 The starting point is chorismate, the branch point intermediate of the Shikimate pathway.4 Chorismate is first isomerized to isochorismate and then converted, in three stages, to o-succinyl-1-benzoate. Two further enzyme-catalyzed reactions, involving ATP and coenzyme A (CoA), lead to a CoA-naphthalene derivative before a recently identified thioesterase removes the CoA moiety to release 1,4-dihydroxy-2-napthanoate.5 Prenylation, followed by methylation, then produces menaquinone. An alternative and distinct route to menaquinone exists in a small subset of bacteria such as Helicobacter pylori.6 The starting point remains chorismate but the intermediates include an inosine derivative, futalosine.
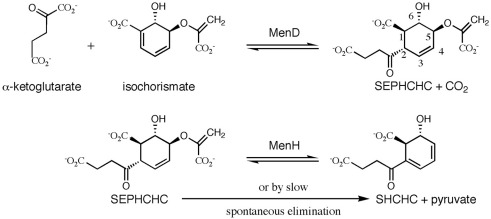
Our interest centers on 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate (SEPHCHC) synthase (EC 2.2.1.9) called MenD, the enzyme that catalyzes the first committed step of the classical menaquinone biosynthetic route (Fig. 1). Initially, the thiamine diphosphate (ThDP)-dependent MenD was misassigned as a bifunctional 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase7 catalyzing decarboxylation of α-ketoglutarate and then ligation of a second substrate, isochorismate, to produce SEPHCHC. Cleavage of the pyruvate moiety then produced 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate. Subsequent studies revealed that MenD is a monofunctional SEPHCHC synthase and the only enzyme known to catalyze addition of a ThDP intermediate to the β-carbon of a second substrate, in a Stetter-like reaction, which is a 1,4-conjugate addition of an aldehyde to a β-unsaturated compound.8 A complication in previous studies was the spontaneous release of pyruvate from SEPHCHC giving the misleading perception that a bifunctional enzyme was present.9,10 Jiang et al. showed that the enzyme encoded by the menH gene is actually responsible for the catalytic cleavage of pyruvate from SEPHCHC (Fig. 1).9
Fig. 1.
The reactions catalyzed by MenD and MenH in menaquinone biosynthesis.
The majority of genes that code for enzymes involved in menaquinone biosynthesis, including menD, have been shown by gene knockout to be essential in Bacillus subtilis.11 Such methods also indicate essentiality of MenD in Haemophilus influenzae12 and Mycobacterium tuberculosis.13 Since these are two important pathogens, then the understanding of MenD function and activity has potential to inform on early-stage drug discovery research.14
The structure of MenD from E. coli in complex with its cofactor ThDP,15 the apo structure of EcMenD,16 and a complex with oxoglutaric acid in the absence of cofactor17 have been determined. In the latter structure, the ligand binds at the site normally occupied by the ThDP diphosphate and so does not provide any information about substrate binding. We now report the medium-resolution crystal structure of the B. subtilis enzyme (BsMenD), solved by targeting the anomalous dispersion signal from a selenomethionine (SeMet) derivative. A comparison of structures and sequences of MenD orthologues guided molecular modeling, which has allowed us to generate hypotheses concerning the functional roles for eight residues in the active site. A detailed kinetic analysis of the wild-type enzyme and eight point mutations serve to address questions concerning recognition and processing of substrate and have allowed us to elucidate the structure–reactivity relationship that governs MenD activity.
Results and Discussion
General comments
BsMenD crystallizes in an orthorhombic system, space group P212121, with four polypeptide chains, labeled A to D in the asymmetric unit. The mass of protein in the tetramer is about 256.4 kDa. The structure was solved by use of single-wavelength anomalous diffraction (SAD) methods and refined to 2.35 Å resolution. The crystallographic statistics and model geometry (Table 1) indicate that the analysis has produced an acceptable medium-resolution model. Continuous and well-defined electron density was observed for each polypeptide chain from Thr2 to Leu580, except for chain C where the final two residues were not observed. Overlay of the chains using Secondary-Structure Matching20 gives root-mean-square deviations (r.m.s.d.'s) in the range 0.22–0.30 Å, indicating that the subunits are, within the errors associated with a medium-resolution structure, essentially identical. We therefore detail only subunit A, unless stated otherwise.
Table 1.
Crystallographic and model geometry statistics for BsMenD
| SeMet derivative | Native | |
|---|---|---|
| Resolution range (Å) | 50–3.0a | 68.8–2.35 |
| Unit cell dimensions: a, b, c (Å) | 100.1, 152.6, 158.1 | 100.2, 153.0, 158.5 |
| Space group | P212121 | P212121 |
| Total/unique reflections | 633,457/93,394 | 720,895/101,958 |
| Redundancy/completeness (%)b | 6.8c/99.7 (99.4) | 7.1/100 (100) |
| Wilson B (Å2) | 23.5 | 41.0 |
| 〈I/σ(I)〉 | 16.5 (7.1) | 17.3 (4.1) |
| Rmerge | 10.4 (27.5) | 10.9 (39.4) |
| Protein residues (A/B/C/D) | 579/579/577/579 | |
| Water molecules/ThDP/Mn2+ | 1075/4/4 | |
| SO42−/ethane-1,2-diol | 8/4 | |
| Rwork/Rfree/DPI (Å)d | 17.3/22.9/0.23 | |
| Average B-factors (Å2) | ||
| Overall/side chain/main chain | ||
| Subunit A | 31.3/31.8/30.8 | |
| Subunit B | 35.9/36.3/35.5 | |
| Subunit C | 35.3/35.7/34.9 | |
| Subunit D | 30.4/30.8/30.1 | |
| Water molecules/ThDP/Mn2+ | 34.7/23.0/25.8 | |
| SO42−SO42-/ethane-1,2-diol | 20.2/28.4 | |
| r.m.s.d. bond lengths (Å)/angles (°) | 0.009/1.32 | |
| Ramachandran plot analysis (%)e | ||
| Favorable/outliers | 96.8/0.6 | |
Overall structure
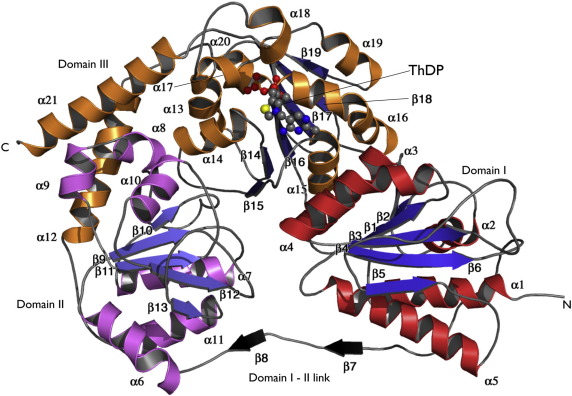
MenD shows the three-domain architecture typical of ThDP-dependent enzymes (Fig. 2).21 Sequence and structural analyses place MenD in the pyruvate oxidase class of these enzymes, where domain I contains the cofactor pyrimidine-binding motif, domain III binds the diphosphate, and the central domain has variable, often ill-defined, function. In BsMenD, domain I spans approximately the first 190 residues. An ordered linker region leads into domain II, which consists of residues 205–345. A smaller linker then leads into domain III, which starts at approximately residue 355 and continues to the C terminus (Fig. 2). Each domain consists of a parallel β-sheet sandwiched between several α-helices; in BsMenD, the central domain has five strands and there are six in domains I and III.
Fig. 2.
The secondary and domain structure of BsMenD. A ribbon diagram of the enzyme subunit showing the elements of secondary structure with atoms of ThDP depicted as spheres according to atom type: C, gray; N, blue; O, red; S, yellow; P, orange. The N- and C-terminal positions are labeled, as are the elements of secondary structure.
Quaternary structure
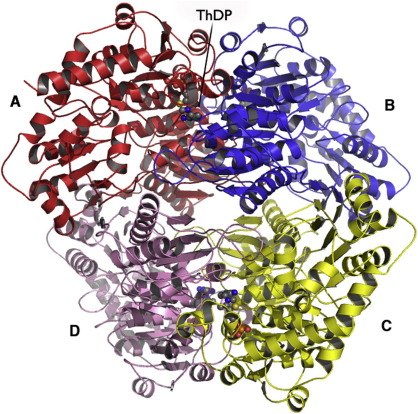
Oligomerization is essential for the function of ThDP-dependent enzymes, with each active site formed from two subunits.21 Most of these enzymes function as either dimers or tetramers. Size-exclusion chromatography indicated that BsMenD consists of a mixture of dimer and tetramer in solution, with the dimer being much more abundant. The crystal structure of BsMenD consists of a dimer-of-dimers (Fig. 3); a similar result was found for EcMenD, which is predominantly dimeric in solution15 and then forms a dimer-of-dimers when crystallized.15–17
Fig. 3.
The BsMenD tetramer. The tetramer is shown as a van der Waals surface with subunits labeled and colored differently. Subunit A is in the same orientation as in Fig. 2.
Analysis using the Protein Interfaces, Surfaces and Assemblies server22 suggests that the dimer-of-dimers is the most stable oligomeric form, burying approximately 23% (5310 Å2) of the surface area of each subunit (23,100 Å2). The principal interface, between subunits A and B (and C and D), buries about 14% of the surface area of each subunit. The A:B interface covers an area of about 3130 Å2, and the C:D interface covers approximately 3160 Å2. Around 30 hydrogen bonds in addition to the salt bridges formed between Arg53 and Asp55 with their counterparts in the second dimer form the most important interactions in this interface (data not shown). Fewer hydrogen bonds link the dimer-of-dimers arrangement where the pairs of subunits involved are A:D and B:C. This interface is formed primarily by residues located in the linker between domains I and II. A short three-amino-acid stretch of β-strand is present at the start of domain II, which forms an extension of the parallel β-sheet of domain II across the interface. In addition, another three-residue stretch (205–207) forms a two-stranded antiparallel β-sheet across the interface. Approximately 7% of the surface area of a subunit is buried at this interface; 1610 and 1620 Å2 for the A:D and B:C interface regions, respectively. The A:C and B:D interfacial regions occlude about 700 and 710 Å2, respectively, each representing approximately 2% of the subunit surface area.
Comparison with EcMenD
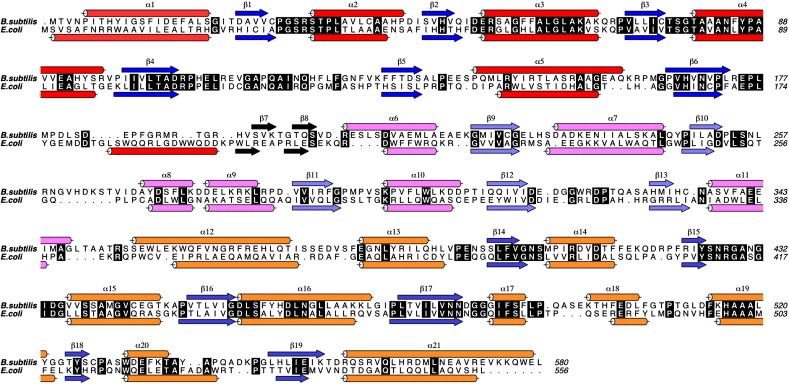
The MenD proteins from different organisms share little sequence identity, typically in the range 20–30%. Such a low level of sequence conservation despite the same function appears to be a common feature of enzymes that are dependent on ThDP.15 BsMenD has 28% overall sequence identity to the E. coli protein. The sequence identity is higher for domains I and III (32% and 33%, respectively) compared to the central domain (16%). Nevertheless, the structures of the two enzymes are closely related, with the majority of secondary-structure elements conserved in each domain (Fig. 4). One exception is the final helix in domain I of EcMenD, which is replaced by a loop and β7 in BsMenD (Fig. 4). This region does not appear to have any functional role.
Fig. 4.
The primary and assigned secondary structure of BsMenD and EcMenD. α-Helices are shown as cylinders and β-strands as arrows, colored according to the domain to which they have been assigned (see Fig. 2). Residues with a black background are strictly conserved.
Subunit A from BsMenD overlays with a subunit from EcMenD [Protein Data Bank (PDB) code: 2jlc] with an r.m.s.d. of approximately 2.0 Å (over 503 aligned Cα). The closeness of the structures extends to the dimer (r.m.s.d. of 2.0 Å over 1006 aligned Cα) and the tetramer (2.4 Å over 1996 Cα). Individual domains align more closely; domain I of BsMenD overlays with an r.m.s.d. of 1.3 Å onto domain I of EcMenD (173 aligned Cα), domain II with an r.m.s.d. of 1.8 Å (117 aligned Cα), and domain III with an r.m.s.d. of 1.7 Å (over 196 aligned Cα). The loop containing residues responsible for binding to the diphosphate group of ThDP (approximately residues Gly486 to Pro509) is disordered in EcMenD in the absence of cofactor16 but ordered in the holo-enzyme.15 The conformation of this cofactor-binding loop in holo-BsMenD is similar to that of the corresponding loop in the holo-Ec structure (data not shown).
The low level of sequence identity between BsMenD and EcMenD serves to highlight the critical determinants of structure and function, and we placed a particular emphasis on conservation when analyzing potential contributions that residues make to enzyme activity. In the following text, unless stated otherwise, we confine our discussion to residues that are strictly conserved between BsMenD and EcMenD.
Cofactor binding and the active site
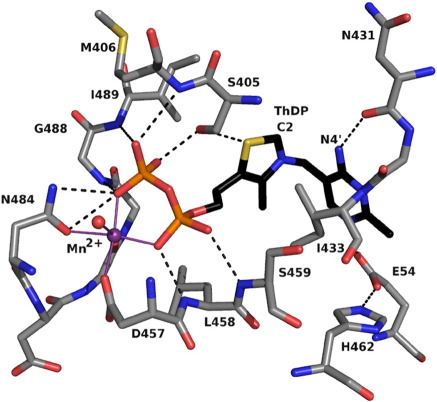
Dimerization is essential for formation of the cofactor binding and active site, with residues from domain I of subunit A and domain III from subunit B forming one site (and vice versa, leading to two active sites per dimer, Fig. 3). A metal cation, assigned as Mn2+, is bound and helps to position the diphosphate group of ThDP and so tether one part of the cofactor to the protein. This is a common feature of ThDP binding and similar to what is observed in EcMenD.15 In BsMenD, the side chains of Asp457 and Asn484, the main-chain carbonyl of Gly486, a water molecule, and two oxygen atoms from the diphosphate group form an octahedral coordination sphere about the metal ion (Fig. 5).
Fig. 5.
ThDP interactions with BsMenD. Atoms are colored as follows: C of ThDP, black; C of protein, gray for subunits A and B; P, orange; N, blue; O, red; S, yellow; Mn2+, purple. Purple continuous lines represent coordination of the transition metal ion, and black broken black lines represent potential hydrogen-bonding interactions.
Almost all ThDP-dependent enzymes have a glutamate interacting with N1′ of the pyrimidine; Glu54 fulfills this role in BsMenD. Mutagenesis of the corresponding residue, Glu55, in EcMenD completely abolished enzyme activity.7 The main-chain amide of Ile433 and carbonyl of Asn431, which participate in hydrogen-bonding interactions with N3′ and N4′, respectively, are from the partner subunit. The side chain of Ile433, a residue strictly conserved in MenD sequences,15 is placed to interact with both the pyrimidine and thiazolium rings helping to hold the cofactor in a V shape, bent at C7′. All ThDP-dependent enzymes that have been structurally characterized have a large hydrophobic side chain in exactly this position. The environment of the cofactor forces N4′ on the pyrimidine ring into close contact with C2 in the thiazolium ring (3.1 Å, Fig. 5), activating the cofactor to contribute to catalysis.
ThDP binding involves contributions from a pair of subunits. The diphosphate is bound between the N-terminal section of α14, strands β16 and β17, and the loop between β17 and α17. The pyrimidine moiety is positioned near the loop linking β15–α15. These are all from one subunit. The partner subunit contributes to binding the pyrimidine with contributions from the C-terminal sections of β1 and β3, the loop between β3 and α4, together with an important contribution from Glu54 at the N terminus of α3. The thiazolium group is sandwiched between Ile433 (on the loop linking β15 with α15) and Phe490 (α17).
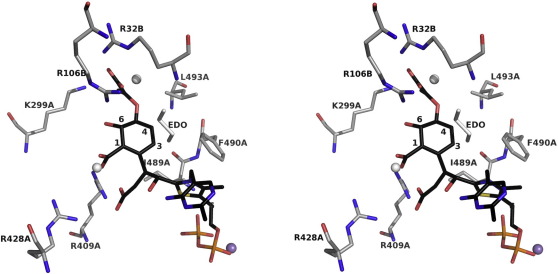
The narrow crevice where substrates bind is likewise formed by a dimer. One subunit contributes three α-helical segments (α10, α14, and α17), which line one side of the cavity. Three non-helical segments (the β1–α2 turn, the loop between β6 and β7, and the loop following β4) are provided by the partner subunit. The active site is polar, primarily basic due to the presence of four arginine residues (32A, 106A, 409B, and 428B) and a lysine (299B). A hydrophobic patch is formed by Ile489B, Phe490B, and Leu493B (Fig. 6). Seven of these eight amino acids are strictly conserved, and one, Lys299, is conserved or replaced by arginine in more than 90% of MenD sequences.
Fig. 6.
A model for the post-decarboxylation covalent adduct formed after ThDP C2 has reacted with α-ketoglutarate and then isochorismate. A stereoview into the active site with atoms colored as follows: C of ThDP, black; C of protein, gray; O, red; N, blue; P, orange; Mn2+, purple. The eight key residues discussed in the text are labeled. Two water molecules are depicted as gray spheres and ethane-1,2-diol is shown as gray sticks, labeled EDO.
Ser405 OG donates a hydrogen bond to one of the phosphate groups of ThDP yet is placed about 3.1 Å from the thiazolium S. This alignment of functional groups may induce a polarization effect on the S atom and contribute to generation of an ylide.
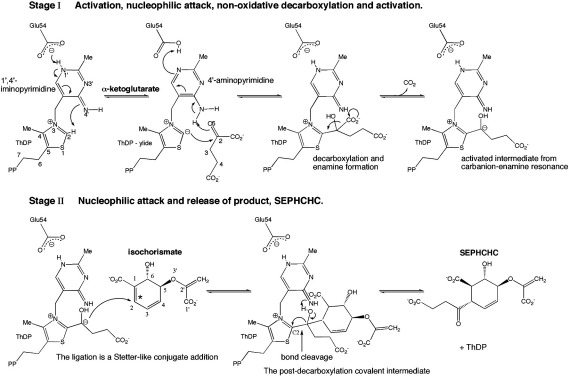
A proposed mechanism
The structure of MenD and our model (Fig. 6) of the activated intermediate–isochorismate complex suggested that the actual enzyme mechanism is driven by the chemical properties of the cofactor ThDP. A two-stage mechanism that corresponds to reactions with each of the substrates can be proposed (Fig. 7).15 The absence of any residue that can act as a general acid/base indicates that the cofactor N4′ is a critical component of the mechanism. Such an observation is consistent with structural and mechanistic studies of another ThDP-dependent enzyme, N2-(2-carboxyethyl)arginine synthase.23
Fig. 7.
A two-stage mechanism for catalysis by BsMenD. An asterisk (⁎) marks the isochorismate C2, which is attacked by the carbanion intermediate. The post-decarboxylation covalent intermediate in stage II is the structure that has been modeled and is shown in Fig. 6.
Our model for activity predicts that five polar residues interact with carboxylates and hydroxyl group of substrates to align reagents for catalysis and that non-polar components of isochorismate interact with the hydrophobic patch. The structural studies, sequence comparisons, molecular modeling, and mechanistic considerations lead us to conclude that the environment provided by MenD for the cofactor and the chemical properties of ThDP provide the functionality for catalysis. A first approximation of how substrates bind can then be used to address the molecular recognition of these substrates.
Mutagenesis and kinetic studies
Attempts to soak ligands such as α-ketoglutarate and salicylic acid into the crystals of BsMenD and thus derive structural data directly relevant to substrate recognition resulted in the loss of diffraction. Co-crystallization experiments were also carried out but either no crystals were observed or, where analyzed, there was no electron density that could be attributed to these ligands (data not shown). A model of the post-decarboxylation covalent adduct formed after reaction of ThDP with first α-ketoglutarate then isochorismate (Figs. 6 and 7) was constructed, guided by the principle that important features relevant to substrate recognition and catalysis should be conserved between BsMenD and EcMenD. We also noted that the positions of two water molecules and ethane-1,2-diol in the active sites serve to identify potential ligand interacting sites and provide a hint of where functional groups of isochorismate might be placed (Fig. 6).
Modeling of substrate binding, sequence, and structure comparisons identified eight residues that are potentially important with respect to substrate recognition. The first stage of the reaction involves the formation of a covalent adduct between the cofactor and α-ketoglutarate; the formation of such species is well established in ThDP-dependent enzymes24 and the adduct structure has been captured crystallographically.25 The driving force for this stage largely comes from the geometry of the cofactor in the binding site, which brings N4′ of the pyrimidine ring and C2 of the thiazolium ring into close proximity. N4′ abstracts the acidic proton from C2, generating a carbanion ylide, which can then attack the first substrate, α-ketoglutarate, to form the first covalent intermediate (Fig. 7). Arg409 and Arg428 (Fig. 5) are well placed to interact with the α-ketoglutaric acid part of this adduct. Mutation of these residues to alanine had a significant effect on the binding of α-ketoglutarate and the rate of reaction (Table 2). The Arg409Ala mutation resulted in a 7-fold increase in Km and 5-fold decrease in kcat. The Arg428Ala mutation gave an increase in Km of more than 20-fold and a 4-fold decrease in kcat. These mutations resulted in comparable kcat/Km values, which are significantly decreased, compared to the wild-type enzyme. Assay of these two mutants with respect to isochorismate indicates that Arg409 has a greater influence on the second substrate binding since the Km is increased 8-fold with no change to Km of the Arg428Ala mutant (Table 3). Although catalytic efficiency is compromised in both cases, ascribed to poorer binding of α-ketoglutarate in the first stage of the reaction, the Arg409Ala mutant is affected to a much larger degree with a reduction in kcat/Km by 2 orders of magnitude. These data suggest then that Arg409 is important for binding of both substrates whereas Arg428 primarily contributes to the binding of α-ketoglutarate. This is consistent with the crystal structure since the side chain of Arg409 is about 1 Å closer to the thiazolium group than Arg428, at the base of the active site cleft directed towards the cavity where the second substrate binds.
Table 2.
Steady-state kinetic parameters with respect to α-ketoglutarate
| Protein | Km (μM) | kcat (min− 1) | kcat/Km (μM− 1 min− 1) | Relative kcat/Km |
|---|---|---|---|---|
| Wild type | 22 ± 5 | 14 ± 2 | 0.64 ± 0.3 | 1 |
| R409A | 159 ± 18 | 2.7 ± 0.3 | 0.16 ± 0.02 | 0.03 |
| R428A | 514 ± 54 | 3.4 ± 0.2 | 0.007 ± 0.001 | 0.01 |
| R32A′ | 22 ± 4 | 4.8 ± 0.4 | 0.22 ± 0.02 | 0.3 |
| R106A′ | 7.7 ± 2.2 | 0.6 ± 0.2 | 0.09 ± 0.03 | 0.1 |
| K299A | 22 ± 4 | 22 ± 2 | 1 ± 0.2 | 1.5 |
| L493A | 18 ± 1 | 10 ± 1 | 0.5 ± 0.1 | 0.8 |
| I489A | 346 ± 10 | 5.7 ± 0.2 | 3.1 ± 0.1 | 0.01 |
| F490A | 274 ± 12 | 1.1 ± 0.1 | 0.0039 ± 0.0002 | 0.006 |
Residues marked with ′ are located in the second subunit forming the active site.
Table 3.
Steady-state kinetic parameters with respect to isochorismate
| Protein | Km (μM) | kcat (min− 1) | kcat/Km (μM− 1 min− 1) | Relative kcat/Km |
|---|---|---|---|---|
| Wild type | 1.1 ± 0.2 | 19 ± 1 | 19 ± 3 | 1 |
| R409A | 8.5 ± 1.2 | 1.4 ± 0.1 | 0.16 ± 0.02 | 0.009 |
| R428A | 1.1 ± 0.5 | 2.8 ± 0.8 | 2.8 ± 0.8 | 0.15 |
| R32A′ | 49 ± 7 | 11 ± 1 | 0.22 ± 0.01 | 0.01 |
| R106A′ | 41 ± 4 | 0.7 ± 0.2 | 0.018 ± 0.01 | 0.00095 |
| K299A | 8.4 ± 1.2 | 25 ± 2 | 3.0 ± 0.3 | 0.16 |
| L493A | 39 ± 1 | 19 ± 1 | 0.49 ± 0.03 | 0.03 |
| I489A | 27 ± 1 | 5.7 ± 0.2 | 0.21 ± 0.01 | 0.01 |
| F490 | 29 ± 3 | 1.7 ± 0.1 | 0.058 ± 0.01 | 0.003 |
Residues marked with ′ are located in the second subunit forming the active site.
Arg32 and Arg106 are in close proximity on one side of the active-site entrance; Lys299 is about 8 Å distant on the other side of the cleft. Arg106 is the closest to the thiazolium group at a distance of about 9 Å. The model of the covalent or activated intermediate suggests that this triumvirate of basic residues is likely to have a small influence, if any, on α-ketoglutarate binding. The kinetic data (Table 2) indeed suggest that this is the case. There is effectively no change to Km with respect to α-ketoglutarate for the Arg32Ala and Lys299Ala mutants, and Km is reduced by a factor of 3 for the Arg106Ala protein. With respect to isochorismate (Table 3), the Lys299Ala mutant displays an increase in Km of about 8-fold but with a comparable kcat value, whereas the mutations to the two arginine residues increase Km between 40- and 50-fold and reduce kcat significantly. The reduction in catalytic efficiency is most pronounced for the Arg106 mutation. These data are consistent with Arg106 performing an important role in recognition and binding of the second substrate.
Three hydrophobic residues, Ile489, Phe490, and Leu493, cluster on one side of the active site (Fig. 6). Of these, Leu493 is most distant, about 9.5 Å from the thiazolium. The kinetic data indicate a minor perturbation of BsMenD activity when Leu493 is mutated to alanine and this is mainly due to an increase in Km for isochorismate. Leu493 does not therefore appear to make a particularly significant or direct contribution to reactivity. The changes to kinetic properties may be due to perturbation of the hydrophobic cluster since the other two residues appear more important. Mutation of Ile489 and Phe490 increases the Km for α-ketoglutarate 16-fold and 12-fold, respectively. The Km for isochorismate is increased by 24- and 25-fold, respectively, and there is a concomitant reduction in catalytic efficiency. Ile489 is adjacent to the site where α-ketoglutarate reacts with the thiazolium group, 3.8 Å distant from the sulfur atom. Phe490 participates in van der Waals interactions with the thiazolium and Ile489. These two residues appear therefore to help bind ThDP. Ile489 is likely to interact with the aliphatic part of the intermediate formed after α-ketoglutarate has reacted with ThDP and, together with Phe490, to interact with the hydrophobic component of the isochorismate ring, C3 and C4 (Fig. 6).
In the second stage of the reaction, the carbanion adduct attacks isochorismate C2 (Fig. 7). Two further arginine residues (Arg32 and Arg106) and a lysine (Lys299) line one side of the putative isochorismate-binding region. A hydrophobic patch consisting of Ile489, Phe490, and Leu493 (Fig. 6) lines the opposite side of the active site. Kinetic characterization of Ala mutants of each of these residues is summarized in Tables 2 and 3, with respect to both substrates. Mutation has little effect on the binding of ThDP (data not shown).
Lys299, the only highlighted residue located in the central domain, appears to have little influence on the reaction. Mutation of any of the arginine residues has a similar effect on the binding of isochorismate; however, alteration to Arg106 has a greater impact upon the rate of reaction. This residue is located at the start of the long loop linking β4 with β5; in addition to its putative role in the reaction, Arg106 is well placed to form a hydrogen bond with the main-chain carbonyl of Pro116, helping to orient this loop, which forms part of the binding pocket. Removal of this interaction could destabilize the loop. Of the hydrophobic residues, mutation of Phe490 has the most deleterious effect, with respect to both substrates, indicating particular importance in defining the overall shape and size of the binding pocket.
Concluding remarks
This combination of single crystal diffraction methods with site-directed mutagenesis and kinetic analyses has allowed us to elucidate the structure–reactivity relationship that governs MenD activity. The enzyme provides the environment to bind the cofactor in a specific manner such that the mechanism is driven by the chemical properties of ThDP. There is no requirement for any amino acid to contribute directly to the mechanism by abstraction or provision of a proton. Rather, the residues in the active site are important for the binding and orientation of substrates allowing MenD to accomplish catalysis. The mutagenesis data are consistent with such a conclusion since they indicate that the most profound effect on catalytic efficiency is derived from alteration of non-polar residues implicated in substrate recognition.
Materials and Methods
Reagents, preparation of recombinant expression systems, and protein purification
Chemicals and materials were sourced from Sigma-Aldrich and VWR International except where stated otherwise. The menD gene was amplified by PCR from B. subtilis genomic DNA (ATCC strain 23857, LGC Standards Office, UK) with 5′-CTCGAGATGACAGTGAACCCAATTAC-3′ and 5′-GGATCCTTACAGTTCCCATTGTTTTTTC-3′ as the forward and reverse primers, respectively (Thermo Scientific). These oligonucleotides include 5′ XhoI and 3′ BamHI restriction sites, respectively, indicated in boldface. The PCR product was ligated into TOPO-BLUNT-II (Invitrogen) and then sub-cloned into a modified pET15b vector (Novagen), which produces a histidine-tagged protein with a tobacco etch virus protease site. The hexa-His tag, tobacco etch virus cleavage site, and additional residues from the choice of cloning sites added a total of 24 extra amino acids at the protein N terminus. The native and SeMet-labeled proteins were obtained using E. coli strains BL21(DE3) and the methionine auxotroph B834 (DE3) (Stratagene), respectively. Cells were grown in 1 L of Luria–Bertani media supplemented with 50 μg mL− 1 of carbenicillin for production of native protein, while for the SeMet protein, bacteria were cultured in minimal media (Molecular Dimensions) supplemented with SeMet following an established protocol.26 Gene expression was induced at 20 °C using 0.5 mM isopropyl-β-d-thiogalactopyranoside, and growth continued for 16 h at room temperature.
The same purification protocol was used for both native and SeMet BsMenD. Cells were harvested by centrifugation for 25 min at 40,000g at 4 °C, re-suspended in lysis buffer (50 mM Tris–HCl, pH 7.5, 250 mM NaCl, and 20 mM imidazole) containing DNase I (0.1 mg) and a single tablet of a cocktail of ethylenediaminetetraacetic acid-free protease inhibitors (Roche), and lysed using a French press at 1000 psi. Insoluble debris was separated by centrifugation at 39,000g for 25 min at 4 °C, and the soluble fraction was loaded onto a 5-mL HisTrap HP column (GE Healthcare) pre-charged with Ni2+. A linear concentration gradient was applied to the column and BsMenD eluted at a concentration of 250 mM imidazole. Fractions were analyzed using SDS-PAGE, and those containing BsMenD were pooled. The protein, still carrying the affinity tag, was further purified using a Superdex 200 26/60 size-exclusion column (GE Healthcare) equilibrated with 50 mM Tris–HCl and 250 mM NaCl, pH 7.5. This column had previously been calibrated with molecular mass standards, blue dextran (> 2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29.5 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa) (GE Healthcare; data not shown). Selected fractions were pooled and dialyzed into 20 mM Tris–HCl and 50 mM NaCl, pH 7.5, and concentrated using Amicon Ultra devices (Millipore) to 8 mg mL− 1 for the native protein and 4 mg mL− 1 for the SeMet sample. The protein concentration was determined spectrophotometrically using a theoretical extinction coefficient of 52,745 M− 1 cm− 1 at 280 nm calculated using ProtParam.27 The full incorporation of SeMet and high level of sample purity for both samples was verified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and SDS-PAGE (data not shown). The native protein expressed at levels in excess of 150 mg L− 1 of culture; yields of the SeMet protein were lower, typically about 60 mg L− 1, due to the lower bacterial cell mass obtained in minimal media.
The native protein typically eluted from the gel-filtration column in two peaks: a major peak with a mass of approximately 150 kDa and a minor peak of with a mass of approximately 290 kDa. The theoretical mass of a BsMenD subunit is approximately 64.1 kDa, and therefore, the peaks correspond to a dimer and tetramer, respectively. Approximately 10% of the total protein was observed in the minor peak, which was discarded, and the major peak was used subsequently.
Crystal growth, structure determination, and refinement
Crystallization and data collection
Sitting and hanging drop vapor diffusion methods were used to screen and optimize crystallization conditions, respectively, based on commercially available screening sets. The BsMenD:ThDP:Mn2+ complex crystallized with the N-terminal hexa-His tag intact whereas the apo-enzyme did not crystallize at all. Holo-enzyme was prepared by incubation with 2 mM MnCl2 and 0.5 mM ThDP, and crystals were obtained using a reservoir (500 μL) comprising 35% polyethylene glycol (PEG) 1.5K, 0.1 M Tris, pH 8, and 0.25 M (NH4)2SO4, for the native protein and 24% PEG 3K, 0.1 M Tris, pH 8, and 0.2 M Li2SO4 for SeMet BsMenD. In both cases, the best crystals were obtained from drops consisting of 1 μL protein and 2 μL of reservoir. Crystals appeared as rectangular prisms, consistently with a notch at one end. The maximum dimensions of native crystals were 0.3 mm × 0.1 mm × 0.1 mm, and the SeMet derivative crystals were smaller at 0.2 mm × 0.1 mm × 0.1 mm.
Crystals were flash cooled in liquid nitrogen, mounted on a goniostat, and maintained at − 173 °C in a flow of cooled nitrogen, and diffraction properties were characterized with a Rigaku Micromax 007 rotating anode R-AXIS IV++ image plate system. The best samples were stored for use at the European Synchrotron Radiation Facility (Grenoble, France). Crystals of both proteins, particularly the SeMet crystals, were radiation sensitive. Native data were collected on beam line ID23-1, and SAD data were measured on beam line BM14, both with ADSC Q315 CCD detectors. The SAD data were collected at the Se K-absorption edge, determined using X-ray Absorption Near Edge Structure spectroscopy to provide the f″ maximum at a wavelength of 0.97864 Å. Data were integrated using XDS28 and scaled with XSCALE (SeMet)28 or SCALA (native),29 and statistics are presented in Table 1.
Structure solution and refinement
Attempts to solve the BsMenD structure by molecular replacement using the EcMenD mode (PDB code: 2jlc) gave ambiguous results; the sequence identity shared between the two is 28%. Since the asymmetric unit was predicted to contain 68 methionine residues, based on four subunits per asymmetric unit, it was judged likely that a SeMet-SAD experiment would provide useful phase information.
Analysis of the SAD data using XPREP30 indicated that the anomalous dispersion signal/noise ratio reduced to a value of 1.0 at 3 Å resolution and these data were input to the program SOLVE.31 Initial phases were obtained by SAD phasing based on the positions of 30 selenium atoms. The initial figure of merit was 0.25 with a Z-score of 62.3. Density modification using RESOLVE32 with an estimated solvent content of 55% improved the figure of merit to 0.75. Automated map interpretation with RESOLVE produced a partial model consisting of approximately 1300 residues split over 52 polypeptide chains. This model was extended using Coot33 with the benefit of the EcMenD structure as a guide. The major secondary-structure elements of a single subunit were completed, and the resulting structure was used as a molecular replacement model for the native data, using Phaser.34 The four subunits present in the asymmetric unit were all located with Z-scores above 12, indicating correct solutions. Further manual rebuilding, the placement of the cofactor ThDP, Mn2+ and SO42− ions and waters using Coot, and incorporation of dual rotamer side chains interspersed with refinement using REFMAC5.35 A strong electron density feature was observed in each active site and this was successfully modeled and refined as ethane-1,2-diol, a precursor of and potential contaminant in PEGs.36 Strict non-crystallographic symmetry restraints were applied during the initial stages of refinement; these were relaxed and then removed as the refinement progressed. Model geometry statistics are presented in Table 1. Subunits A and D are involved in more contacts with symmetry-related molecules, leading to significantly lower B-factors for these chains (Table 1). Figures were prepared with Chemdraw (Adept Scientific), PyMOL,37 and ALINE.38
Mutagenesis and kinetic studies
The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to introduce eight point mutations into BsMenD. The primers used to generate the mutants are listed in Supplemental Table 1. Expression and purification of the mutants followed the procedures described above for the native protein. Preparation and purification of isochorismate followed previous protocols.39 Protein activity was measured by monitoring the decrease in absorption at 278 nm due to the consumption of isochorismate.8 All kinetic assays were carried out in 50 mM Tris–HCl, pH 7.8, with 20 mM NaCl and 5 mM MgSO4 at 23 °C. Circular dichroism measurements indicated that in each case, the mutated protein was still correctly folded (data not shown).
PDB accession numbers
Coordinates and structure factors have been deposited with accession code 2x7j.
Acknowledgements
This research was supported by awards from the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets, BBS/B/14434), The Wellcome Trust (grant numbers 082596 and 083481), and the Research Grants Council of the Hong Kong Special Administrative Region of the People's Republic of China (GRF601209). We thank the European Synchrotron Radiation Facility, Grenoble, France, for synchrotron beam time and excellent staff support.
Edited by G. Schulz
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2010.06.025
Appendix A.
Oligodeoxynucleotide primers used in site-directed mutagenesis modification of BsMenD.
References
- 1.Meganathan R. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic reactions. Vitam. Horm. 2001;61:173–218. doi: 10.1016/s0083-6729(01)61006-9. [DOI] [PubMed] [Google Scholar]
- 2.Soballe B., Poole R.K. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology. 1999;145:1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay P.K. Vitamin K-dependent γ-glutamylcarboxylation: an ancient posttranslational modification. Vitam. Horm. 2008;78:157–184. doi: 10.1016/S0083-6729(07)00008-8. [DOI] [PubMed] [Google Scholar]
- 4.Knaggs A.R. The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 2003;20:119–136. doi: 10.1039/b100399m. [DOI] [PubMed] [Google Scholar]
- 5.Widhalm J.R., van Oostende C., Furt F., Basset G.J. A dedicated thioesterase of the Hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc. Natl Acad. Sci. USA. 2009;106:5599–5603. doi: 10.1073/pnas.0900738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiratsuka T., Furihata K., Ishikawa J., Yamashita H., Itoh N., Seto H., Dairi T. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science. 2008;321:1670–1673. doi: 10.1126/science.1160446. [DOI] [PubMed] [Google Scholar]
- 7.Bhasin M., Billinsky J.L., Palmer D.R. Steady-state kinetics and molecular evolution of Escherichia coli MenD [(1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase], an anomalous thiamin diphosphate-dependent decarboxylase-carboligase. Biochemistry. 2003;42:13496–13504. doi: 10.1021/bi035224j. [DOI] [PubMed] [Google Scholar]
- 8.Stetter H. Catalyzed addition of aldehydes to activated double bonds—a new synthetic approach. Angew. Chem., Int. Ed. 1976;15:639–647. [Google Scholar]
- 9.Jiang M., Cao Y., Guo Z.F., Chen M., Chen X., Guo Z. Menaquinone biosynthesis in Escherichia coli: identification of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate as a novel intermediate and re-evaluation of MenD activity. Biochemistry. 2007;46:10979–10989. doi: 10.1021/bi700810x. [DOI] [PubMed] [Google Scholar]
- 10.Jiang M., Chen X., Guo Z.F., Cao Y., Chen M., Guo Z. Identification and characterization of (1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase in the menaquinone biosynthesis of Escherichia coli. Biochemistry. 2008;47:3426–3434. doi: 10.1021/bi7023755. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akerley B.J., Rubin E.J., Novick V.L., Amaya K., Judson N., Mekalanos J.J. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl Acad. Sci. USA. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 14.Hunter W.N. Structure-based ligand design and the promise held for antiprotozoan drug discovery. J. Biol. Chem. 2009;284:11749–11753. doi: 10.1074/jbc.R800072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson A., Fyfe P.K., Hunter W.H. Specificity and reactivity in menaquinone biosynthesis: the structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase) J. Mol. Biol. 2008;384:1353–1368. doi: 10.1016/j.jmb.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priyadarshi A., Saleem Y., Nam K.H., Kim K.S., Park S.Y., Kim E.E., Hwang K.Y. Structural insights of the MenD from Escherichia coli reveal ThDP affinity. Biochem. Biophys. Res. Commun. 2009;380:797–801. doi: 10.1016/j.bbrc.2009.01.168. [DOI] [PubMed] [Google Scholar]
- 17.Priyadarshi A., Kim E.E., Hwang K.Y. Structural and functional analysis of Vitamin K2 synthesis protein MenD. Biochem. Biophys. Res. Commun. 2009;388:748–751. doi: 10.1016/j.bbrc.2009.08.093. [DOI] [PubMed] [Google Scholar]
- 18.Cruickshank D.W.J. Remarks about protein structure precision. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 19.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 21.Duggleby R.G. Domain relationships in thiamine diphosphate-dependent enzymes. Acc. Chem. Res. 2006;39:550–557. doi: 10.1021/ar068022z. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Caines M.E., Elkins J.M., Hewitson K.S., Schofield C.J. Crystal structure and mechanistic implications of N2-(2-carboxyethyl)arginine sythase, the first enzyme in the clavulanic acid biosynthesis pathway. J. Biol. Chem. 2004;279:5685–5692. doi: 10.1074/jbc.M310803200. [DOI] [PubMed] [Google Scholar]
- 24.Kluger R., Tittmann K. Thiamin diposphate catalysis: enzymic and nonenzymic covalent intermediates. Chem. Rev. 2008;108:1797–1833. doi: 10.1021/cr068444m. [DOI] [PubMed] [Google Scholar]
- 25.Berthold C.L., Toyota C.G., Moussatche P., Wood M.D., Leeper F., Richards N.G., Lindqvist Y. Crystallographic snapshots of oxalyl-CoA decarboxylase give insights into catalysis by nonoxidative ThDP-dependent decarboxylases. Structure. 2007;15:853–861. doi: 10.1016/j.str.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Hall D.R., Gourley D.G., Leonard G.A., Duke E.B.H., Anderson L.A., Boxer D.H., Hunter W.N. The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli; a novel combination of domain folds. EMBO J. 1999;18:1435–1446. doi: 10.1093/emboj/18.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press Inc.; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 28.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 29.Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 30.Bruker . Bruker AXS Inc.; Madison, WI: 2004. XPREP. [Google Scholar]
- 31.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Ray W.J., Puvathingal J.M. A simple procedure for removing contaminating aldehydes and peroxides from aqueous solutions of polyethylene glycols and of nonionic detergents that are based on the polyoxyethylene linkage. Anal. Biochem. 1985;146:307–312. doi: 10.1016/0003-2697(85)90544-5. [DOI] [PubMed] [Google Scholar]
- 37.DeLano W.L. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System.http://pymol.sourceforge.net [Google Scholar]
- 38.Bond C.S., Schuttelkopf A.W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 39.Rusnak F., Liu J., Quinn N., Berchtold G.A., Walsh C.T. Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization and substrate specificity of isochorismate synthase. Biochemistry. 1990;29:1425–1435. doi: 10.1021/bi00458a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligodeoxynucleotide primers used in site-directed mutagenesis modification of BsMenD.