Summary
Mycobacterium tuberculosis has 10 universal stress proteins, whose function is unknown. However, proteomic and transcriptomic analyses have shown that a number of usp genes are significantly upregulated under hypoxic conditions and in response to nitric oxide and carbon monoxide, as well as during M. tuberculosis infection of macrophage cell lines. Six of these USPs are part of the DosR regulon and this, along with their expression pattern and the phenotypes of usp mutants in other bacterial species, suggests a potential role in the persistence and/or intracellular survival of Mtb. Knock-out mutants of individual usp genes encoding the USPs Rv1996, Rv2005c, Rv2026c and Rv2028c were generated and their growth and survival under hypoxic and other stress conditions examined. Although the majority of usp genes are highly induced in hypoxic conditions, mutation did not affect the long term survival of Mtb under these conditions, or in response to a range of stress conditions chosen to represent the environmental onslaughts experienced by the bacillus during an infection, nor during infection of mouse and human – derived macrophage cell lines. The possibility remains that these USPs are functionally redundant in Mtb.
Keywords: DosR, Hypoxia, Stationary phase, hspX
1. Introduction
Over a third of the world’s population is thought to be latently or persistently infected with Mycobacterium tuberculosis, which represents a huge potential reservoir of infection.1 There is an urgent need to fully understand the host/pathogen interaction, in particular during this latent state, in order to develop improved vaccine, diagnostic and treatment strategies with which to curtail this disease. In addition, persistent bacilli may be responsible for the need for the long antibiotic treatment regimes required to treat TB effectively.2 Infection with M. tuberculosis (Mtb) generally occurs upon inhalation of the bacilli in airborne droplets. The bacilli are then taken up by alveolar macrophages and it is here that they survive and propagate where most other pathogens perish. Understanding the ability of the tubercle bacillus to persist during latent infection has been the object of much research in attempts to define both the initiating factor(s) and the bacterial genes required for non-replicating persistence. It is thought that Mtb will experience hypoxic conditions in the granulomas that form during chronic infection as they are avascular, inflammatory and necrotic, conditions which lead to low oxygen environment.3–5 Mtb can survive for long periods in a non-replicating anaerobic state.6 Wayne and co-workers developed a hypoxic model of non-replicating persistence,7 in which the shift-down from active replication to non-replicating persistence occurs via oxygen depletion, in order to mimic the hypoxic in vivo latency conditions. In addition, nutrient depletion models8,9 may mimic other conditions encountered in vivo, and a recently published in vivo model utilises hollow fibres to form what the authors describe as granulomas.10
The DosR/DosS/DosT regulatory system has been described in a number of Mycobacterium species11–15 and is expressed in response to hypoxia and nitric oxide stress by controlling the expression of a set of ∼48 genes termed the DosR regulon.15 More recently, this regulatory system has been shown to respond to carbon monoxide, which is generated in macrophages in response to Mtb infection16,17 It is thought to be important for adaptation to hypoxia, NO and CO as well as a long term survival in the host.13,14,16–20 The identification of the function of these genes may have allowed the elucidation of the role of this regulon in hypoxia-induced persistence, but 44 of the 48 genes encode conserved hypothetical proteins. However, 6 of these conserved hypothetical proteins are from a protein family called universal stress proteins (USPs) and we hypothesized that determining the function of these proteins would help to define non-replicating persistence, along with determining the function of an as yet poorly understood protein family.
The USP domain is a conserved domain of 140–160 amino acids and is widespread throughout the Archaea, bacteria, plants and fungi, yet it is absent in humans, making the USPs potential drug targets. With few exceptions such as Mycoplasma species, USPs are present in all Eubacteria and Archaea, and while some bacteria, such as Mycobacterium leprae, have a single USP most have multiple USPs.21 The domain name originates from the UspA of Escherichia coli, which was named due to its upregulation in response to a wide variety of stresses. Mutation of the E. coli uspA gene results in a survival defect under a variety of growth-arrested conditions, while overexpression induces a growth arrested state.22–24 E. coli has six usp genes, which have as yet undefined roles in functions ranging from oxidative stress to adhesion and motility.25
In the enteric bacterium Salmonella typhimurium a uspA mutant showed reduced survival following nutrient starvation and increased sensitivity to oxidative stress and it makes an important contribution to virulence.26 In the opportunistic pathogen Pseudomonas aeruginosa USPs have been shown to play a role in bacterial survival under anaerobic stationary phase; conditions thought to resemble those present in the cystic fibrosis lung colonized by P. aeruginosa.27,28 The structures of a number of bacterial USP proteins have been resolved, with ATP present in the crystal structure of MJ0577 from Methanococcus jannaschii,29 while that of UspA from Haemophilus influenzae had no bound ATP.30 This has led to the idea that the USP domain family falls into two groups, those proteins represented by the ATP binding structure of MJ0577 and those represented by the non-ATP binding structure of UspA,21,30,31 with the putative ATP binding USPs having a G2×G9×GS motif that forms the ATP binding site of MJ0577.
Mtb has 10 USPs in total and interestingly, 6 of these are DosR-regulated and their expression is also upregulated in macrophages15,32 leading us to hypothesize that they may be playing a role in persistence and/or intracellular survival. The USPs of Mtb are upregulated and overlap in their response to many different stresses, including low pH,33 nitric oxide,32 UV light and mitomycin C34 (Table 1). This work aimed to investigate the role of USPs from Mtb in the survival of this bacterium in response to a variety of stresses. We have constructed, complemented and phenotypically characterized 4 USP mutants of Mtb; 3 are DosR-regulated (Rv1996, Rv2005c, Rv2028c) and while Rv2026c is not DosR-regulated it is adjacent in the genome to DosT (Rv2027c), one of the two histidine kinases that interact with DosR and is thought to be primarily responsible for sensing hypoxia. Surprisingly we found no discernable phenotype in response to a wide variety of stresses and growth conditions, raising the possibility that USPs are functionally redundant in Mtb.
Table 1.
Summary of published expression studies in which data from usp gene expression is reported and the conditions chosen for the stress screening assays performed on usp mutants in this current study. The experimental conditions were chosen to represent the range of environmental onslaughts encountered by the bacillus during the disease state, from the period spent outside the host during transmission (cold shock), to the cell wall damaging surfactants in the lung (SDS) to the nitrosative and oxidation stresses experienced in the macrophage. Concentrations of stress agents were chosen through a search of the literature or through the testing of stress agents.
| Stress condition | Exact condition tested in current work | USP Induction |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rv1636 (TB15.3) | Rv1996 | Rv2005c | Rv2026c | Rv2028c | Rv2319c | Rv2623*† (TB31.7) | Rv2624c† | Rv3134c† | Rv1028 | ||
| Low O213 (2 h at 0.2% O2) | Survival under hypoxic and normoxic SP over 130 days (see methods section) | U | U | U | U | U | |||||
| Low O265 (1% O2 in chemostat) | U | U | U | U | U | U | |||||
| Low O2 − P66 (22–26 days of anaerobic growth) | U | ||||||||||
| DosR-regulated15 (dosR mutant versus wildtype. m = motif upstream) | Um | Um | U | U | U | Um | |||||
| ΔsigC versus wildtype in Mtb CDC155167 | U (MT2699) | ||||||||||
| ΔhspR versus wt68 | U | ||||||||||
| ΔrelAMTB versus wildtype during starvation in Tris-buffered saline plus Tween69 | D (6 h) | U (4 h/6 h) | |||||||||
| NO/low O214 (DETA/NO 50 μm for 40 min/0.2% O2 for 2 h) | U | U | U | U | U | U | |||||
| NO70 (100 μm NOR-3 and SPER-NO for 4 h) | U | U | U | U | U | ||||||
| Palmitic acid32 (0.05 M for 1 h) | U | ||||||||||
| H2O232 (5 mM for 40 min) | |||||||||||
| H2O2(4 mM for 12h) [See Gene Expression Omnibus Accession number GDS326 (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS326)] | U | U | U | U | U | U | U | U | |||
| UV/Mitomycin C (0.2 μg/ml Mitomycin C/variable UV exposure for up to 12 h) [See Gene Expression Omnibus Accession number GDS326 (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS326)] | U | U | U | U | U | U | U | U | |||
| Low pH33 (pH 5.5 for 15 min) | |||||||||||
| PBS starvation 4 h8 | D | D | U | ||||||||
| PBS starvation 24 h8 | U | ||||||||||
| PBS starvation 96 h8 | D | D | D | U | |||||||
| Shaking versus standing cultures71 (7 days–promoter activity of chosen genes determined) | U | U | |||||||||
| Low O271 (1.3% – promoter activity of chosen genes determined as above) | U | U | |||||||||
| Intra versus extracellular growth71 (48 h infection in J774A.2 macrophages versus tissue culture medium) | U | U | |||||||||
| IFN- γ activated MΦ32 (Murine bone-marrow derived macrophages infected at an M.O.I. of 2–5. 4, 24 and 48 h post-infection) | U | U | U | U | U (48 h only) | U | U | U | |||
| NOS2−/− MΦ32 (Murine bone-marrow derived macrophages from NOS2−/− mice infected at an M.O.I. of 2–5. 24 h post-infection) | U | U | U | ||||||||
| Murine infection72 (Balb/c and SCID mice infected intranasally with 103 CFU. 7, 14 and 21 days post-infection) | U at 7/14/21 days in Balb/c and at 21 days in SCID | U (14 days/Balb/c) | U (21 days/Balb/c) | ||||||||
| In vitro survival63(TraSH essentiality screen) | E | E | E | E§ | |||||||
| SDS 0.05 and 0.1%–3 days | |||||||||||
| 55 °C heat shock, 5 h and 72 h | |||||||||||
| Osmotic shock, 2.5 M NaCl (72 h) | |||||||||||
| Exposure to Isoniazid 0.1, 1 μgml−1 (72 h) | |||||||||||
| Cold shock 4 °C for up to 72 h | |||||||||||
| Ethanol 5, 10% (72 h) | |||||||||||
All regulatory data from published microarray studies, except for proteomic studies (P). U = Up-regulated, D = down-regulated, MΦ = macrophage, E = essential for optimal growth in vitro, SP = stationary phase.
Expressed in the murine lung, concurrent with the onset of Th1 immunity.72
In transposon screen for intracellular survival, mutants in Rv2623 and Rv2624c grew better in un-activated macrophages and were attenuated in pre-activated macrophages, whilst an Rv3134c mutant grew best in post-activated cells.73
Essential in vivo74
2. Materials and methods
2.1. Growth of M. tuberculosis strains
M. tuberculosis H37Rv cultures were grown, at 37 °C with 150 rpm shaking, in Middlebrook 7H9 medium (Difco) plus 10% oleic acid, albumin, dextrose and catalase (OADC, Difco) and 0.05% Tween 80 (Sigma). For oxygen-sufficient cultivation 50 ml cultures were grown in 250 ml Erlenmeyer flasks. The number of bacilli was estimated either using optical density (at 600 nm) or by determination of colony forming units following plating on Middlebrook 7H10 medium (Difco), plus OADC.
2.2. Growth of M. tuberculosis strains under hypoxic conditions
Oxygen starved Mtb cultures were grown in 125 ml flasks in a volume of 75 ml 7H9 + OADC with 150 rpm shaking, giving a headspace ratio (HSR) of 1.67 as described for the growth of Mycobacterium smegmatis to a hypoxic stationary phase.35,11 Each culture was sealed using a rubber SubaSeal septum (number 53, Sigma) to prevent air exchange and samples of culture were removed using a 19G sterile needle through the SubaSeal to prevent the introduction of oxygen into the culture. Sealed cultures grew exponentially but leveled off at an O.D. 600nm of ∼0.2. Preliminary experiments in which the HSR were varied inferred that with an HSR of 1.67 cultures entered stationary phase due to oxygen-starvation, as was the case with M. smegmatis.35
2.3. Determination of colony forming units (CFUs)
In 96-well plates, a 10-fold dilution series of the cultures were prepared in PBS + 0.05% Tween 80. Growing cultures were diluted in 100 μl, transferring 10 μl between each well, to serially dilute down to the 10−8 dilution. 20 μl spots of each dilution were dropped onto dry 7H10 plates in triplicate and allowed to dry before incubating at 37 °C for 2 weeks, at which point the colonies were counted.
2.4. Mutant construction
Two different approaches were used for constructing usp mutants.
-
(i)
sacB counter selection method. Mtb H37Rv ΔRv2026c and ΔRv2028c mutants were generated using a sacB counter selection.36 ˜1 kb regions of DNA flanking the Rv2026c and Rv2028c genes were PCR-amplified from the genomic template. The Rv2026c primers were as follows: Upstream Flanking Sequence (UFS) F: tctagaatgacggttgccgtcagcggaacg and R: tctagaggctgtcgcagcagacatttcacg, both of which have a 5′ XbaI site and Downstream Flanking Sequence (DFS) F: tctagagtcgtccgtcccagctagtactgg, with a 5′ XbaI site and R: gatatcttgcgcgaggtccagggcggggtc, with a 5′ EcoRV site. The Rv2028c primers were: UFS F: actagaccgcgaatcatcactttgaccatg, with a 5′ SpeI site and R: gggccctttgtgtgattggttcatggcgag with a 5’ApaI site and DFS F: tctagaggtcagcagtatctgtgactgtgc with a 5′ XbaI site and R: catatgaagccggtcgaaccggcggggcgt with a 5′ NdeI site. The UFS and DFS regions were cloned either side of the hygromycin cassette in the suicide vector pSMT100 to make a marked mutant of Rv2026c and upstream of the hygromycin to make an in-frame deletion of Rv2028c37 and transformed into E. coli DH5α and plasmids from transformants screened by PCR analysis for successful constructs. Following electroporation of the constructs into Mtb H37Rv and plating onto 7H10 plus hygromycin (100 μg/ml), single crossovers were purified by three rounds of re-streaking onto fresh hygromycin-containing plates. Gene replacement events (double crossovers) were selected for by growing colonies in liquid 7H9 media in the presence of 5% sucrose for 1 week and plated onto 7H10 plus 5% sucrose (100 μg/ml hygromycin was also included for the marked Rv2026c mutation), prior to re-streaking. Confirmation of gene deletion was carried out by PCR using primers outside of the upstream and downstream flanking regions, and observing a shift in size of the PCR product for the mutants. The PCR products were cloned into pGEM-T (Promega) and sequenced to confirm the mutation.
-
(ii)
Specialised transduction. The mycobacteriophage-based method of specialized transduction,38 which utilizes conditionally replicating shuttle phasmids created from pHAE159 and pYUB854, was used to create the ΔRv1996 and ΔRv2005c mutants. Initially, upstream (UFS) and downstream (DFS) sequences flanking the usp gene to be mutated were amplified from Mtb genomic DNA using the following PCR conditions: 35 cycles of 95 °C for 30s, 55 °C for 30 s and 72 °C for 1 min. The primers used had an AflII site on the 5′ ends of UFS F, an XbaI site on the 5′ end of UFS R, HindIII on DFS F and SpeI on DFS R to allow directional insertion into the pYUBB854 vector. The primers were as follows: Rv1996 UFS F: ggtaccgtactcgccgccaccactg, R: tctagagaaaccgcgaatactcaaat and DFS F: aagcttggcgccgatcgaatggagaaacct and R: actagtcgaagatcactg-cggcgtcaac and Rv2005c UFS F: cttaaggtctaccaggaggcgct, R: actagtgcgtggccttgtcgac and DFS F:tctagaccacgtcgctacatcgg and R: gacgtcgaagtggtggaa. Following PCR purification, the PCR product was ligated into the pGEM-Tamp vector (Promega), and then transformed into competent E. coli DH5α cells39 and plated out LB agar plus 100 μg/ml ampicillin. Following overnight incubation of isolated white colonies in LB broth plus ampicillin, PCR analysis was used as above to identify successful ligations. The cloned UFS or DFS fragment was then sub-cloned into appropriately restriction enzyme digested pYUB854Hyg(AflII/XbaI for UFS fragments and HindIII/SpeI for DFS fragments)38 and the recombinant pYUB854 derived plasmids transformed into E. coli HB101, and plasmids from transformants screened by PCR analysis for successful constructs. The temperature sensitive phage pHA159 and the pYUB854hyg vector, containing both the UFS and DFS fragments for a particular usp gene disruption, were then digested with PacI and following gel extraction, were ligated using T4 DNA ligase to create a shuttle phasmid, which was then in vitro packaged using the Giga XL in vitro packaging mix (Stratagene) and transformed into E. coli HB101 cells according to the manufacturer’s instructions. Successful phasmids were identified by PacI digestion, following antibiotic selection on LB plus hygromycin (100 μg/ml) plates, purification and growth overnight in LB broth plus hygromycin (100 μg/ml). The phasmid was then transduced into M. smegmatis mc2155 at the permissive temperature of 30 °C, which allows replication and lysis, and a high titre lysate was produced as described previously.38 Transduction into Mtb was then carried out with the high titre lysate at an M.O.I. of 10:1 at the non permissive temperature of 37 °C, which results in delivery of the substrate. Cells only and phage only controls were included. Following incubation for 2–3 weeks on 7H10 plus OADC, glycerol and hygromycin, colonies were picked, grown in liquid medium (7H9 plus OADC, glycerol, Tween 80 and hygromycin) for 2 weeks and then confirmation of gene deletion was carried out as above via PCR using primers outside the upstream and downstream flanking regions and observing a shift in size for the mutants.
2.5. Complementation of the mutants
Complementation analyses of Rv2005c and Rv1996 were performed by cloning the full length genes carrying the predicted promoter regions of the genes into the integration proficient vector pMV306kan.40,41 Initially, the surrounding area was amplified from Mtb genomic DNA using gene specific primers with XbaI and HindIII sites attached to the 5′ end of the forward and reverse primers respectively to allow directional insertion into the pMV306 vector. The primer sequences were Rv1996F: tctagagccgacgatgacagcgt and R: tcctcgggtattacggcg and Rv2005c F: tctagattgaggacctaagcccgtt and R: gttggtcggtgagtccat and the PCR program and preliminary ligation stage in the pGEM-T vector was the same as used in the generation of the UFS and DFS fragments noted above. The pGEM-T clone plus the insert and pMV306kan vector were then digested with the appropriate restriction enzyme, the insert and pMV306kan vector gel purified using the Qiagex gel extraction kit (Qiagen) and ligated using T4 DNA ligase according to the manufacturer’s instructions. Transformation was then carried out in E. coli HB101 cells, plasmids were extracted using the Qiagen miniprep kit and PCR analysis was used to screen for successful constructs. The vector, plus the vector only control, was then transformed into the Mtb mutants using electroporation.42 Prior to use in experimental procedures, complementation was confirmed by PCR analysis for the targeted gene on DNA isolated from the mutant, wildtype and complemented strains. Mtb ΔRv2026c and ΔRv2028c::hyg were complemented with a single copy of the gene under the control of a tetracycline inducible promoter.43 The full length Rv2026c and Rv2028c genes were amplified by PCR and a ribosome binding site included in the forward primer. The primer sequences were Rv2026F: ggatcctttaagaaggagtatacatatgtctgctgcgacagcgaaa and R: cccgggctagctgggacggacgacgat and Rv2028F: ggatcctttaagaaggagatatacatatgaaccaatcacacaaacc and R: gcatgctcacagatactgctgaccgac. Fragments were cloned into the pMind-Lux vector using a Klenow blunted SpeI site in the vector and BamHI, replacing the LuxAB reporter present in pMind-Lux. The region containing the tetracycline repressor, tetracycline regulatable promoter and the usp gene was removed from this vector using KpnI and HindIII, and cloned into the single copy, integrating vector pMV361 using Klenow blunt ended DraIII and EcoRI sites, which remove the hsp60 promoter in pMV361. RT-PCR was used to determine that Rv2026c and Rv2028c could be induced from the tetracycline regulatable promoter, restoring transcription of the genes in the mutant backgrounds.
2.6. Screening for survival phenotypes under stress
Stress agents (Table 1) were diluted in PBS + 0.05% Tween 80 and 500 μl aliquots added to a 24-well plate. Mtb strains were grown to mid-exponential or stationary phase as indicated and 10 μl aliquots of culture added to each well of the plate. Typically this corresponded to between 1 and 5 × 107 cells. CFU determination was carried out at 0 h and 3 days post-inoculation, unless otherwise noted.
2.7. Cell culture and infection model
The murine macrophage-like cell line J774A.2 and the human monocyte-derived cell line THP-1 were obtained from the American Type Culture Collection (Manassas, VA) and maintained in RPMI medium plus l-glutamine and 10% heat inactivated fetal bovine serum (h.i. FBS, Life Tech). Viable cell number was determined using a haemocytometer and trypan blue staining (Sigma). Prior to infection, the cells were seeded in 24-well plates at 5 × 105 cells/ml. THP-1 cells were pre-activated with PMA at 20 ng/ml overnight. Infection was carried out at a multiplicity of infection (M.O.I.) of 10:1 by overlaying the monolayers with bacteria re-suspended in RPMI and washing 3 times with prewarmed medium after 2 h to remove any extracellular bacilli. The infected monolayers were then incubated at 37 °C with 5% CO2. Successful intracellular infection was confirmed by Kinyoun staining, using the TB cold stain kit (BDH) on formaldehyde-fixed infected monolayers. All treatments were carried out in triplicate and the controls included non-infected cells.
3. Results
3.1. The universal stress proteins (USPs) of M. tuberculosis
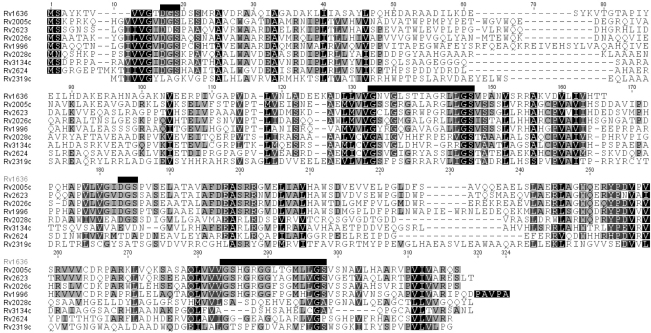
M. tuberculosis contains 10 USPA domain (PFAM PF00582)-containing proteins, all of which appear to be stand alone proteins with either single (Rv1636) or tandem domains with the exception of the KdpD (Rv1028), which contains a USP domain of unknown function as part of a sensor kinase protein. All 10 Mtb USPs possess a striking degree of similarity to each other within those regions identified as being conserved across all species, although this does not translate to a dramatic degree of similarity across the entire proteins. Figure 1 shows an alignment of 9 of the 10 Mtb USPs, (excluding KdpD). All of the tandem domain Mtb USPs have a G2×G9×GS conserved motif, which is part of a proposed ATP binding site29,30 and a conserved D (in a DGS motif) in USPA domain 1, except Rv2139 which has only a partially conserved motif (G12xGS). Four of the USPs also have these features fully conserved in UspA domain 2 (Rv2005c, Rv2623, Rv2026c and Rv1996). So all of the Mtb USPs, with the possible exception of Rv2139, fall into the putative ATP binding class of USPs.30
Figure 1.
Clustal alignment of the USPs of M. tuberculosis. The 9 tandem domain proteins show a high degree of similarity across two key conserved motifs (DGS and G2×G9×GS, indicated above the sequence by short and long horizontal bars, respectively), but are less similar across the other regions of the proteins. Rv1636 is the only single domain USP present in Mtb. Rv2319c lacks some of the highly conserved residues present in all of the other USPs. Rv2005c, Rv2623, Rv2026c and Rv1996 in particular possess a high degree of similarity with each other in their second USP domain.
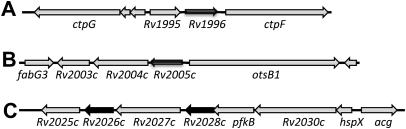
The genes encoding universal stress proteins in Mtb are distributed across the chromosome and are surrounded by genes of very different or unknown function, so that it is difficult to discern a functional role based on the identification and context of the neighboring genes. The genome context of the tandem USPs studied here is shown in Figure 2. The currently available mycobacterial genome sequences were searched using a Blast search with all of the Mtb USP protein sequences to try to identify homologues in the related species. All of the Mtb USPs were determined to be conserved in Mycobacterium bovis and M. microti, with no alterations in amino acid sequence. Interestingly, M. leprae, the causative agent of leprosy in humans, possesses only one USP. In comparison to Mtb, the M. leprae genome has undergone extensive reductive evolution, with the loss of approximately 1700 genes, around 25% of the total genome.44 Blast searches of the M. leprae predicted proteins, using the Mtb USPs as query sequences, revealed that of the Mtb USPs, Rv1636 is the only one with a homologue in M. leprae. The amino acid sequences of this protein, (Ml1390), and of Rv1636 show an 89% identity with each other.
Figure 2.
Genome context of the M. tuberculosis usp genes studied in this work.
3.2. Construction of usp mutants
To investigate the functions of the Mtb USPs, knock-out mutants were constructed for four of the tandem domain USPs; Rv1996, Rv2005c, Rv2026c and Rv2028c in Mtb H37Rv. Three are DosR-regulated (Rv1996, Rv2005c, Rv2028c) and Rv2026c is adjacent in the genome to Rv2027c (DosT), one of two histidine kinases that interact with DosR and is thought to be responsible for hypoxia sensing. Deletion mutants of Mtb H37Rv lacking Rv2026c and Rv2028c were generated by a 2 step allelic exchange mutagenesis and the mutants confirmed by PCR analysis.36,37,45 In addition, marked deletion mutants of Rv1996 and Rv2005c were constructed by a mycobacteriophage-based method of specialized transduction46 and the mutant constructs confirmed by PCR. Each of the mutants was complemented in single copy using pMV361 for Rv2026c::Hyg and ΔRv2028c and pMV306 for Rv1993::Hyg, and Rv2005c::Hyg.
3.3. Individual usp genes are nonessential for growth in vitro
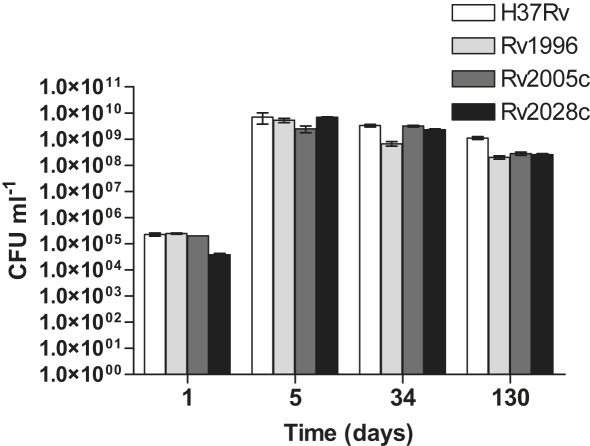
The Mtb H37Rv usp mutant strains were initially tested for any growth defects in broth cultures and on 7H11 plates, plus OADC and glycerol, and plus and minus Tween 80. None of the mutants tested appeared to have any discernable phenotype in liquid or solid media, with colony size and morphology appearing similar to that of the wildtype H37Rv strain (results not shown) and the growth rate and final cell density shown by each of the usp mutant strains was very similar to the wildtype, as were the complemented mutants. Due to the upregulation of the DosR regulon under hypoxic conditions,15 the fact that mutation of dosR compromises bacterial survival11,14,47,48 and that six USPs, including Rv1996, Rv2005c, and Rv2028c, are DosR-regulated, the survival of the mutants under hypoxic stationary phase conditions was investigated over a period of 130 days, but no survival defect was observed (Figure 3). Similarly, no survival differences were observed over periods of up to 80 days during normoxic stationary phase (data not shown). So none of the four usp genes mutated showed survival phenotypes that have been commonly associated with usp mutants from other bacteria.24,26–28,49
Figure 3.
Hypoxic survival of M. tuberculosis H37Rv usp mutants. The Mtb tandem domain usp mutants were cultured to hypoxic stationary phase and survival followed for 130 days. The viability (CFUs) was measured at several time-points to determine if the mutants possess any survival defect under these conditions. Error bars represent the standard deviations of 3 independent cultures.
3.4. Individual usp genes are nonessential during in vitro stress challenge
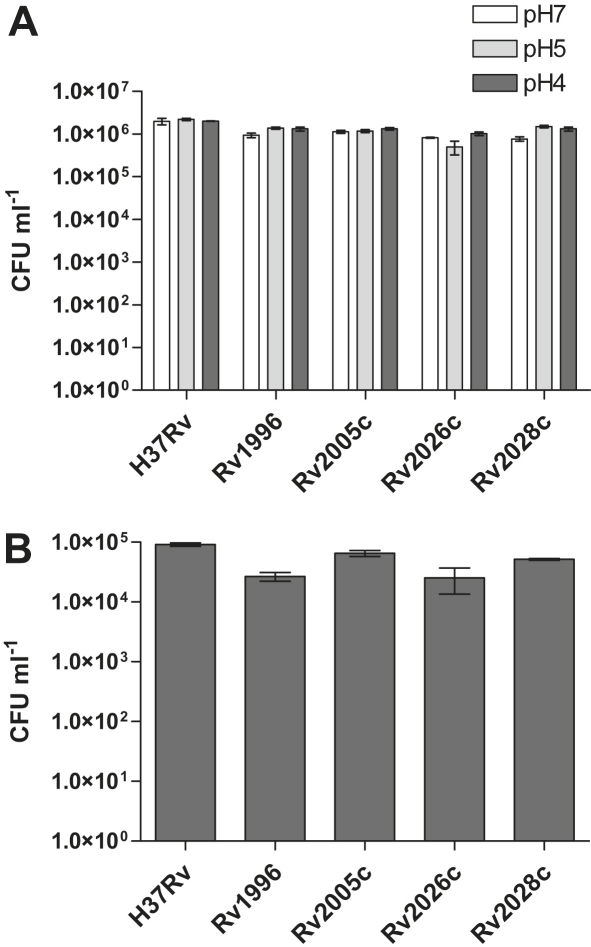
Mining of publically available Mtb microarray data identified a range of stress conditions that lead to the upregulation of one or more USPs (Table 1). Multiple studies have shown that 6 usp genes are upregulated by low oxygen and NO in a DosR dependent manner; 7 usp genes are upregulated by H2O2 and by DNA damaging agents UV light and mitomycin C, which is interesting in the context of the importance of a number of USPs from E. coli having a putative role in DNA protection.50,51 Therefore, these conditions together with others (Table 1), chosen to represent the predicted environmental conditions experienced in the disease state, were used to screen the usp mutants and complemented strains for survival defects compared to wildtype. The screens were performed in 24-well plates and CFU platings used to determine the survival of the cultures following the stress challenge. In all cases the survival of the usp mutants was found to be no different from that of the wildtype. None of the stress agents tested yielded a phenotype for any of the mutants. A representative selection of the results is shown in Figure 4.
Figure 4.
The M. tuberculosis usp mutants exhibited no survival defect in response to a range of stress conditions. A number of other stress conditions were also tested (see Table 1), but no survival phenotype was observed when comparing the usp mutants to the wildtype H37Rv. As examples, the results for A) low pH and B) Nitrosative stress (5 mM GSNO) are shown. Error bars represent the standard deviations of 3 independent cultures.
3.5. Rv2026c is not involved in the expression of DosR-regulated genes
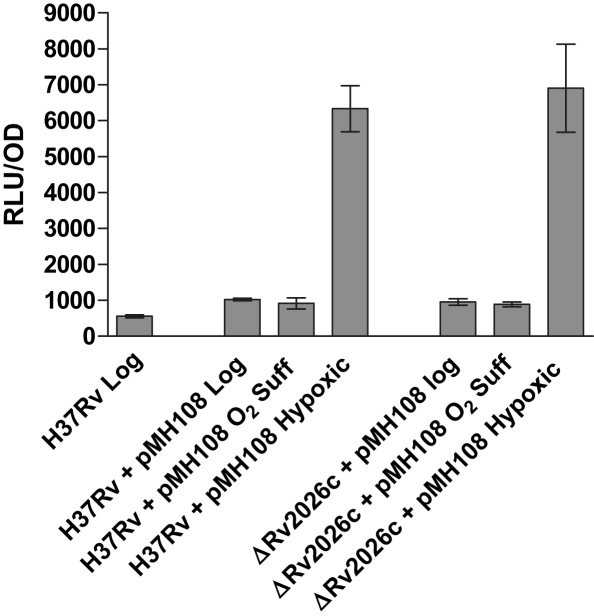
Rv2026c is not a part of the DosR regulon, and its expression is not induced by hypoxia or NO, and while it is expressed during starvation in PBS (Table 1), the ΔRv2026c mutant showed a comparable survival defect to the wildtype under these conditions (data not shown). RT-PCR experiments showed that Rv2026c was transcribed during exponential phase, normoxic and hypoxic stationary phase and as expected mutation of dosR did not affect expression levels (not shown). Rv2026c is adjacent to dosT (Rv2027c) (Figure 2C), encoding the DosT histidine kinase partner of DosR, and its apparent constitutive expression led us to consider whether this USP might be involved in DosR mediated induction of genes. Both the sensor kinases DosT and DosS activate the response regulator DosR, resulting in induction of the DosR regulon, with the evidence being that DosT preferentially responds to hypoxia,52 and a dosT mutant was only able to induce DosR regulon expression to 40–45% of normal levels (Roberts et al., 2004). The possible role of Rv2026c in the DosR pathway was tested using a luciferase reporter assay with a plasmid construct containing the hspX promoter region upstream of firefly luciferase gene.20 The marked induction of hspX expression under hypoxic conditions occurred in wildtype H37Rv, as expected, and the ΔRv2026c mutant showed no difference in hspX expression indicating that Rv2026c is not involved in the expression of DosR-regulated genes (Figure 5).
Figure 5.
hspX promoter activity in M. tuberculosis H37Rv and ΔRv2026c. The plasmid pMH108 carrying the hspX promoter region upstream of the firefly luciferase genes was transformed into Mtb H37Rv and ΔRv2026c. The promoter activity was measured in cultures grown under oxygen-sufficient conditions to mid-exponential phase and stationary phase and in cultures grown to hypoxic stationary phase. Relative light units (RLU) were determined and normalised using the OD of the cultures. The hspX promoter was seen to be active in both the wildtype and the ΔRv2026c mutants strain. Error bars represent the standard deviations of 3 independent cultures.
3.6. Growth and survival of usp deletion mutant strains in macrophage models of infection
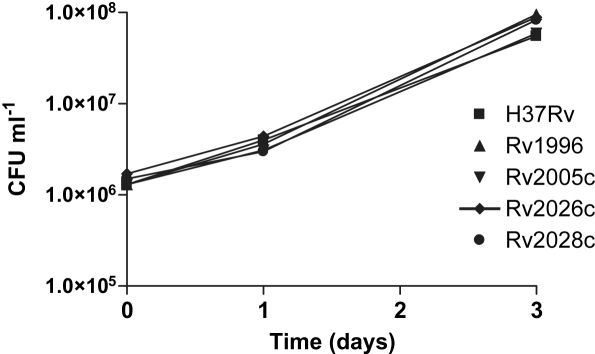
As all of M. tuberculosis USPs, with the exception of Rv1636 and kdpD, are upregulated during macrophage infection,32 we determined whether any of our usp mutants exhibited a survival deficiency in macrophages. Initially, we looked at survival in the murine macrophage-like cell line J774 (Figure 6), and by the human monocyte-derived cell line THP-1, which was pre-activated with PMA (results not shown). No significant differences in growth or survival were noted with any of the mutants tested. This was also the case when the cells were pre-activated overnight with interferon gamma (at 10 U/ml; results not shown). Therefore, we can conclude that none of the usp mutants tested has a discernable intracellular survival phenotype.
Figure 6.
Intracellular survival of the usp mutants in the murine macrophage-like cell line J774 plus IFN-gamma. The usp mutants were tested for a survival defect in a macrophage cell line by determining CFU/ml over a 3-day time course. A similar curve was obtained for the inactivated macrophage infection.
4. Discussion
This paper reports the construction of isogenic Mtb mutants, with disruptions in four genes encoding for the universal stress proteins Rv1996, Rv2005c, Rv2026c and Rv2028c, and their phenotypic analysis. In bacteria there is a consistent link between USPs and survival under growth arrested conditions. Mutation of uspA gene of E. coli leads to premature death in stationary phase.22,23 A S. typhimurium uspA mutant is less virulent, more sensitive to oxidative stress and survives carbon or phosphorous starvation poorly,26 while work on USPs from the major periodontal pathogen Porphyromonas gingivalis indicates a role in anaerobic biofilm survival.49 In the opportunistic pathogen P. aeruginosa, the USP PA3309 is essential for survival during pyruvate fermentation and is expressed in anaerobic biofilms,28 whereas the USP PA4352 is important for survival during anaerobic stationary phase under nitrate limitation and in the presence of the uncoupler CCCP.27 Therefore, USPs are important for stationary phase survival of P. aeruginosa under exactly the types of survival conditions thought to exist in the CF lung. There are parallels with Mtb in which 6 USP genes are upregulated by low oxygen conditions (Table 1). We tested the survival of Mtb usp mutants under a variety of non-growing and other stress conditions selected in part based upon published expression data (Table 1). Our results show that individual usp genes encoding Rv1996, Rv2005c, Rv2026c and Rv2028c can be mutated in Mtb without any effect on growth or survival under normoxic or hypoxic stationary phase conditions. Mutation of the gene encoding the response regulator DosR, which regulates Rv1996, Rv2005c and Rv2028c, leads to a hypoxic survival defect in Mtb, as do mutations of dosR genes in M. bovis BCG and M. smegmatis,11,47 but clearly the phenotype does not result from a failure to express one of the four usp genes mutated here, despite the fact three are upregulated by hypoxia (Table 1). The genes encoding Rv1996, Rv2005c, Rv2026c and Rv2028c are also upregulated in activated human53,54 and mouse macrophages.32 However, in our studies, we found that there was no significant difference in intracellular growth between the wildtype and usp mutants in murine or THP-1 macrophage cell lines, either non-activated or activated with IFNγ. We have not tested the mutant in bone-marrow derived macrophages or in a mouse model of infection, however, so it remains to be determined if our Mtb dosR mutant is deficient in models more closely representing the in vivo situation.
While the exact function of the Mtb tandem domain USPs studied here still remains to be determined, we believe that these results are interesting as they suggest that the functions of the Mtb USPs are either partially or completely redundant. A redundant function for the USPs would result in the disruption of the single usp genes being compensated for by continuing expression of the remaining homologues. Our investigation of the published microarray data has indicated that the in vitro expression patterns of these genes do overlap, suggestive of potentially overlapping functions. However, despite the overlaps in expression, the USPs do show some evidence of differential regulation. One possibility is that the individual members or small subsets of the Mtb USP family serve different functions in specific phases of an infection, although they are nonessential when knocked out. A similar result to that described here has previously been reported for the Mtb rpf genes, which are nonessential unless knocked out in multiples of more than one.55–58 However, whereas the USPs of E. coli each seem to play a role in defense against DNA damaging agents they do not show redundancy in this role as individual mutants in uspA, uspC, uspD and uspE each show increased sensitivity to UV light but the evidence suggests that they operate and are required in the same pathway.50,51
The diverse genomic contexts of the Mtb usp genes may enable prediction of their role based upon the function of neighboring genes impossible. Biochemical characterization of these USPs will be important to elucidate their individual functions. UspA from E. coli is a serine threonine phosphoprotein whereas phosphorylation in vivo is dependent on the phosphoprotein TypA.59,60 MJ0577 from M. jannaschii had a tightly bound ATP in its crystal structure,29 whereas in the structure of UspA from H. influenzae did not have an ATP in its structure.30 UspG of E. coli is post-translationally modified during its overexpression in E. coli and in vitro experiments showed it to have intrinsic autophosphorylation and autoadenylation activity.61 A recent publication on another tandem domain usp from Mtb indicates it can also bind ATP. As this work was being written up for publication a comprehensive study of the tandem domain USP Rv2623 from Mtb was published.62 Rv2623 is also part of the DosR regulon and indeed is regulated by similar environmental conditions as the hypoxia-induced USPs studied here (Table 1). The ΔRv2623 mutant was unable to establish a chronic infection in mice and guinea pigs, exhibiting a hypervirulent phenotype with increased bacterial load and mortality and the overproduction of Rv2623 led to an attenuation of bacterial growth in vitro. Furthermore, consistent with our findings here, the Rv2623 mutant did not show any in vitro growth differences to the wildtype and was no more sensitive than the wildtype to a range of environmental stress conditions, including DNA damaging agents, oxidative stress and heat shock and acid shock.62 The lack of in vitro growth phenotypes for Rv2623 and Rv2026c contradicts the TRaSH screening experiments for identifying essential Mtb genes in which both USPs were found to be essential for in vitro survival (Table 1).63 While many of the genes identified in Mtb using TRaSH screenings are conserved in M. leprae, neither Rv2623 nor Rv2026c have homologues in M. leprae and, while an innovative approach, TRaSH does have limitations (see Ref.64). Clearly, there could be additional environments in vitro which we have not examined under which these proteins are necessary for survival and investigation of their role in long term survival in an appropriate animal model may also illuminate phenotypic differences.
The ability of Mtb to survive a range of rapidly changing conditions that other species would not be able to withstand could therefore explain why it has evolved to possess more USPs than most other species, and why these proteins have apparently redundant functions. However, the testing of the redundancy hypothesis requires the evaluation of Mtb strains harboring multiple disruptions of the usp genes, which is currently under way.
Acknowledgements
Vectors: The integrating vector pMV306, multi-copy vector pMV361 and the specialized transduction vectors pHAe159 and pYUB854 were kind gifts from the laboratory of Professor W.R. Jacobs Jr. (Albert Einstein College of Medicine, USA). The suicide delivery vector pSMT100 was obtained from Dr. B.D. Robertson (Imperial College London). pMH108 was a kind gift from Clifton Barry 3rd’s laboratory (Tuberculosis Research Unit, National Institutes for Allergy and Infectious diseases, Rocky Mountain Laboratories, USA)
Funding
This research was funded by the Wellcome Trust and the United Kingdom Biotechnology and Biological Sciences Research Council.
Competing Interests
None.
Ethical approval
Not required.
References
- 1.Fenton M.J. Macrophages and tuberculosis. Curr Opin Hematol. 1998;5:72–78. doi: 10.1097/00062752-199801000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison D.A. Understanding the chemotherapy of tuberculosis – current problems. J Antimicrob Chemother. 1992;29:477–493. doi: 10.1093/jac/29.5.477. [DOI] [PubMed] [Google Scholar]
- 3.Canetti G. Springer; New York: 1955. The Tubercule bacillus in the pulmonary lesion of man. [Google Scholar]
- 4.Imboden P., Schoolnik G.K. Construction and characterization of a partial Mycobacterium tuberculosis cDNA library of genes expressed at reduced oxygen tension. Gene. 1998;213:107–117. doi: 10.1016/s0378-1119(98)00192-9. [DOI] [PubMed] [Google Scholar]
- 5.Russel W.F., Dressler S.H., Middlebrook G., Denst J. Implications of the phenomenon of open cavity healing for the chemotherapy of pulmonary tuberculosis. Am Rev Tuberc. 1955;71:441–446. [PubMed] [Google Scholar]
- 6.Wayne L.G., Sohaskey C.D. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 7.Wayne L.G., Hayes L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 9.Smeulders M.J., Keer J., Speight R.A., Williams H.D. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karakousis P.C., Yoshimatsu T., Lamichhane G., Woolwine S.C., Nuermberger E.L., Grosset J. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Toole R., Smeulders M.J., Blokpoel M.C., Kay E.J., Lougheed K., Williams H.D. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J Bacteriol. 2003;185:1543–1554. doi: 10.1128/JB.185.5.1543-1554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boon C., Li R., Qi R., Dick T. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J Bacteriol. 2001;183:2672–2676. doi: 10.1128/JB.183.8.2672-2676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman D.R., Voskuil M., Schnappinger D., Liao R., Harrell M.I., Schoolnik G.K. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park H.D., Guinn K.M., Harrell M.I., Liao R., Voskuil M.I., Tompa M. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiloh M.U., Manzanillo P., Cox J.S. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–330. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A., Deshane J.S., Crossman D.K., Bolisetty S., Yan B.S., Kramnik I. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J Biol Chem. 2008;283:18032–18039. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balazsi G., Heath A.P., Shi L., Gennaro M.L. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol Syst Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A., Toledo J.C., Patel R.P., Lancaster J.R., Jr., Steyn A.J. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Y., Crane D.D., Simpson R.M., Zhu Y.Q., Hickey M.J., Sherman D.R. The 16-Kda alpha-crystalline (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc Natl Acad Sci U S A. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvint K., Nachin L., Diez A., Nystrom T. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol. 2003;6:140–145. doi: 10.1016/s1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 22.Nystrom T., Neidhardt F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol. 1992;6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 23.Nystrom T., Neidhardt F.C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 24.Nystrom T., Neidhardt F.C. Isolation and properties of a mutant of Escherichia coli with an insertional activation of the uspA gene, which encodes a universal stress protein. J Bacteriol. 1993;175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachin L., Nannmark U., Nystrom T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. 2005;187:6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W.T., Karavolos M.H., Bulmer D.M., Allaoui A., Hormaeche R.D., Lee J.J. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb Pathog. 2007;42:2–10. doi: 10.1016/j.micpath.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Boes N., Schreiber K., Hartig E., Jaensch L., Schobert M. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J Bacteriol. 2006;188:6529–6538. doi: 10.1128/JB.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber K., Boes N., Eschbach M., Jaensch L., Wehland J., Bjarnsholt T. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J Bacteriol. 2006;188:659–668. doi: 10.1128/JB.188.2.659-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarembinski T.I., Hung L.W., Mueller-Dieckmann H.J., Kim K.K., Yokota H., Kim R. Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc Natl Acad Sci U S A. 1998;95:15189–15193. doi: 10.1073/pnas.95.26.15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sousa M.C., McKay D.B. Structure of the universal stress protein of Haemophilus influenzae. Structure. 2001;9:1135–1141. doi: 10.1016/s0969-2126(01)00680-3. [DOI] [PubMed] [Google Scholar]
- 31.O’Toole R., Williams H.D. Universal stress proteins and Mycobacterium tuberculosis. Res Microbiol. 2003;154:387–392. doi: 10.1016/S0923-2508(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 32.Schnappinger D., Ehrt S., Voskuil M.I., Liu Y., Mangan J.A., Monahan I.M. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher M.A., Plikaytis B.B., Shinnick T.M. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boshoff H.I., Reed M.B., Barry C.E., 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 35.Keer J., Smeulders M.J., Williams H.D. A purF mutant of Mycobacterium smegmatis has impaired survival during oxygen-starved stationary phase. Microbiology. 2001;147:473–481. doi: 10.1099/00221287-147-2-473. [DOI] [PubMed] [Google Scholar]
- 36.Pelicic V., Reyrat J.M., Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 37.Stewart G.R., Snewin V.A., Walzl G., Hussell T., Tormay P., O’Gaora P. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med. 2001;7:732–737. doi: 10.1038/89113. [DOI] [PubMed] [Google Scholar]
- 38.Bardarov S., Bardarov S., Jr., Pavelka M.S., Jr., Sambandamurthy V., Larsen M., Tufariello J. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J. Cold Spring Harbor Laboratory Press; 1989. Molecular cloning. [Google Scholar]
- 40.Kong D., Kunimoto D.Y. Secretion of human interleukin 2 by recombinant Mycobacterium bovis BCG. Infect Immun. 1995;63:799–803. doi: 10.1128/iai.63.3.799-803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stover C.K., de la Cruz V.F., Fuerst T.R., Burlein J.E., Benson L.A., Bennett L.T. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 42.Hatfull GF, Jacobs WR, editors. Molecular genetics of mycobacteria. Washington, DC. ASM Press.
- 43.Blokpoel M.C., Murphy H.N., O’Toole R., Wiles S., Runn E.S., Stewart G.R. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;396:190–198. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 45.Pelicic V., Jackson M., Reyrat J.M., Jacobs W.R., Jr., Gicquel B., Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardarov S., Kriakov J., Carriere C., Yu S., Vaamonde C., McAdam R.A. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boon C., Dick T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol. 2002;184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rustad T.R., Harrell M.I., Liao R., Sherman D.R. The enduring hypoxic response of Mycobacterium Tuberculosis. PLoS One. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W., Honma K., Sharma A., Kuramitsu H.K. A universal stress protein of Porphyromonas gingivalis is involved in stress responses and biofilm formation. FEMS Microbiol Lett. 2006;264:15–21. doi: 10.1111/j.1574-6968.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 50.Diez A., Gustavsson N., Nystrom T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol Microbiol. 2000;36:1494–1503. doi: 10.1046/j.1365-2958.2000.01979.x. [DOI] [PubMed] [Google Scholar]
- 51.Gustavsson N., Diez A., Nystrom T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol. 2002;43:107–117. doi: 10.1046/j.1365-2958.2002.02720.x. [DOI] [PubMed] [Google Scholar]
- 52.Honaker R.W., Leistikow R.L., Bartek I.L., Voskuil M.I. The unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect Immun. 2009 doi: 10.1128/IAI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cappelli G., Volpe E., Grassi M., Liseo B., Colizzi V., Mariani F. Profiling of Mycobacterium tuberculosis gene expression during human macrophage infection: upregulation of the alternative sigma factor G, a group of transcriptional regulators, and proteins with unknown function. Res Microbiol. 2006;157:445–455. doi: 10.1016/j.resmic.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Volpe E., Cappelli G., Grassi M., Martino A., Serafino A., Colizzi V. Gene expression profiling of human macrophages at late time of infection with Mycobacterium tuberculosis. Immunology. 2006;118:449–460. doi: 10.1111/j.1365-2567.2006.02378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tufariello J.M., Jacobs W.R., Jr., Chan J. Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo. Infect Immun. 2004;72:515–526. doi: 10.1128/IAI.72.1.515-526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Downing K.J., Betts J.C., Young D.I., McAdam R.A., Kelly F., Young M. Global expression profiling of strains harbouring null mutations reveals that the five rpf-like genes of Mycobacterium tuberculosis show functional redundancy. Tuberculosis (Edinb) 2004;84:167–179. doi: 10.1016/j.tube.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 57.Downing K.J., Mischenko V.V., Shleeva M.O., Young D.I., Young M., Kaprelyants A.S. Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro. Infect Immun. 2005;73:3038–3043. doi: 10.1128/IAI.73.5.3038-3043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kana B.D., Gordhan B.G., Downing K.J., Sung N., Vostroktunova G., Machowski E.E. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freestone P., Nystrom T., Trinei M., Norris V. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J Mol Biol. 1997;274:318–324. doi: 10.1006/jmbi.1997.1397. [DOI] [PubMed] [Google Scholar]
- 60.Freestone P., Trinei M., Clarke S.C., Nystrom T., Norris V. Tyrosine phosphorylation in Escherichia coli. J Mol Biol. 1998;279:1045–1051. doi: 10.1006/jmbi.1998.1836. [DOI] [PubMed] [Google Scholar]
- 61.Weber A., Jung K. Biochemical properties of UspG, a universal stress protein of Escherichia coli. Biochemistry. 2006;45:1620–1628. doi: 10.1021/bi051301u. [DOI] [PubMed] [Google Scholar]
- 62.Drumm J.E., Mi K., Bilder P., Sun M., Lim J., Bielefeldt-Ohmann H. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-binding: requirement for establishing chronic persistent infection. PLoS Pathog. 2009;5:e1000460. doi: 10.1371/journal.ppat.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 64.Warner D.F., Mizrahi V. Mycobacterial genetics in target validation. Drug Discov Today Technol. 2004;1:93–98. doi: 10.1016/j.ddtec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Bacon J., James B.W., Wernisch L., Williams A., Morley K.A., Hatch G.J. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 66.Starck J., Kallenius G., Marklund B.I., Andersson D.I., Akerlund T. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology. 2004;150:3821–3829. doi: 10.1099/mic.0.27284-0. [DOI] [PubMed] [Google Scholar]
- 67.Sun R., Converse P.J., Ko C., Tyagi S., Morrison N.E., Bishai W.R. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- 68.Stewart G.R., Wernisch L., Stabler R., Mangan J.A., Hinds J., Laing K.G. The heat shock response of Mycobacterium tuberculosis: linking gene expression, immunology and pathogenesis. Comp Funct Genomics. 2002;3:348–351. doi: 10.1002/cfg.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahl J.L., Kraus C.N., Boshoff H.I., Doan B., Foley K., Avarbock D. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohno H., Zhu G., Mohan V.P., Chu D., Kohno S., Jacobs W.R., Jr. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 2003;5:637–648. doi: 10.1046/j.1462-5822.2003.00307.x. [DOI] [PubMed] [Google Scholar]
- 71.Florczyk M.A., McCue L.A., Purkayastha A., Currenti E., Wolin M.J., McDonough K.A. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect Immun. 2003;71:5332–5343. doi: 10.1128/IAI.71.9.5332-5343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Talaat A.M., Lyons R., Howard S.T., Johnston S.A. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rengarajan J., Bloom B.R., Rubin E.J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sassetti C.M., Rubin E.J. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]