Abstract
Summary
This study was conducted to test whether there exists an association between vitamin D-binding protein (DBP) gene and compression strength index (CSI) phenotype. Candidate gene association analyses were conducted in total sample, male subgroup, and female subgroup, respectively. Two single-nucleotide polymorphisms (SNPs) with significant association results were found in males, suggesting the importance of DBP gene polymorphisms on the variation in CSI especially in Caucasian males.
Introduction
CSI of the femoral neck (FN) is a newly developed phenotype integrating information about bone size, body size, and bone mineral density. It is considered to have the potential to improve the performance of risk assessment for hip fractures because it is based on a combination of phenotypic traits influencing hip fractures rather than a single trait. CSI is under moderate genetic determination (with a heritability of ~44% found in this study), but the relevant genetic study is still rather scarce.
Methods
Based on the known physiological role of DBP in bone biology and the relatively high heritability of CSI, we tested 12 SNPs of the DBP gene for association with CSI variation in 405 Caucasian nuclear families comprising 1,873 subjects from the Midwestern US. Association analyses were performed in the total sample, male and female subgroups, respectively.
Results
Significant associations with CSI were found with two SNPs (rs222029, P=0.0019; rs222020, P=0.0042) for the male subgroup. Haplotype-based association tests corroborated the single-SNP results.
Conclusions
Our findings suggest that the DBP gene might be one of the genetic factors influencing CSI phenotype in Caucasians, especially in males.
Keywords: Association, Compression strength index, DBP, Haplotype, SNP
Introduction
Osteoporosis contributes to millions of fractures annually across the world [1] and represents a major public health threat in the elderly. Hip fractures are the most severe clinical outcome of osteoporosis due to its high prevalence [1, 2], serious impact on quality of life [3, 4], and excessive therapeutic costs [5]. The leading factor contributing to an increased risk of hip fractures is reduced bone strength at the proximal femur [6]. Clinically, bone mineral density (BMD) is often used to assess bone strength and predict fracture risk [1, 7]. Recent studies have shown, however, that only 50–70% of total bone strength can be attributed to BMD [8, 9]. Other factors, such as geometric structure of the femoral neck (FN), also contribute to hip strength [10]; greater femoral neck width (FNW) is associated with increased ability to resist fractures, independent of areal BMD [11]. In addition, lower body weight is associated with unfavorable changes in some of the geometric parameters of the femoral neck, which subsequently compromises hip strength [12]. Since these varied factors may influence bone strength and the overall risk of hip fractures in nonadditive ways [13], the concept of compression strength index (CSI) of the FN was developed to improve the performance of risk assessment of hip fractures [13]. CSI is a function of bone size, body size, and BMD and is based on theoretical considerations from a biomechanical viewpoint [13]. One standard deviation decrease of CSI was associated with a 2.56-fold (95% confidence interval 0.25–0.60) increase in the relative risk of hip fractures [13].
CSI was calculated as the ratio of the cross product of BMD and FNW to weight. Since the heritabilities of BMD, FNW, and weight are high [14–16], it is highly probable that CSI will be influenced by genetic factors. However, the heritability estimation of CSI and the genes responsible for CSI variation in humans has yet to be determined.
Vitamin D-binding protein (DBP), alternatively known as group-specific component, plays various roles in multiple biological and metabolic pathways, such as transporting vitamin D metabolites, regulation of bone development, binding to fatty acids, and modulation of immune and inflammatory responses [17, 18]. Particularly, DBP plays an important role in osteoclast differentiation [19], and the critical role of DBP in maintaining the skeleton has been validated in experimental animal models [20]. Furthermore, (TAAA)n repeat polymorphisms in the DBP gene have been associated with bone fracture risk, as well as with BMD of the spine and FN [19]. We are not aware of any previously published studies that have explored the relationship between DBP and CSI.
In this study, we evaluated the heritability of CSI and then investigated the association between DBP gene polymorphisms and CSI variation using high-density single-nucleotide polymorphisms (SNPs) in 1,873 subjects from 405 Caucasian nuclear families.
Subjects and methods
Subjects
This study was approved by the Institutional Review Board of all involved institutions. All participants signed informed-consent documents before entering the study. The study subjects came from an expanding database created for ongoing studies in the Midwestern USA to search for genes underlying common complex diseases/traits in humans. The detailed study design and recruitment procedures have been published before [21]. Briefly, people with chronic diseases and conditions that might potentially affect bone mass, structure, or metabolism were excluded. All 1,873 participants were US Caucasians of European origin and came from 405 nuclear families comprising 740 parents and 389 male and 744 female offspring. The average family size was 4.63±1.78 (mean ± SD, standard deviation), ranging from 3 to 12. The whole sample yielded 1,512 sib pairs and 2,266 parent–offspring pairs in total.
Measurements
BMD and bone area at the FN were measured by Hologic QDR 2000+ or 4500 dual X-ray absorptiometry (DXA; Hologic, Bedford, MA, USA). Both machines were calibrated daily. The coefficient of variation (CV) values of the DXA measurements were 1.87% on the Hologic 2000+ and 1.98% on the Hologic 4500 for FN BMD and 2.70% on the Hologic 2000+ and 2.78% on the Hologic 4500 for bone area. The absolute error of measurements by the two different machines is only ~2% in the study by Reid et al. [22]. In our study, approximately 92% of all the subjects were measured on the Hologic 4500 scanner and all data obtained from different machines were transformed to a compatible measurement using the transformation formula described by Genant et al. [23]. This transformation is based on linear regression rules and has been shown to be highly reliable and efficient in calibrating both BMD and bone area measured by different scanners [23] and extensively used in several studies [24–26]. In addition, members of the same nuclear family were measured on the same machine, ensuring minimum or no effect on our association analysis because of measurements by different scanners.
CSI was calculated as , where BMD refers to areal BMD of the FN [13]; FNW is the periosteal diameter of the FN and can be approximated by dividing the areal bone size of the FN by the width of the region of interest (in Hologic DXA systems, the width of the FN region is standardized at 1.5 cm) [27].
During the same visit for the DXA measurement, information on age, ethnic background, medical history, etc. were collected via a questionnaire. Weight and height were measured by a nurse using a calibrated balance beam scale and a standard wall-mounted ruler (see Table 1 for the descriptive characteristics of the study subjects).
Table 1.
Basic characteristics of study subjects
| Age group | Age (years) | Height (m) | Weight (kg) | FN BMD (g/cm2) | FN area (cm2) | FNW (cm) | CSI (g/kg m) |
|---|---|---|---|---|---|---|---|
| Male | |||||||
| <20 (12) | 19.58±0.27 | 1.80±0.07 | 80.60±16.06 | 0.95±0.12 | 5.65±0.27 | 3.77±0.18 | 4.58±0.87 |
| 20–29 (107) | 24.03±2.85 | 1.81±0.07 | 84.54± 14.34 | 0.98±0.15 | 5.87±0.52 | 3.91±0.35 | 4.56±0.71 |
| 30–39 (76) | 35.46±3.08 | 1.81±0.07 | 89.15±15.51 | 0.89±0.11 | 5.81±0.39 | 3.87±0.26 | 3.93±0.64 |
| 40–49 (130) | 45.65±2.83 | 1.79±0.07 | 89.73±14.79 | 0.87±0.13 | 5.85±0.47 | 3.90±0.32 | 3.84±0.59 |
| 50–59 (126) | 54.69±2.80 | 1.77±0.06 | 91.52±15.23 | 0.84±0.13 | 5.89±0.42 | 3.92±0.28 | 3.64±0.64 |
| 60–69 (81) | 64.97±2.96 | 1.77±0.06 | 91.59±15.60 | 0.79±0.12 | 5.93±0.43 | 3.96±0.28 | 3.45±0.59 |
| 70–79 (66) | 73.93±2.25 | 1.74±0.06 | 86.60±14.07 | 0.76±0.12 | 6.15±0.63 | 4.10±0.42 | 3.73±0.76 |
| >80 (24) | 82.59±2.34 | 1.73±0.08 | 80.21±13.71 | 0.76±0.14 | 6.06±0.45 | 4.04±0.30 | 3.89±0.72 |
| Female | |||||||
| <20 (24) | 19.47±0.28 | 1.66±0.07 | 64.48±10.72 | 0.89±0.18 | 4.94±0.43 | 3.29±0.28 | 4.58±0.77 |
| 20–29 (151) | 24.89±2.90 | 1.67±0.06 | 69.52±17.31 | 0.89±0.12 | 4.93±0.30 | 3.28 ±0.20 | 4.36±0.77 |
| 30–39 (192) | 35.20±2.90 | 1.65±0.06 | 68.89±15.78 | 0.83±0.11 | 4.96±0.35 | 3.30±0.23 | 4.11±0.75 |
| 40–49 (256) | 45.27±2.85 | 1.65±0.07 | 72.42±15.51 | 0.83±0.13 | 5.05±0.40 | 3.36±0.27 | 3.93±0.71 |
| 50–59 (165) | 54.02±2.86 | 1.63±0.06 | 73.91±16.63 | 0.78±0.12 | 5.07±0.38 | 3.38±0.25 | 3.65±0.66 |
| 60–69 (94) | 65.18±2.96 | 1.62±0.05 | 74.48±16.02 | 0.73±0.11 | 5.10±0.41 | 3.39±0.27 | 3.44±0.68 |
| 70–79 (70) | 74.35±3.06 | 1.59±0.06 | 71.71±13.38 | 0.69±0.12 | 5.13±0.37 | 3.42±0.25 | 3.33±0.64 |
| >80 (15) | 82.80±1.85 | 1.55±0.06 | 65.95±13.80 | 0.62±0.17 | 5.05±0.52 | 3.37±0.34 | 3.19±0.72 |
The numbers in the brackets are the sample size in each age group. In our association study sample, a total of 1,589 out of the 1,873 subjects have CSI data available. All data here are presented as means ± SD of the raw phenotypic values without adjustment
FN femoral neck, FNW femoral neck width, CSI compression strength index
Genotyping
Genomic DNA was extracted from whole blood using a commercial isolation kit (Gentra Systems, Minneapolis, MN, USA) following the procedure detailed in the kit. DNA concentration was assessed by a DU530 UV/VIS spectrophotometer (Beckman Coulter, Inc., Fullerton, CA, USA). The SNPs of interest were identified by searching public databases including dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), JSNP (http://snp.ims.u-tokyo.ac.jp/), HGVbase (http://hgv-base.cgb.ki.se/), SNP Consortium (TSC; http://snp.cshl.org/), and SNPper (http://snpper.chip.org/bio/snpper-enter). Initially, we selected 17 SNPs in or around the DBP gene on the basis of the following criteria: (1) validation status (validated experimentally in human populations), especially in Caucasians; (2) an average density of one SNP per 3 kb; (3) degree of heterozygosity, that is, minor allele frequencies (MAFs) >0.05; (4) functional relevance and importance; (5) reported to dbSNP and verified by various sources. Fourteen SNPs were successfully genotyped using the high-throughput Bead-Array SNP genotyping technology of Illumina Inc. (San Diego, CA, USA) and 12 SNPs were eventually analyzed after two SNPs were abandoned due to MAFs < 0.05. The average genotyping error rate estimated through blind sample duplication was ~0.01%.
Statistical analysis
The heritability of CSI was evaluated by a maximum likelihood variance decomposition method implemented in program SOLAR [16, 28] v4.1.0. Such an approach partitions the phenotypic variance of CSI into additive genetic and environmental variance components, making it possible to test whether correlations among family members for CSI are solely the result of the shared environment or also include genetic effects [29]. The sample for heritability evaluation came from our previous whole-genome linkage studies [26, 30].
PedCheck [31] was used to check Mendelian consistency of SNP genotype data and any inconsistent genotypes were removed. Then, the error checking option embedded in Merlin [32] was run to identify and discard the genotypes flanking excessive recombinants, thus further reducing genotyping errors. Allele frequency for each SNP was estimated by maximum likelihood methods implemented in SOLAR. And the Hardy–Weinberg equilibrium (HWE) was tested using the PEDSTATS procedure implemented in Merlin in the founders of the sample.
Linkage disequilibrium (LD) and haplotype analyses for the DBP gene were performed in the 703 unrelated parents of the study sample. The parental group consisted of 340 males and 363 females, ranging in age from 40.7 to 87.9. Population haplotypes and their frequencies were inferred using PHASE v2.1.1 software [33]. Pairwise |D′| for all studied SNPs were calculated and LD structures plotted by the program GOLD [34]. HaploBlockFinder [35] was used to identify block structures and select haplotype-tagging SNPs (htSNPs) for the DBP gene. To infer haplotypes defined by the tagging SNPs of each block of DBP for all the study subjects, we adopted the algorithm of integer linear programming implemented in PedPhase v2.0, which is based on LD assumption and is capable of recovering phase information at each marker locus with high speed and accuracy, even in the presence of 20% missing data [36].
We conducted family-based association tests (FBAT) [37] for single SNPs and haplotypes with the CSI as a quantitative trait in all subjects, as well as in male and female subjects, separately. We adopted the additive genetic model implemented in the software package FBAT V1.5.5 for our association analyses because this model works well even when the true genetic model is not an additive one. The allelic association in this framework examines transmission of the markers of interest from parents to the affected offspring. Haplotype association tests were performed by the haplotype-based association test (HBAT) which is robust to population admixture, phenotype distribution misspecification, and ascertainment bias. Empirical global P values for single-SNP and haplotype analyses were obtained using the Monte-Carlo permutation procedures implemented in HBAT (10,000 permutations were conducted). To correct for the false-positive results caused by multiple testing, the program SNP spectral decomposition (SNPSpD) [38] was used to establish the experiment-level significance threshold needed to keep the type I error rate below 5% on the basis of the spectral decomposition of matrices of pairwise LD between SNPs [38]. This method provides efficient and reasonable corrections for multiple testing problems by estimating the effective test number [38]. We also randomly selected one subject from each family to generate an unrelated testing sample for detecting the magnitude of SNP effects by linear regression analyses in Minitab software (Minitab Inc., State College, PA, USA).
Age, age-squared, sex, sex × age, and age-squared × sex were tested for significant effect on CSI, and significant factors (P<0.05) were adopted in the linear model to adjust the raw CSI values in the total sample by use of Minitab. In sex-specific analyses, the CSI residuals were obtained by adjusting original CSI data for age. All the CSI residuals were tested for normality by Kolmogorov–Smirnov test implemented in the software Minitab before subsequent analyses. Due to their departure from a normal distribution, the residuals were transformed by the Box–Cox transformation procedure implemented in Minitab.
In addition, we also tested whether DBP polymorphisms were related to FN BMD, FNW, and weight using the previously mentioned protocols in order to find the difference between the abilities of CSI and FN BMD and FNW and weight to assess hip fractures.
Results
Heritability estimation
Descriptive characteristics of the study subjects, stratified by sex and age, are presented in Table 1. The data show that the means of CSI generally decrease with the increase of age in both sexes. And the means of CSI are much higher in men than in women in the age groups of 70–79 and >80 years. The heritability of adjusted CSI under the polygenic model was ~0.44 (standard error = 0.03, P=5.54×10−64).
LD and haplotype analysis
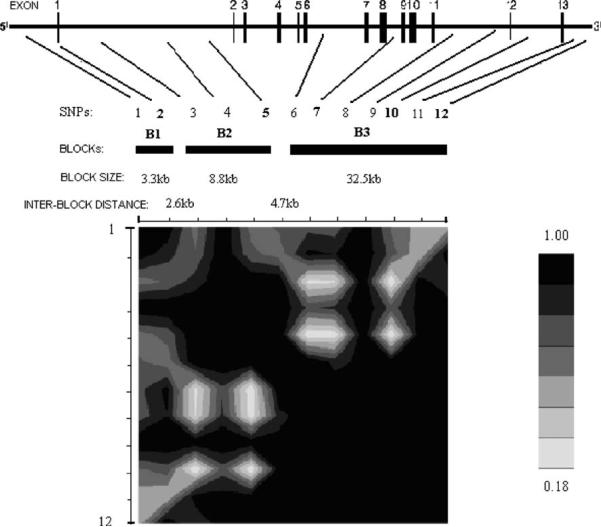
The DBP gene has a length of ~52 kb and is located on chromosome 4q13.3. Information about the 12 analyzed SNPs within DBP is summarized in Table 2. On average, these SNPs were spaced ~4 kb apart and covered the full transcript length of DBP. They can capture 19 SNPs of a total of 35 SNPs in or around DBP gene from Hapmap with MAF > 0.05 and R2>0.8. So the coverage should be about 54% (=19/35). Three blocks with high pairwise LD were reconstructed (Fig. 1); these blocks ranged in size from 3.3 to 32.5 kb. Block 1 contained SNP 1 and SNP 2 and spanned the promoter region. Block 2 included SNPs 3, 4, and 5 and mainly covered intron 1. Block 3 contained SNPs 6–12 and extended from intron 6 to the 3′ untranslated region. Five htSNPs were identified to represent common haplotypes for each block of DBP (Fig. 1).
Table 2.
Basic characteristics of the studied SNPs in the DBP gene
| Number | Gene and SNP ID | Allele variantsa | MAF | Chromosomal positionb (bp) | SNP type |
|---|---|---|---|---|---|
| 1 | rs16847036 | G/A | 0.050 | 72873089 | Promoter |
| 2 | rs3733359 | A/G | 0.050 | 72868638 | Exon 1 |
| 3 | rs222029 | G/A | 0.162 | 72863826 | Intron 1 |
| 4 | rs1352843 | G/A | 0.156 | 72861818 | Intron 1 |
| 5 | rs222020 | G/A | 0.155 | 72855136 | Intron 1 |
| 6 | rs222003 | C/G | 0.070 | 72844952 | Intron 6 |
| 7 | rs222035 | A/C | 0.425 | 72840538 | Intron 8 |
| 8 | rs222040 | G/A | 0.421 | 72835796 | Intron 11 |
| 9 | rs222042 | A/G | 0.072 | 72831480 | Intron 11 |
| 10 | rs705117 | G/A | 0.140 | 72826979 | Intron 12 |
| 11 | rs17467825 | G/A | 0.284 | 72824381 | 3′ UTR |
| 12 | rs1491711 | C/G | 0.333 | 72821116 | 3′ UTR |
SNP single-nucleotide polymorphism, MAF minor allele frequency
The first allele is the minor allele
Positions are based on NCBI web site
Fig. 1.
LD structure of the DBP gene. The analyzed SNPs are marked in sequence by their locations. The SNP IDs correspond to those given in Table 2. Haplotype-tagging SNPs (htSNPs) are in bold. Blocks with all pairwise |D′| values >0.8 are illustrated and numbered B1–B3. Block size and interblock distances are indicated
Association of DBP polymorphisms with CSI
Significant empirical global P values from single-SNP association analyses for CSI are summarized in Table 3. Among the 12 SNPs, two markers, SNP 3 (rs222029) and SNP 5 (rs222020), showed nominally significant associations (P<0.05) with CSI variation in the total sample by an FBAT test. In sex-specific analyses, the significant association of SNPs 3 (rs222029, P=0.0019) and 5 (rs222020, P=0.0042) with CSI were only present in men. The experiment-wide significant threshold p value was set at 0.0049 by the program SNPSpD. Thus, the association of SNP 3 (rs222029) and SNP 5 (rs222020) with CSI variation in the male sample remained significant after correction for multiple testing. The contributions of these two SNPs to CSI variation were 1.80% and 1.60%, respectively, in our male random sample as determined by R2 value in the linear regression analyses (Table 3).
Table 3.
SNP-based and haplotype-based association results of DBP polymorphisms
| SNPsa and haplotypesb | CSI P valuesc | R2 %d | FN BMD P valuesc | FNW P valuesc | Weight P valuesc | |
|---|---|---|---|---|---|---|
| Total | SNPs from block 2 | |||||
| SNP 3 | 0.0106 | 1.50 | 0.0925 | 0.1876 | 0.5208 | |
| SNP 5 | 0.0110 | 1.20 | 0.0580 | 0.1671 | 0.7209 | |
| Haplotypes from Block 2 | ||||||
| AAA, 0.836 | 0.0110 (0.0115) | 0.0580 (0.0816) | 0.1671 (0.1957) | 0.7209 (0.6044) | ||
| GGG, 0.114 | 0.0110 (0.0911) | 0.0580 (0.2897) | 0.1671 (0.0945) | 0.7209 (0.9664) | ||
| Male | SNPs from block 2 | |||||
| SNP 3 | 0.0019 | 1.80 | 0.0708 | 0.6369 | 0.3361 | |
| SNP 5 | 0.0042 | 1.60 | 0.1195 | 0.4454 | 0.3063 | |
| Haplotypes from Block 2 | ||||||
| AAA, 0.836 | 0.0042 (0.0041) | 0.1195 (0.0847) | 0.4454 (0.4870) | 0.3063 (0.3137) | ||
| GGG, 0.114 | 0.0042 (0.0353) | 0.1195 (0.0815) | 0.4454 (0.2783) | 0.3063 (0.7938) | ||
| Female | SNPs from block 2 | |||||
| SNP 3 | 0.3774 | 0.70 | 0.4515 | 0.5051 | 0.8633 | |
| SNP 5 | 0.3643 | 0.40 | 0.2554 | 0.5464 | 0.7958 | |
| Haplotypes from Block 2 | ||||||
| AAA, 0.836 | 0.3643 (0.4848) | 0.2554 (0.4059) | 0.5464 (0.5506) | 0.7958 (0.8819) | ||
| GGG, 0.114 | 0.3643 (0.8418) | 0.2554 (0.9987) | 0.5464 (0.5147) | 0.7958 (0.9224) | ||
SNP numbers corresponded to those given in Table 2. For simplicity, the results for all the other polymorphisms of the DBP gene are not listed because they are not significant
In block 2, we only list results for two haplotypes with estimated frequencies more than 5% (0.836 and 0.114, respectively). The SNP in bold was tag SNP
P values of family-based association test are shown, which were generated by 10,000 permutations. P values<0.05 are shown in bold. Significant P values after correction for multiple testing by use of SNPSpD approach are indicated by bold italics. Besides htSNP-based haplotype analysis, we also performed all-SNP-based haplotype analysis in block 2 using an “hbat” command. The results of all-SNP haplotype tests are shown in brackets.
R2 indicates the magnitude of SNPs effects on CSI, which was obtained using linear regression analyses.
The haplotype association results based on htSNPs were largely consistent with our results from single-SNP association tests. We have presented the association results for the most common haplotypes (frequency >5%) for block 2 in Table 3. We observed a nominally significant association between CSI and block 2, represented by SNP 5 (rs222020) and containing SNPs 3–5, with P values of 0.0110 and 0.0042 in the total and male samples, respectively. The association of block 2 with CSI in the male sample remained significant after correction for multiple testing. In addition, we performed association analyses based on the haplotypes constructed by all three of the SNPs in block 2 using the “hbat” command in the FBAT package. The major haplotype of block 2 (frequency of 83.6%) containing the three loci combination “A-A-A” consistently showed an experiment-wide significant association signal (P=0.0041) in the male sample. Compared with those with homozygous G-G-G haplotype, the subjects with homozygous A-A-A haplotype had higher CSI. The analyses of haplotype effects in the random sample showed that the A-A-A conferred 4.4% higher CSI than the alternative haplotype G-G-G did.
Association of DBP polymorphisms with FN BMD, FNW, and weight
For FN BMD, FNW, and weight, there was no evidence of association (all P values>0.05) with DBP polymorphism for any single-SNP and haplotype.
Discussion
In this study, we performed both SNP-based and haplotype-based analyses to assess the association of the DBP gene with CSI in a large sample of nuclear families using FBAT software. The rationale for performing this study was based on the known physiological role of DBP in bone biology [39–41] and the relatively high heritability of CSI.
To the best of our knowledge, this is the first study to test for associations between DBP and CSI. The significant associations with CSI were found to be male specific in our sample. The most promising associations between haplotypes and CSI were detected in block 2 in the male subgroup. Importantly, associations between DBP and FN BMD and FNW and weight were not significant. Consequently, it is evident that important genetic factors influencing osteoporosis may be missed if BMD or FNW or weight is the only phenotypic trait analyzed for the genetic dissection of osteoporosis. In addition, SNP 3 (rs222029), which produced the most significant association signal in males and is contained in block 2, lies in a transcription factor binding site revealed by a web-based functional analysis and selection tool for SNP (http://fastsnp.ibms.sinica.edu.tw/pages/input_SNPListAnalysis.jsp). Therefore, the variation of SNP 3 (rs222029), or functional mutations in strong LD with it, may influence DBP gene expression by affecting the binding capacity of transcription factors. Further molecular studies are required to validate this hypothesis. Finally, these sex-specific association results were not unexpected given previous studies demonstrating that DBP polymorphisms affect plasma levels of DBP, bone density, and fracture risk in males [19] and that FN BMD correlated with serum DBP levels in elderly Caucasian men [42]. Besides BMD, another major contributory factor to fracture risk is muscle strength [43]. There is evidence showing that muscle strength significantly correlated with 25 (OH) vitamin D only in males [44]. Therefore, the importance of DBP as vitamin D transport molecule on fractures might be more obvious in males.
A bone fracture occurs when stress within the bone exceeds its ultimate strength. The stress effect depends on the geometry, mechanical properties, and spatial distribution of the bone, as well as the direction, magnitude, and position of the force applied [45]. Based on the structural engineering principles [13], CSI integrates density and geometric information embedded in DXA scans with mechanical loading effects of total body weight on the skeletal system. Previous studies show that FN BMD, FN geometric parameters, and weight are highly correlated with each other [46–48]. They may share common genetic factors and molecular pathways contributing to their variation in human populations. Only one phenotype alone may not be able to predict hip fractures well. This is similar to the situation of metabolic syndrome (Mets), which was defined by multiple concurrent diseases including abdominal obesity, insulin resistance, dyslipidemia, and elevated blood pressure [49]. To investigate the genetic factors that may have a common contribution to a clustering of hip fracture-related phenotypes and maximize the chance to identify hip fracture-related genetic factors, it is helpful to use an alternative measurement that characterizes the common features of these highly correlated phenotypes that are significant to fractures. Thus, this newly developed CSI phenotype combining FN BMD with FNW and weight was used to find the shared genetic information among its correlated components.
The FBAT software used in our association study is an extension of the transmission disequilibrium test and can deal with incomplete nuclear families [37]. The statistical power of our study was estimated by an online Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/qtlassoc.htm) under a relatively conservative significance level of P=0.001. Assuming that a marker is in strong LD (|D′| = 0.9) with a functional mutation that accounts for 4% variation in phenotype, our study design can reach >90% power in both females and males, under additive models. The 90% power was estimated under a rather ideal situation. We note that the power under various conditions may be affected by multiple other factors, such as LD between the markers tested and the functional allele and the phenotypic variation the functional allele may account for. When assuming a marker is in a weaker LD (|D′| = 0.75) with a functional mutation that accounts for 1% variation in phenotype, the power reduces to 43% and 72% in male and female, respectively.
Several other issues need to be addressed here. First, the FNW (the average diameter of the FN) was obtained by dividing the bone area by the width of the region of interest, usually 1.5 cm. A previous study [27] has assessed the reliability of this approximation of bone geometry, with the conclusion that FN diameter estimated from bone area had a single measured CV of 10.6% compared with 9.6% (n=30) obtained by direct measurement with the DXA ruler tool. Second, CSI of the FN studied here was derived from DXA measurement, which is not sensitive to the mineralization quality of bones. Although deterioration in the quality of mineralization with age may contribute to an increased susceptibility to fractures in the elderly, age alone explains only a small part of the total variance in material strength of cadaveric cortical femoral bone [50]. Therefore, the CSI phenotype based on structural measures can provide good assessment of hip fracture risk [13]. Third, due to the shape of the fan beam X-ray in our QDR 4500 scanner, an inherent magnification of scanned bone area at FN were generated and hence caused the projected area of FN to decrease linearly with distance above the X-ray source. This change was estimated as 1.6% cm−1 by simulating FN as round solid aluminum rod in Cole's study [51]. However, there is no good method to correct the magnification error for an individual measured by this scanner because the soft tissue thickness that could be used to correct this error is not available [52]. Thus, hardware and software modifications for the Hologic fan beam DXA machines would be necessary to allow scanning at different table heights to calculate soft tissue thickness [52] and thus overcome the limitation.
In summary, our findings suggest the importance of DBP gene polymorphisms on the variation in CSI in Caucasians, especially in men. It is necessary to perform further statistical genetic and functional studies to replicate and confirm these results in other populations in order to assess the generality of the findings.
Acknowledgements
The study was partially supported by grants from NIH (R01 AR050496, R21 AG027110, R01 AG026564, P50 AR055081, and R21 AA015973). The study was also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, HuNan Province, Xi'an Jiaotong University, and the Ministry of Education of China.
Footnotes
Conflicts of interest None.
References
- 1.Cummings SR, Melton LJ. Epidemiology and outcomes of osteoporotic fractures. Lancet. 2002;359:1761–1767. doi: 10.1016/S0140-6736(02)08657-9. [DOI] [PubMed] [Google Scholar]
- 2.Lau EM. Epidemiology of osteoporosis. Best Pract Res Clin Rheumatol. 2001;15:335–344. doi: 10.1053/berh.2001.0153. [DOI] [PubMed] [Google Scholar]
- 3.Braithwaite RS, Col NF, Wong JB. Estimating hip fracture morbidity, mortality and costs. J Am Geriatr Soc. 2003;51:364–370. doi: 10.1046/j.1532-5415.2003.51110.x. [DOI] [PubMed] [Google Scholar]
- 4.March LM, Cameron ID, Cumming RG, et al. Mortality and morbidity after hip fracture: can evidence based clinical pathways make a difference? J Rheumatol. 2000;27:2227–2231. [PubMed] [Google Scholar]
- 5.Campion EW, Jette AM, Cleary PD, et al. Hip fracture: a prospective study of hospital course, complications, and costs. J Gen Intern Med. 1987;2:78–82. doi: 10.1007/BF02596300. [DOI] [PubMed] [Google Scholar]
- 6.Lauritzen JB. Hip fractures. Epidemiology, risk factors, falls, energy absorption, hip protectors, and prevention. Dan Med Bull. 1997;44:155–168. [PubMed] [Google Scholar]
- 7.Melton LJ, 3rd, Atkinson EJ, O'Fallon WM, et al. Long-term fracture prediction by bone mineral assessed at different skeletal sites. J Bone Miner Res. 1993;8:1227–1233. doi: 10.1002/jbmr.5650081010. [DOI] [PubMed] [Google Scholar]
- 8.Ammann P, Rizzoli R. Bone strength and its determinants. Osteoporos Int. 2003;14(Suppl 3):S13–S18. doi: 10.1007/s00198-002-1345-4. [DOI] [PubMed] [Google Scholar]
- 9.Stone KL, Seeley DG, Lui LY, et al. BMD at multiple sites and risk of fracture of multiple types: long-term results from the Study of Osteoporotic Fractures. J Bone Miner Res. 2003;18:1947–1954. doi: 10.1359/jbmr.2003.18.11.1947. [DOI] [PubMed] [Google Scholar]
- 10.Black DM, Bouxsein ML, Marshall LM, et al. Proximal femoral structure and the prediction of hip fracture in men: a large prospective study using QCT. J Bone Miner Res. 2008;23:1326–1333. doi: 10.1359/JBMR.080316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng XG, Lowet G, Boonen S, et al. Assessment of the strength of proximal femur in vitro: relationship to femoral bone mineral density and femoral geometry. Bone. 1997;20:213–218. doi: 10.1016/s8756-3282(96)00383-3. [DOI] [PubMed] [Google Scholar]
- 12.Brownbill RA, Ilich JZ. Hip geometry and its role in fracture: what do we know so far? Curr Osteoporos Rep. 2003;1:25–31. doi: 10.1007/s11914-003-0005-8. [DOI] [PubMed] [Google Scholar]
- 13.Karlamangla AS, Barrett-Connor E, Young J, et al. Hip fracture risk assessment using composite indices of femoral neck strength: the Rancho Bernardo study. Osteoporos Int. 2004;15:62–70. doi: 10.1007/s00198-003-1513-1. [DOI] [PubMed] [Google Scholar]
- 14.Prentice A. The relative contribution of diet and genotype to bone development. Proc Nutr Soc. 2001;60:45–52. doi: 10.1079/pns200072. [DOI] [PubMed] [Google Scholar]
- 15.Peacock M, Koller DL, Lai D, et al. Sex-specific quantitative trait loci contribute to normal variation in bone structure at the proximal femur in men. Bone. 2005;37:467–473. doi: 10.1016/j.bone.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayoumi RA, Al-Yahyaee SA, Albarwani SA, et al. Heritability of determinants of the metabolic syndrome among healthy Arabs of the Oman family study. Obesity (Silver Spring) 2007;15:551–556. doi: 10.1038/oby.2007.555. [DOI] [PubMed] [Google Scholar]
- 17.Gomme PT, Bertolini J. Therapeutic potential of vitamin D-binding protein. Trends Biotechnol. 2004;22:340–345. doi: 10.1016/j.tibtech.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 18.White P, Cooke N. The multifunctional properties and characteristics of vitamin D-binding protein. Trends Endocrinol Metab. 2000;11:320–327. doi: 10.1016/s1043-2760(00)00317-9. [DOI] [PubMed] [Google Scholar]
- 19.Papiha SS, Allcroft LC, Kanan RM, et al. Vitamin D binding protein gene in male osteoporosis: association of plasma DBP and bone mineral density with (TAAA)(n)-Alu polymorphism in DBP. Calcif Tissue Int. 1999;65:262–266. doi: 10.1007/s002239900695. [DOI] [PubMed] [Google Scholar]
- 20.Safadi FF, Thornton P, Magiera H, et al. Osteopathy and resistance to vitamin D toxicity in mice null for vitamin D binding protein. J Clin Invest. 1999;103:239–251. doi: 10.1172/JCI5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong DH, Liu YZ, Liu PY, et al. Association analysis of estrogen receptor alpha gene polymorphisms with cross-sectional geometry of the femoral neck in Caucasian nuclear families. Osteoporos Int. 2005;16:2113–2122. doi: 10.1007/s00198-005-2011-4. [DOI] [PubMed] [Google Scholar]
- 22.Reid DM, Mackay I, Wilkinson S, et al. Cross-calibration of dual-energy X-ray densitometers for a large, multi-center genetic study of osteoporosis. Osteoporos Int. 2006;17:125–132. doi: 10.1007/s00198-005-1936-y. [DOI] [PubMed] [Google Scholar]
- 23.Genant HK, Grampp S, Gluer CC, et al. Universal standardization for dual x-ray absorptiometry: patient and phantom cross-calibration results. J Bone Miner Res. 1994;9:1503–1514. doi: 10.1002/jbmr.5650091002. [DOI] [PubMed] [Google Scholar]
- 24.Huang QY, Xu FH, Shen H, et al. Genome scan for QTLs underlying bone size variation at 10 refined skeletal sites: genetic heterogeneity and the significance of phenotype refinement. Physiol Genomics. 2004;17:326–331. doi: 10.1152/physiolgenomics.00161.2002. [DOI] [PubMed] [Google Scholar]
- 25.Shen H, Zhang YY, Long JR, et al. A genome-wide linkage scan for bone mineral density in an extended sample: evidence for linkage on 11q23 and Xq27. J Med Genet. 2004;41:743–751. doi: 10.1136/jmg.2004.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong DH, Shen H, Xiao P, et al. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res. 2006;21:424–437. doi: 10.1359/JBMR.051202. [DOI] [PubMed] [Google Scholar]
- 27.Rivadeneira F, Houwing-Duistermaat JJ, Beck TJ, et al. The influence of an insulin-like growth factor I gene promoter polymorphism on hip bone geometry and the risk of nonvertebral fracture in the elderly: the Rotterdam Study. J Bone Miner Res. 2004;19:1280–1290. doi: 10.1359/JBMR.040405. [DOI] [PubMed] [Google Scholar]
- 28.Gu D, Rice T, Wang S, et al. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luke A, Guo X, Adeyemo AA, et al. Heritability of obesity-related traits among Nigerians, Jamaicans and US black people. Int J Obes Relat Metab Disord. 2001;25:1034–1041. doi: 10.1038/sj.ijo.0801650. [DOI] [PubMed] [Google Scholar]
- 30.Liu XG, Liu YJ, Liu J, et al. A bivariate whole genome linkage study identified genomic regions influencing both BMD and bone structure. J Bone Miner Res. 2008;23:1806–1814. doi: 10.1359/JBMR.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 33.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis GR, Cookson WO. GOLD—graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K, Jin L. HaploBlockFinder: haplotype block analyses. Bioinformatics. 2003;19:1300–1301. doi: 10.1093/bioinformatics/btg142. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Jiang T. Computing the minimum recombinant haplotype configuration from incomplete genotype data on a pedigree by integer linear programming. J Comput Biol. 2005;12:719–739. doi: 10.1089/cmb.2005.12.719. [DOI] [PubMed] [Google Scholar]
- 37.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 38.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benis KA, Schneider GB. The effects of vitamin D binding protein-macrophage activating factor and colony-stimulating factor-1 on hematopoietic cells in normal and osteopetrotic rats. Blood. 1996;88:2898–2905. [PubMed] [Google Scholar]
- 40.Schneider GB, Benis KA, Flay NW, et al. Effects of vitamin D binding protein-macrophage activating factor (DBP-MAF) infusion on bone resorption in two osteopetrotic mutations. Bone. 1995;16:657–662. doi: 10.1016/8756-3282(95)00118-w. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto N, Naraparaju VR, Orchard PJ. Defective lymphocyte glycosidases in the macrophage activation cascade of juvenile osteopetrosis. Blood. 1996;88:1473–1478. [PubMed] [Google Scholar]
- 42.Rapado A, Hawkins F, Sobrinho L, et al. Bone mineral density and androgen levels in elderly males. Calcif Tissue Int. 1999;65:417–421. doi: 10.1007/s002239900726. [DOI] [PubMed] [Google Scholar]
- 43.Sipila S, Heikkinen E, Cheng S, et al. Endogenous hormones, muscle strength, and risk of fall-related fractures in older women. J Gerontol A Biol Sci Med Sci. 2006;61:92–96. doi: 10.1093/gerona/61.1.92. [DOI] [PubMed] [Google Scholar]
- 44.Bischoff HA, Stahelin HB, Tyndall A, et al. Relationship between muscle strength and vitamin D metabolites: are there therapeutic possibilities in the elderly? Z Rheumatol. 2000;59(Suppl 1):39–41. doi: 10.1007/s003930070037. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Peel N, Clowes JA, et al. Use of DXA-based structural engineering models of the proximal femur to discriminate hip fracture. J Bone Miner Res. 2009;24:33–42. doi: 10.1359/jbmr.080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irdesel J, Ar I. The relationship between the proximal femur morphometry and bone mineral density in Turkish women. Minerva Med. 2006;97:153–159. [PubMed] [Google Scholar]
- 47.Xu H, Long JR, Yang YJ, et al. Genetic determination and correlation of body weight and body mass index (BMI) and cross-sectional geometric parameters of the femoral neck. Osteoporos Int. 2006;17:1602–1607. doi: 10.1007/s00198-006-0141-y. [DOI] [PubMed] [Google Scholar]
- 48.Harris SS, Dawson-Hughes B. Weight, body composition, and bone density in postmenopausal women. Calcif Tissue Int. 1996;59:428–432. doi: 10.1007/BF00369205. [DOI] [PubMed] [Google Scholar]
- 49.Cornier MA, Dabelea D, Hernandez TL, et al. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith CB, Smith DA. Relations between age, mineral density and mechanical properties of human femoral compacta. Acta Orthop Scand. 1976;47:496–502. doi: 10.3109/17453677608988727. [DOI] [PubMed] [Google Scholar]
- 51.Cole JH, Scerpella TA, van der Meulen MC. Fan-beam densitometry of the growing skeleton: are we measuring what we think we are? J Clin Densitom. 2005;8:57–64. doi: 10.1385/jcd:8:1:057. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths MR, Noakes KA, Pocock NA. Correcting the magnification error of fan beam densitometers. J Bone Miner Res. 1997;12:119–123. doi: 10.1359/jbmr.1997.12.1.119. [DOI] [PubMed] [Google Scholar]