Abstract
Ras-associated binding (Rab) protein 3A is a neuronal guanosine triphosphate (GTP)-binding protein that binds synaptic vesicles and regulates synaptic transmission. A mouse mutant, earlybird (Ebd), with a point mutation in the GTP-binding domain of Rab3A (D77G), exhibits anomalies in circadian behavior and homeostatic response to sleep loss. Here, we show that the D77G substitution in the Ebd allele causes reduced GTP and GDP binding, whereas GTPase activity remains intact, leading to reduced protein levels of both Rab3A and rabphilin3A. Expression profiling of the cortex and hippocampus of Ebd and Rab3a-deficient mice revealed subtle differences between wild-type and mutant mice. Although mice were backcrossed for three generations to a C57BL/6J background, the most robust changes at the transcriptional level between Rab3a−/− and Rab3a+/+ mice were represented by genes from the 129/Sv-derived chromosomal region surrounding the Rab3a gene. These results showed that differences in genetic background have a stronger effect on gene expression than the mutations in the Rab3a gene. In behavioral tests, the Ebd/Ebd mice showed a more pronounced mutant phenotype than the null mice; Ebd/Ebd have reduced anxiety-like behavior in the elevated zero-maze test, reduced response to stress in the forced swim test and a deficit in cued fear conditioning (FC), whereas Rab3a−/− showed only a deficit in cued FC. Our data implicate Rab3A in learning and memory as well as in the regulation of emotion. A combination of forward and reverse genetics has provided multiple alleles of the Rab3a gene; our studies illustrate the power and complexities of the parallel analysis of these alleles at the biochemical, molecular and behavioral levels.
Keywords: Behavior, GTPase, microarray, mouse mutant, Rab3A
Ras-associated binding (Rab) protein 3A plays a regulatory role in neurotransmitter release and calcium-triggered synaptic vesicle exocytosis (Sudhof 2004; Takai et al. 1996; Takai et al. 2001; Zerial & McBride 2001). Rab3A is the key member of the functionally redundant Rab3 family of small G proteins named Rab3A, Rab3B, Rab3C and Rab3D (Schluter et al. 2002; Schluter et al. 2004) and is the most abundant among these paralogs in the brain (Geppert et al. 1994). Rab3A functions by shuffling between a vesicle-associated guanosine triphosphate (GTP)-bound state and vesicle-dissociated GDP-bound state under regulation by Rab GDP dissociation inhibitor (Rab GDI) (Matsui et al. 1990; Sasaki et al. 1990; Ullrich et al. 1993), GTPase activating protein (Rab3 GAP) (Fukui et al. 1997; Nagano et al. 1998), GDP/GTP exchange protein (Rab3 GEP) (Wada et al. 1997) and guanine nucleotide exchange factors (Burton et al. 1994; Luo et al. 2001). Several Rab3A effectors have been identified, including rabphilin3A (Li et al. 1994; Shirataki et al. 1992; Shirataki et al. 1993) and RIM1α/2α (Wang & Sudhof 2003; Wang et al. 1997; Wang et al. 2000), which bind only to GTP-Rab3A but not to GDP-Rab3A.
Electrophysiological studies of mice with a targeted loss-of-function mutation in Rab3a revealed altered short-term plasticity in synapses of the hippocampal CA1 region. The loss of Rab3A affected the late step of calcium-triggered vesicle exocytosis following transportation and docking (Geppert et al. 1994; Geppert et al. 1997; Schluter et al. 2004). Rab3A is also required for hippocampal CA3 mossy fiber long-term potentiation (LTP) and normal long-term depression (LTD), two forms of synaptic plasticity (Geppert et al. 1997). Rab3A mutants exhibit impairments in protein kinase A-dependent forms of corticoamygdala LTP and latephase LTP at hippocampal CA3-CA1 synapses (Huang et al. 2005). These phenotypes may be partly mediated by the interaction of Rab3A with its effector protein RIM1a (Betz et al. 2001; Powell et al. 2004; Schoch et al. 2002).
Behavioral assessments of Rab3a-null mutant mice have revealed normal hippocampus-dependent learning and memory in contextual fear conditioning (FC) as well as in the Morris water-maze test (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004), although D’Adamo et al. reported moderately impaired platform reversal learning in the water maze in reference memory and episodic-like memory tasks (D’Adamo et al. 2004). In addition, Hensbroek et al. found reduced exploratory behavior, whereas D’Adamo et al. reported increased exploratory behavior in Rab3a knockout mice (D’Adamo et al. 2004; Hensbroek et al. 2003). Lastly, D’Adamo et al. reported reduced anxiety-like behavior in the same mice (D’Adamo et al. 2004), which was not detected by Powell et al. (Powell et al. 2004)
In addition to the targeted loss-of-function allele of Rab3a, a second allele, earlybird (Rab3aEbd), was identified independently using a forward genetic approach (Kapfhamer et al. 2002). Ebd/Ebd mice carry a point mutation that causes the replacement of aspartic acid with glycine in Rab3A’s GTP-binding pocket, a highly conserved domain in all GTPases. Behavioral assessment of multiple rest-activity parameters in the Rab3a−/− and Ebd/Ebd mice revealed shortened circadian period of activity and anomalies in sleep homeostasis (Kapfhamer et al. 2002).
As the first step toward understanding the mechanisms that underlie the behavioral anomalies in Ebd mice, we examined the biochemical properties of the mutant form of Rab3A (Rab3AD77G). We also compared Ebd and Rab3a−/− alleles at the transcriptome level by microarray analysis and at the behavioral level through a battery of tests. Our results show that disrupted GTP and GDP binding to Rab3A in Ebd mice and loss-of-function of Rab3A in Rab3a−/− mice affect learning and memory as well as emotional regulation, without significant changes in overall gene expression.
Materials and methods
Animals
Knockout mice for Rab3a (C57BL/6J; 129Sv-Rab3atm1Sud; stock code 002443) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) and backcrossed for three generations (N4) to C57BL/6J (B6). Rab3aEbd mice were identified in an ENU-mutagenesis screen of C57BL/6J mice (Kapfhamer et al. 2002), and after a cross to C3H/HeJ (C3), mice were backcrossed for three generations (N4) to C57BL/6J. Mutant and wild-type mice were maintained on a light/dark (12:12) cycle with lights on at 0700 h. Food and water were available ab libitum using standard mouse hus-bandry procedures. All animal experiments were carried out according to the National Institutes of Health guidelines for the use of animals and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Wild-type, heterozygous and homozygous mutant mice were generated by intercrossing Ebd/+ and Rab3a+/− heterozygous mice. At time of weaning (3 weeks), tail biopsies were taken to extract genomic DNA for genotyping. For Ebd mice, genotyping was performed by allelic discrimination on ABI Prism 7900HT (Applied Biosystems, Foster City, CA, USA) using a custom-designed single nucleotide polymorphism (SNP) assay that has a probe flanking the point mutation [A/G]. Genotypes were called automatically or manually using sds 2.2 software (Applied Biosystems). The knockout line was genotyped as previously described (Kapfhamer et al. 2002). To assess the 129/Sv (129) or C3H-derived portion of the chromosome surrounding the Rab3a locus, the following 12 microsatellite markers were used for genotyping: D8Mit190.1, D8Mit46, D8Mit128, D8Mit25, D8Mit29, D8Mit73, D8Mit178.1, D8Mit304, D8Mit104, D8Mit348, D8Mit45.1 and D8Mit84. Among them, D8Mit46, D8Mit128 and D8Mit25 showed no detectable polymorphism between C57BL/6J and 129/Sv strains.
Tissue collection, RNA extraction and cDNA generation
For the analysis of both the Ebd and Rab3a−/− alleles, wild-type and homozygous mutant mice were sacrificed at Zeitgeber time 9. Cerebral cortex and hippocampus were dissected, frozen on dry ice and then transferred to −80 °C. Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) followed by cleanup using RNeasy mini kit (Qiagen Inc., Valencia, CA, USA). For all of these RNA samples, 1–2 μg was reverse transcribed using a High Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer’s instructions.
Cloning and expression of wild-type and Ebd mutant Rab3A
The cDNAs from the cortex of wild-type and Ebd mutant mice were used as template to amplify Rab3a by AccuPrime Pfx SuperMix (Invitrogen Life Technologies) using forward primer 5′-AGAGAGGGTAAGATGGCTTCC-3′ and reverse primer 5′-AATAGGGTAGTCGGGGATGG-3′. The polymerase chain reaction (PCR) products were inserted into the pCR2.1 vector using a TA Cloning Kit (Invitrogen Life Technologies) following the manufacturer’s instructions. The wild-type and Ebd Rab3A cDNAs were then subcloned in-frame into the pcDNA3.1/His C vector (Invitrogen Life Technologies). Positive clones were confirmed by sequencing. HEK 293 (human embryonic kidney epithelial cell line) cells (kindly provided by Dr Adam Crystal) were transfected by Lipofectamine 2000 (Invitrogen Life Technologies) with pcDNA3.1/His C constructs harboring his-tagged wild-type and Ebd mutant Rab3A. Stable cell lines were selected by culturing cells in the presence of 0.5 mg/ml of geneticin. Cells were lysed by BD Xtractor, and his-tagged protein was purified by BD TALON affinity resin (BD Biosciences, Palo Alto, CA, USA).
Western blot analysis
The mouse brain tissues and the cells were homogenized in CelLytic MT (Sigma, St. Louis, MO, USA) in the presence of 2% proteinase inhibitor cocktail P-8340 (Sigma). The homogenates were centrifuged at 10 000 g at 4 °C for 20 min. The supernatants were applied to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), and the protein was transferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon-P, MiliPore, Bedford, MA, USA). The blot was blocked at room temperature for 1 h in 5% fat-free milk dissolved in Tris buffered saline supplemented with 0.05% Tween 20. The primary antibodies used were anti-His antibody (Amersham Biosciences, Piscataway, NJ, USA), anti-Rab3A (clone 42.2, Synaptic Systems, Gottingen, Germany), antirabphilin3A (polyclonal rabbit antibody, Synaptic Systems), anti-β-tubulin (clone 3B11, Synaptic Systems), anti-Rab GDI and anti-Rab GAP p130 (kindly provided by Professor Yoshimi Takai). The secondary antibodies were horseradish peroxidase (HRP)-conjugated sheep anti-mouse immunoglobulin (Ig)G and HRP-conjugated monkey anti-rabbit IgG (Amersham Biosciences). The protein amount was quantified by using ImageJ (http://rsb.info.nih.gov/ij/) and normalized to β-tubulin.
Co-immunoprecipitation
Co-immunoprecipitation was carried out according to published procedure (Nagano et al. 2002) with modifications. Briefly, HEK 293 cells were transfected with His-tagged Rab3A (wild type or mutant) and Myc-tagged rabphilin3A (a kind gift from Professor Yoshimi Takai). Cells were lysed in lysis buffer containing 20 mm Tris–Cl, pH 7.5, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, 0.5 mm PMSF and 0.5% protease inhibitor cocktail (P-8340, Sigma). After clearing the lysates with control IgG, Rab3A or rabphilin3A was immunoprecipitated using anti-His-G (Invitrogen Life Technologies) or anti-Myc (9B11, Cell Signaling, Danvers, MA, USA) monoclonal antibodies and protein A agarose (Invitrogen Life Technologies). SDS–PAGE and Western blots were performed as described above. To detect the interaction between Rab3A and rabphilin3A, Rab GDI and Rab GAP, cortex dissected from Ebd and knockout and their wild-type littermates was homogenized in lysis buffer described above. Immunoprecipitation was carried out using anti-Rab3A and then immunoblotted using antirabphilin3A, anti-Rab GDI and anti-Rab GAP.
[α-32P]GTP overlay
The GTP-binding activity was examined by [a-32P]GTP overlay assay according to Klinz et al. (Klinz 1994). Wild-type and Ebd mutant Rab3A proteins were resolved on SDS–PAGE without preheating and then immunoblotted onto a PVDF membrane (Immobilon-P, MiliPore). The membrane was incubated at room temperature for 1 h in binding buffer (50 mm Tris–Cl, pH 7.5, 20 μm MgCl2, 1 mm DTT, 0.2% Tween-20) containing 1 μCi/ml of [α-32P]GTP (Amersham Biosciences) and then washed six times for 10 min each with wash buffer (50 mm Tris–Cl, pH 7.5, 25 mm MgCl2, 1mm DTT, 0.2% Tween-20). The blot was air-dried and exposed to X-ray film at −80 °C for 12–24 h.
[γ-35S]GTP and [3H]GDP binding assays
The binding assays were carried out according to Giovedi et al. (Giovedi et al. 2004). Briefly, samples (1 pmol/25 μl) were incubated in binding buffer (20 mm Tris–Cl, pH 7.5, 10 mm EDTA, 5 mm MgCl2,1 mm DTT) containing 0, 1, 3, 10, 30, 100, 300 and 1000 nm [γ-35S]GTP or [3H]GDP (PerkinElmer Life Sciences, Boston, MA, USA) for 30 min at 30 °C. Non-specific binding was determined in the presence of 300 μm GTP and GDP. The reaction was stopped by adding 1 ml of ice-cold stopping solution (25 mm Tris–Cl, pH 7.5, 20 mm MgCl2, 100 mm NaCl), followed by rapid filtration on 0.45-μm nitrocellulose filters (Whatman, Maidstone, England). The radioactivity retained on the filters was determined by scintillation counting.
The [3H]GDP dissociation assay
The assay was carried out as described (Giovedi et al. 2004; Kikuchi et al. 1995) with modification. Briefly, samples (2 pmol/24 μl) were incubated in loading buffer (20 mm Tris–Cl, pH 7.5, 10 mm EDTA, 5 mm MgCl2, 1 mm DTT, 1 μm [3H]GDP) at 30 °C for 20 min. Then 1 μl of 375 mm MgCl2 was added, and samples were put on ice to stabilize the complex. The dissociation reaction was started by adding 75 μl of dissociation solution (20 mm Tris–Cl, pH 7.5, 200 mm NaCl, 2 mm MgCl2,1 mm DTT, 67 μm GTP) and incubating at 30 °C for 0, 15, 30, 60 or 120 min. The dissociation reaction was stopped by filtration on 0.45-μm nitrocellulose filters, which were then subjected to scintillation counting.
GTPase assays
We employed two GTPase assays. The first (charcoal method) was carried out as described (Kikuchi et al. 1995). Wild-type and mutant Rab3A (25 pmol in 24-μl reaction) were loaded with [α-32P]GTP by incubating proteins at 30 °C for 10 min in a loading buffer containing 20 mm Tris–Cl, pH 7.5, 100 mm NaCl, 1 mm EDTA, 1 mm DTT and 10 nm [α-32P]GTP. The reaction was started by adding 1 μl of MgCl2 to a final concentration of 5 mm. The reaction was stopped by adding 750 μl of ice-cold stopping solution (50 mm NaH2PO4,5% charcoal) at various time-points. Samples were centrifuged twice at 10 000 g at room temperature. The amount of [32P] released from [α-32P]GTP was determined by applying 400 μl of supernatant to scintillation counting. The second GTPase assay [thin layer chromatography (TLC) method] was performed as described (Giovedi et al. 2004) with modifications. Proteins (20 pmol in 20-μl reaction) were loaded with [α-32P]GTP in loading buffer containing 10 nm [α-32P]GTP. The reaction was started by bringing MgCl2 to 5 mm. At indicated time, the reaction was stopped by adding 5 × stopping solution (1% SDS, 10 mm DTT, 10 mm EDTA, 2.5 mm GTP and 2.5 mm GDP) followed by incubating at 65 °C for 20 min. Samples were spotted to PEI-cellulose TLC sheets (Sigma) and air-dried for 20 min at room temperature. The TLC sheets were developed with 4m formic acid (pH 3.5 by NH4OH). Then the sheets were air-dried and exposed to X-ray film at −80 °C. The amount of GTP and GDP was quantified using NIH ImageJ (http://rsb.info.nih.gov/ij/).
Microarray analysis
Arrays
We used Affymetrix expression set MOE430A/B (Affymetrix, Santa Clara, CA, USA), which consists of 45 037 probe sets representing 39 015 transcripts for 34323 genes. All protocols were conducted as described in the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, total RNA was isolated from the cortex and hippocampus of individual animals from Ebd line (+/+ and Ebd/Ebd, n = 3 each) and the knockout line (Rab3a+/+ and Rab3a−/−, n = 6 each). Five microgram of total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were fragmented to 200 nucleotides or less, heated at 99 °C for 5 min and hybridized for 16 h at 45 °C to MOE430A/B microarrays. The microarrays were then washed with low (6× SSPE) and high (100 m m MES, 0.1m NaCl) stringency buffers and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect fluorescence signal at 3 μm resolution after excitation at 570 nm. The average signal from two sequential scans was calculated for each microarray feature. Affymetrix Microarray Suite 5.0 was used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (tau = 0.015) was used to assign a call (or flag) of absent, present or marginal for each assayed gene, and a P value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for non-specific signal by subtracting the mismatch probe value) was calculated using the One-step Tukey’s Biweight Estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays: after exclusion of the highest and lowest 2%, the average feature signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor. The data files from MAS 5.0 were imported to GeneSpring 7 (Silicon Genetics, Redwood City, CA, USA) for further analysis. To minimize multiple testing problems in statistical analysis, the gene list was reduced by discarding those scored as ‘absent’ in two or more replicate samples. This resulted in 20 975 genes that were subjected to significance analysis of microarray (for information see http://www-stat.stanford.edu/~tibs/SAM/) and PaGE (Grant et al. 2005) to generate lists of statistically significant genes. The genes that showed a fold change more than 1.5 were considered as differentially expressed genes.
Real-time quantitative PCR and RT-PCR
Real-time PCR was carried out on ABI Prism 7900HT sequence detection system (Applied Biosystems) by relative quantification (ΔΔCt method) using GAPDH or 18S rRNA as the endogenous control. The template cDNAs were generated as described above and pooled from seven to nine animals per genotype. For each cDNA sample, we performed three or four technical repeats on the 384-well plates. The primer and TaqMan probe sets for target genes were either bought as Assays-on-Demand or ordered as Assays-by-Design by using the Assays-by-design File Builder (Applied Biosystems). The information for these assays (assay IDs and consensus sequences) is available upon request. Data files containing the Ct values from Applied Biosystems SDS 2.2 software were analyzed according to User Bulletin #2 for ABI Prism 7700 Sequence Detection System (Applied Biosystems). Reaction time (RT)-PCR was used to investigate the mRNA isoforms of Peroxiredoxin II (Prdx2) gene. The cDNAs were generated as described above, and PCR was done with Taq DNA polymerase (Roche, Indianapolis, IN, USA). To detect the longer (Prdx2-L) and shorter (Prdx2-S) isoforms, a common left primer was used: 5′-AGGACTTCCGAAAGCTAGGC-3′. The right primers for Prdx2-L and Prdx2-S are 5′-TGGATCTGGCGTTAAAGAGG-3′ and 5′-TTGACTGTGATCTGGCGAAG-3′, respectively. The PCR products were quantified by measuring the intensities of the bands using ImageJ (http://rsb.info.nih.gov/ij/).
Behavioral assessment
All behavioral testing was performed between 1400 and 1700 h with the experimenter blind to the subjects’ genotype. A total of 7–23 male and female mice per genotype, aged 6–10 weeks, were used for behavioral assessment. The behavioral tests were performed in the following sequence: rotarod, zero maze, forced swim test (FST), Morris water maze, FC and acoustic startle/prepulse inhibition (PPI).
Rotarod
The test was performed according to published procedure (Tarantino et al. 2000). Mice were placed on the accelerating rotarod (Model 7650, Ugo Basile, Camerio VA, Italy), which accelerated at a constant rate from 4 to 40 rpm in 5 min, for a maximum time of 5 min. Mice were allowed to perform the test until they fell from the rod and given three trials with 45- to 60-min intertrial intervals. Rotarod performance reported here reflects the average performance of the second and third trials.
Zero maze
Behavior in the zero maze was assessed as previously reported (Tarantino et al. 2000). Mice were placed in one of the closed quadrants and were allowed to investigate the maze for 5 min in a single session. Mice were scored on several anxiety-related variables as identified in previous studies including time spent in open quadrants and number of transitions between quadrants. The animals were tracked during the session by a Poly Track Video System (San Diego Instruments, San Diego, CA, USA) suspended approximately 130 cm above the maze.
FST
The FST is a validated model for studying the depressive-like behavior in rodents (Borsini & Meli 1988; Wieland & Lucki 1990). A single-day trial was used in which mice are individually placed in plexiglass cylinders (46 cm tall, 20.5 cm diameter) containing 23–25 °C water to a depth of 20 cm, for a total of 6 min. Mice were manually scored for total immobility time and time to first float (latency) during the entire 6-min trial, with ‘immobility’ defined as movement limited to that required to maintain the mouse’s head above water. The water was changed between subjects. In a comparable forced swim set-up, C57BL/6J inbred mice showed sensitivity to different types of antidepressant drugs, including desipramine (Lucki et al. 2001).
Morris water maze
The Morris water maze can be used to assess spatial learning and memory in mice (D’Hooge & De Deyn 2001). Mice were placed into a 110-cm-diameter pool of opaque water (colored with white tempura non-toxic paint) at room temperature and trained over a 7-day period to locate a single hidden platform (area 10 cm2) submerged 0.5 cm below the water surface. The maze was surrounded by several visual cues that remained constant in the room. Mice were subjected to four training trials per day, in which they were placed into each of four quadrants (NW, NE, SE and SW) and given a maximum 60 seconds to locate the platform, with an interval of 10 min between trials. Time to platform location (latency) was recorded for each trial. If a mouse failed to locate the platform, it was guided to the platform by the experimenter. Once on the platform, a 20-second acquisition period was provided before the mouse was removed from the maze and dried off. At the end of the training period, learning and memory was further assessed by a probe trial in which the platform was removed, and the path of the mouse through the maze was recorded using a video tracking system (San Diego Instruments). The recording was analyzed with Poly Track Video Tracking System software (San Diego Instruments), and the percent of total distance traveled and percent of total time spent in the platform quadrant were measured.
FC
The basic apparatus and procedure were as previously described (Lattal & Abel 2001). Mice were trained in a rectangular (16″L × 6″W × 8 3/8″H) FC chamber (Medical Associates; Columbus, OH, USA) for 120 seconds before the onset of a 30 seconds white noise tone [(conditioned stimulus (CS)]. At 148 seconds, a 1.5-mA foot shock [(unconditioned stimulus (US)] was administered through the floor of the apparatus for 2 seconds. The mice were removed from the chamber at 300 seconds. Mice were tested for contextual conditioning in the same chamber for 300 seconds, 24 h after training. For the cued conditioning test, the mouse was placed into a new chamber environment with the floor covered by a smooth, blue panel. Following 120 seconds in the altered context, the CS tone was administered for 180 seconds. ‘Freezing’ behavior was scored at intervals of 5 seconds during training (baseline freezing), immediately following cessation of the US (immediate freezing), during the 300 seconds in the same context (contextual conditioning), during the first 120 seconds of the cued trial (altered context freezing) and during the last 180 seconds of the cued trial (cued conditioning). Freezing was defined as complete lack of movement, except for respiration. Percent freezing was calculated as (no. of recorded freezes/no. of possible freezes) × 100.
Acoustic startle and PPI of acoustic startle
Acoustic startle response (ASR) and PPI of acoustic startle were measured using SR-Laboratory Systems (San Diego Instruments) according to published procedure (Tarantino et al. 2000). After a 5-min acclimation period under continuous 70-dB white noise, mice were subjected to the following trials (×5) in pseudo-randomized order: 40-ms 90dB, 100dB, 110dB and 120dB, with average startle amplitude across the five 120-dB trials calculated as ASR. PPI was measured immediately after the Acoustic Startle test. The PPI session consisted of 15 trials spaced 15 s apart. Each trial consisted of a 40-ms 120dB stimulus delivered 60 ms after a 20 ms prepulse stimulus of either 70, 90 or 95dB. The ASRs for the 120 bB stimuli preceded by a 70dB prepulse were averaged and used to normalize the values obtained from the 90dB and 95dB prepulse trials. % PPI was calculated as: 100 - [(average ASR after 90 or 95dB prepulse/average ASR after 70bB prepulse) × 100]. % PPI values from the 90dB and 95dB prepulse trials were averaged to generate the final % PPI.
Statistical analysis
Statistical analysis was performed using either excel software, or vassarstats (http://faculty.vassar.edu/lowry/VassarStats.html). Strain effects among three genotypes were determined for most behavioral measures using a one-way analysis of variance (anova), followed by Tukey post hoc tests when appropriate. The effects of genetic backgrounds and the interaction between mutations and backgrounds were examined by two-way anova.
Results
Decreased GTP and GDP binding of Rab3AD77G disrupts its interaction with effectors and causes reduced protein levels of Rab3AD77G and Rabphilin3A
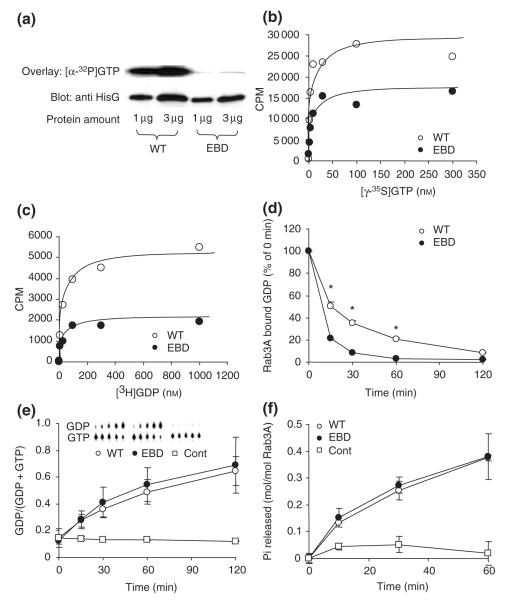
To compare Rab3a-null and Ebd mutants at the molecular and behavioral level, it was necessary to evaluate biochemical properties of the Ebd mutation. In the Rab3A protein, the Ebd mutation changes Asp77 in the highly conserved GTP-binding domain into Gly (Kapfhamer et al. 2002). On the basis of studies in other small GTPases (John et al. 1993; Jung et al. 1994; Ostermeier & Brunger 1999; Rak et al. 2003), we predicted that the Ebd mutation affects nucleotide binding. To test this prediction, we carried out biochemical assays for GTP/GDP binding, GDP dissociation and GTPase activity using wild-type and mutant Rab3AD77G expressed in HEK293 cells. To examine the binding of GTP, two different assays were employed: an [α-32P]GTP overlay assay and a [γ35S]GTP binding assay. In the [α-32P]GTP overlay assay, wild-type and Rab3AD77G proteins were blotted to PVDF membranes that were examined for [α-32P]GTP binding and protein amount. Wild-type Rab3A showed much stronger [α-32P]GTP binding than Rab3AD77G, indicating reduced GTP affinity for Rab3AD77G (Fig. 1a). The [γ-35S]GTP binding assay was carried out by employing a saturation binding experiment: wild-type and Rab3AD77G proteins were incubated with various concentrations of [γ-35S]GTP, and the proteinbound radioactivity was recovered by filtration through a nitrocellulose membrane and quantified by scintillation counting. Rab3AD77G bound less [γ-35S]GTP than wild-type Rab3A (Fig. 1b). This result, consistent with the result from the overlay assay, indicates that the Ebd mutation in Rab3A decreases GTP binding.
Figure 1. Biochemical analysis of wild-type (WT) and earlybird mutant (EBD) ras-associated binding (Rab) protein 3A.
(a) [α-32P]guanosine triphosphate (GTP) overlay assay was performed by blotting wild-type and mutant proteins to PVDF membranes, which were either incubated with [α-32P]GTP to examine GTP binding (upper panel) or subjected to immunoblotting to visualize protein content (lower panel). (b) A saturation GTP binding assay was performed by incubating Rab3A proteins with various concentrations of [γ-35S]GTP. Figure shown is a representative result of three independent experiments. (c) A [3H]GDP binding assay was performed in a similar manner as [γ-35S]GTP binding assay described in (b). (d) The GDP dissociation assay was started by adding an extra amount of GTP and GDP after loading proteins with [3H]GDP. The radioactivity was normalized to time zero and plotted as a function of time. *P < 0.01, student’s t-test. (e) GTPase activity assessed using thin layer chromatography (TLC) method. The GDP amount generated was plotted against time of incubation. The control experiments contain no protein. The intersect shows a representative TLC image. Data shown are means ± SD of three independent experiments. (f) GTPase activity assessed using the charcoal method. The amount of Pi released was plotted against time of incubation. No protein was included in the control experiments. Data represent means ± SD of two independent experiments in duplicates.
We next examined GDP binding in Rab3AD77G. A saturation binding assay using [3H]GDP as a radioligand showed that Rab3AD77G bound less [3H]GDP (Fig. 1c). In a [3H]GDP dissociation assay, Rab3AD77G showed faster GDP dissociation than wild-type Rab3A (Fig. 1d), providing independent evidence that GDP binding affinity is decreased in Rab3AD77G.
To examine the intrinsic GTPase activity, we used TLC to measure the conversion of [α-32P]GTP to [α-32P]GDP by wildtype and mutant proteins. The amount of [α-32P]GDP released from Rab3AD77G was comparable to that from wild-type Rab3A (Fig. 1e). Similar results were obtained from a separate method measuring α-32P release upon hydrolysis of [α-32P]GTP by wild-type and mutant Rab3A proteins (Fig. 1f). These results indicate that GTPase activity is not affected by the D77G mutation.
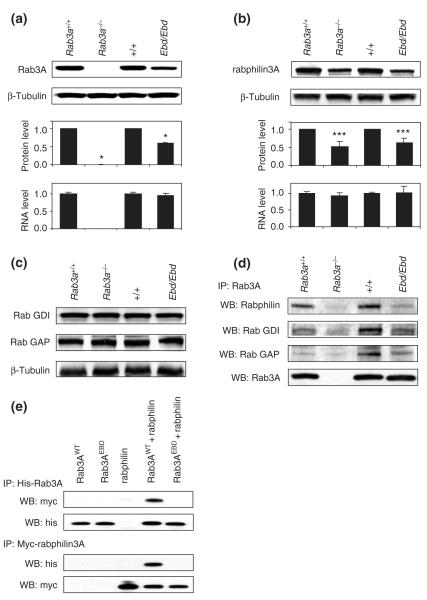
In addition to reduced GTP/GDP binding, Ebd mutant mice have reduced levels of Rab3A protein (Kapfhamer et al. 2002) (Fig. 2a). To address whether the decreased Rab3A protein in Ebd mice is due to decreased mRNA levels or changes in protein stability or degradation, we measured the mRNA level by real-time quantitative (Q) PCR using a probe that spans the exon 1–exon 2 junction so that it detects mutant and wild-type Rab3A mRNA equally well. In this assay, both wild-type and Ebd mice showed comparable Rab3A mRNA levels in brain tissues, whereas Rab3a−/− mice exhibited no detectable Rab3A mRNA (Fig. 2a). This result was further confirmed in independent samples by microarray analysis (see below). Therefore, we conclude that the point mutation in Ebd does not affect transcription of the Rab3a gene or the stability of Rab3A mRNA. Presuming that the point mutation is unlikely to affect the translation of Rab3A mRNA, the decreased Rab3A protein level in Ebd mice is most likely due to the decreased stability of the mutant Rab3A protein.
Figure 2. Protein stability of ras-associated binding (Rab) protein 3A and its effectors and their interactions in mutant mice.
(a) Rab3A protein levels were examined by immunoblotting with anti-Rab3A in homogenates from the hippocampus of earlybird (Ebd) and Rab3a null mice. The upper two panels show the representative Western blots for indicated wild-type and mutant strains of mice. The protein levels were quantified using ImageJ and normalized to β-tubulin. The mRNA levels of Rab3A measured by real-time quantitative polymerase chain reaction. Data are means ± SD. *P < 0.001, student’s t-test. (b) Rabphilin3A protein levels (upper and middle panels) and mRNA levels (lower) in the hippocampus of Ebd and Rab3a-null mice were examined similarly as described in (a). (c) Protein levels of Rab GDI and Rab GAP in the cortex of Ebd and Rab3a-null mice were examined by Western blot using anti-Rab GDI1 and anti-Rab GAP p130 antibodies. Beta-tubulin was also examined as a control for sample input. (d) Co-immunoprecipitation in the cortex homogenates. Rab3A was immunoprecipitated with anti-Rab3A antibodies, and Rab3A-bound rabphilin3A, Rab GDI and Rab GAP were detected using respective antibodies in Western blot. Anti-Rab3A antibodies were used to examine the sample input (lower panel). (e) Co-immunoprecipitation in the lysates of HEK 293 cells co-transfected with His-tagged Rab3A and Myc-tagged rabphilin3A.
It was previously reported that rabphilin3A, a Rab3A effector, was decreased in Rab3a−/− mice (Li et al. 1994; Schluter et al. 2004). Our experiments confirmed this finding (Fig. 2b). Furthermore, we found that rabphilin3A protein was decreased by about 40% in the cortex and hippocampus of Ebd mice (Fig. 2b). Q-PCR and microarray analysis (data not shown) showed that the rabphilin3A mRNA level was not affected in the brain tissues of either Rab3a knockout or Ebd mice (Fig. 2b), indicating that the reduced rabphilin3A protein is due to decreased protein stability. The Ebd mutation and the loss of Rab3A do not lead to decreased protein levels of at least two other Rab3A effectors, Rab GDI and Rab GAP (Fig. 2c).
The interaction between Rab3AD77G and its effectors, including rabphilin3A, Rab GDI and Rab GAP, was examined in vivo by co-immunoprecipition in cortex homogenates (Fig. 2d) and in vitro by co-immunoprecipition analysis in HEK293 cells co-transfected with Myc-tagged rabphilin3A and His-tagged wild-type or mutant Rab3A (Fig. 2e). Both sets of experiments showed that the Ebd mutation leads to decreased binding of Rab3AD77G to rabphilin3A, Rab GDI and RabGAP. Furthermore, our experiment with the Rab3AD77G protein shows that binding of rabphilin3A to Rab3A is required for the stability of rabphilin3A, as previously found in studies of Rab3A knockout mice (Li et al. 1994; Schluter et al. 2004).
Microarray experiments revealed subtle effects of Ebd and Rab3a-null mutations on gene expression in cortex and hippocampus
To compare the null and Ebd alleles at the molecular level and to investigate if they cause overall changes in gene expression, we performed transcription profiling by microarray analysis in two tissues (cortex and hippocampus) from four genotypes (+/+, Ebd/Ebd, Rab3a+/+ and Rab3a−/−).
Though Rab3A has wide-spread expression in central nervous system (Stettler et al. 1994), we used cortex and hippocampus as a tissue source for microarray analysis, because they can be easily dissected and samples can be collected from individual mice with no need for pooling. Furthermore, behavioral analysis of Ebd and Rab3A−/− mutants by us and others (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004) revealed a wide range of behavioral anomalies in which the cortex and hippocampus play important roles.
The Ebd allele was generated on a C57BL/6J background and crossed for one generation to C3H/HeJ, whereas the Rab3a−/− allele was generated on a 129/Sv background and maintained on a mixed B6 × 129 background. To minimize the effect of genetic background on molecular and behavioral phenotypes, we backcrossed both alleles for three generations to C57BL/6J. This three-generation backcross to C57BL/6J (N4 generation) would result in partial congenic lines consisting of approximately 94% C57BL/6J genome and approximately 6% C3H/HeJ in Ebd mice or 129/Sv in Rab3a knockout mice.
Global profiling of gene expression in Rab3A mutants and their wild-type littermates
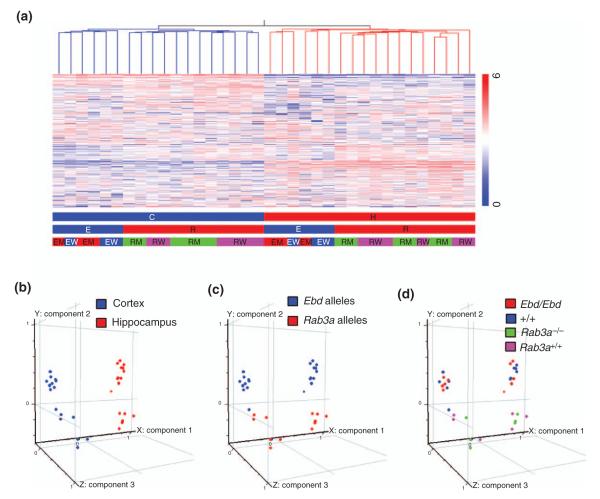
There are three different parameters in the 36 samples of our microarray experiment. (a) Genotype at the Rab3a locus (Ebd/Ebd vs. +/+ and Rab3a−/− vs. Rab3a+/+); (b) brain region (cortex vs. hippocampus) and (c) genetic background (B6.C3; N4 in Ebd/Ebd and +/+ mice vs. B6.129; N4 in Rab3a−/− and Rab3a+/+ mice). We first asked how do these factors affect overall gene expression? To visualize overall similarities in expression patterns, we performed hierarchical clustering by Pearson Correlation of the genes that are present or marginal in at least two/three of the three or six replicates across all 36 samples. The condition tree indicates that the major factor that affects gene expression in our experiment is brain region specificity, because samples from the same brain region cluster together (Fig. 3a). Mice with different genetic backgrounds (B6.C3; N4 in Ebd/Ebd and +/+ mice vs. B6.129; N4 in Rab3a−/− and Rab3a+/+ mice) have more dissimilar patterns of expression than mutant vs. wildtype (Ebd/Ebd vs. +/+ Rab3a−/− vs. Rab3a+/+). This finding was confirmed by Principle Components Analysis on all 36 conditions (Fig. 3b–d). The first component captured 24% of the overall variability, the second component 10% and the third 6%. This result indicates that both mutations cause subtle changes in gene expression compared with brain region or genetic background of the mice.
Figure 3. Global profiling of the 36 microarray samples from cortex and hippocampus of earlybird (Ebd) and Rab3a-null mutants and their wild-type littermates.
(a) Hierarchical clustering of the 20975 genes across 36 samples. Pearson correlation was used to construct the gene tree and the condition tree (only the condition tree is shown). Normalized expression values are displayed according to the vertical color bar. The horizontal bars beneath indicate the sample information: C, cortex; H, hippocampus; E, Ebd line; R, Rab3a null line; EM, Ebd/Ebd;EW, +/+ (wild-type controls of Ebd/Ebd); RM, Rab3a−/−; RW, Rab3a+/+. (b–d) Principle Component Analysis on these 36 samples. (b) Different brain regions are distributed along the first component that contributed 24.09% to the overall variance in the gene expression among these 36 samples. (c) Genetic backgrounds are distributed along the second component that had a contribution of 10.08%. (d) The four different genotypes do not show clear distribution pattern, indicating that Ebd or Rab3a-null mutation had subtle effect on gene expression.
The Ebd mutation of Rab3A has a subtle effect on gene expression
In Ebd alleles, no differentially expressed genes were identified with a false discovery rate (FDR) less than 10%. At a FDR around 50%, three genes in the cortex and two genes in the hippocampus were identified as having significantly altered expression (Table 1); however, their differential expression in Ebd/Ebd vs. +/+ was not confirmed by Q-PCR (data not shown). These genes may simply be false positives due to the high FDR (50%) in the microarray analysis, or they might have fallen beyond the resolution of Q-PCR due to the small fold change in their expression (mostly <twofold). Ultimately, the small number of genes identified as differentially expressed and the small fold changes of these genes indicate that the Ebd mutation has subtle, if any, effect on gene expression.
Table 1.
Differentially expressed genes in the Earlybird and Rab3a−/− mice
| Gene Name | Affy ID | FC# | Location## | Description |
|---|---|---|---|---|
| cem/cew* | ||||
| Morc2a | 1456398_at | 1.62 | 11:3610702 | Microrchidia 2A |
| Casp8 | 1424552_at | 1.53 | 1:59046582 | Caspase 8 |
| Dad1 | 1454860_x_at | 0.62 | 14:49105685 | Defender against cell death 1 |
| hem/hew** | ||||
| Ptgds | 1423860_at | 0.53 | 2:25474022 | Prostaglandin D2 synthase (brain) |
| Ttr | 1455913_x_at | 0.39 | 18:21036130 | Transthyretin |
| crm/crw*** | ||||
| C030044C12Rik | 1429097_at | 0.66 | 8:82332807 | RIKEN cDNA C030044C12 gene |
| Atp11c | 1437600_at | 0.66 | X:55703999 | Atpase, class VI, type 11C |
| 5330410G16Rik | 1452825_at | 0.63 | 8:68939012 | RIKEN cDNA 5330410G16 gene |
| Chd9 | 1459450_at | 0.63 | 8:89421405 | Chromodomain helicase DNA binding protein 9 |
| Rasd2 | 1427344_s_at | 0.62 | 8:74420202 | RASD family, member 2 |
| Zfp617 | 1449546_a_at | 0.62 | 8:70422797 | Similar to hypothetical protein FLJ38281 |
| 6430500C12Rik | 1453402_at | 0.57 | 8:38069818 | AV338754 RIKEN full-length enriched, cDNA clone 6430500C12 |
| Slc5a5 | 1436239_at | 0.55 | 8:69338006 | Solute carrier family 5 (sodium iodide symporter), member 5 |
| Trmt1 | 1436494_x_at | 0.51 | 8:83273987 | TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae) |
| 1447892_at | 0.43 | 18:23434305 | Similar to glyceraldehyde-3-phosphate dehydrogenase | |
| 1420287_at | 0.39 | 8:83678096 | Transcribed locus | |
| C030036D22Rik | 1443744_at | 0.36 | 18:72562266 | RIKEN cDNA C030036D22 gene |
| Large | 1460113_at | 0.35 | 8:71338357 | Like-glycosyltransferase |
| Usp38 | 1428592_s_at | 0.29 | 8:79520109 | Ubiquitin specific protease 38 |
| 1444260_at | 0.2 | 8:68465175 | Weakly similar to XP_227873.2 similar to RIKEN cDNA 9330196J05 | |
| Prdx2 | 1430979_a_at | 0.14 | 8:83555736 | Peroxiredoxin 2 |
| Rab3a | 1422589_at | 0.01 | 8:69208775 | RAB3A, member RAS oncogene family |
| hrm/hrw**** | ||||
| Lpl | 1431056_a_at | 2.4 | 8:67335311 | Lipoprotein lipase |
| Xrn1 | 1442141_at | 1.72 | 9:95600697 | 5′-3′ exoribonuclease 1 |
| Mfap3l | 1428804_at | 1.63 | 8:59024530 | Microfibrillar-associated protein 3-like |
| Slc13a4 | 1433734_at | 1.54 | 6:35199160 | Solute carrier family 13 (sodium/sulfate symporters), member 4 |
| Large | 1443212_at | 0.65 | 8:71338357 | Like-glycosyltransferase |
| C030044C12Rik | 1429097_at | 0.63 | 8:82332807 | RIKEN cDNA C030044C12 gene |
| Pgpep1 | 1418329_at | 0.62 | 8:69104250 | Pyroglutamyl-peptidase I |
| Zfp330 | 1451216_at | 0.61 | 8:81326159 | Zinc finger protein 330 |
| 5330410G16Rik | 1452825_at | 0.58 | 8:68939012 | RIKEN cDNA 5330410G16 gene |
| Slc5a5 | 1436239_at | 0.55 | 8:69338006 | Solute carrier family 5 (sodium iodide symporter), member 5 |
| Trmt1 | 1436494_x_at | 0.54 | 8:83273987 | TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae) |
| Zfp617 | 1449546_a_at | 0.48 | 8:70422797 | Similar to hypothetical protein FLJ38281 |
| Usp38 | 1428592_s_at | 0.39 | 8:79520109 | Ubiquitin specific protease 38 |
| Prdx2 | 1430979_a_at | 0.11 | 8:83555736 | Peroxiredoxin 2 |
| Rab3a | 1422589_at | 0.02 | r8:69208775 | RAB3A, member RAS oncogene family |
Fold Change.
start site for genes or probesets are from NCBI m34 mouse assembly.
comparison between the cortex of Ebd/Ebd mice (cem) and their wild-type littermates (cew).
comparison between the hippocampus of Ebd/Ebd mice (hem) and their wild-type littermates (hew).
comparison between the cortex of Rab3a−/− (crm) and Rab3a+/+ mice (crw).
comparison between the hippocampus of Rab3a−/− (hrm) and Rab3a+/+ mice (hrw).
Differential gene expression in the Rab3a−/− mice due to congenic genomic sequence from 129/Sv background
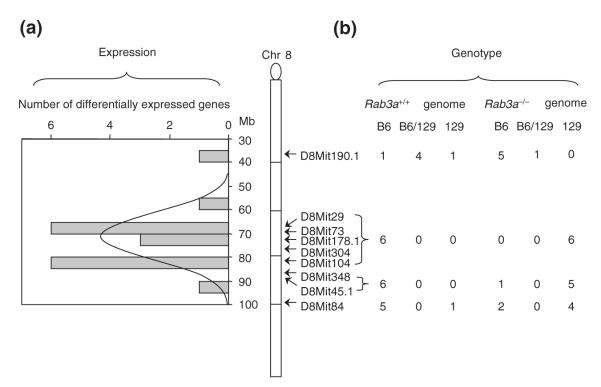
In the Rab3a−/− mice, 23 genes including Rab3a were differentially expressed between Rab3a−/− and wild-type littermates in either the cortex or the hippocampus (Table 1). Among them, 18 map to the chromosomal region surrounding the Rab3a locus on mouse chromosome 8 (Fig. 4a). We examined the expression patterns of three differentially expressed genes (Lpl, Slc5a5 and Prdx2) by Q-PCR or RT-PCR and confirmed the microarray results (see Supplementary material, Fig. S1). The expression patterns of these three genes in Rab3a+/+ and Rab3a−/− strains were comparable to those in C57BL/6J and 129/Sv strains, respectively (see Supplementary material, Fig. S1). Furthermore, even the tissue-specific expression pattern of Lpl and the strain-specific splicing of Prdx2 in Rab3a−/− corresponded to those detected in the 129/Sv strain (see Supplementary material, Fig. S2). Clustering of differentially expressed genes on chromosome 8 could be explained by differential gene expression between the 129/Sv chromosomal segment surrounding the genetically disrupted Rab3a gene and the C57Bl/6J wild-type chromosome. We thus genotyped the mice used for the microarray experiment at several chromosome 8 loci with a set of microsatellite markers which detect polymorphisms between the C57BL/6J and 129/Sv strains. In Rab3a−/− mice, the 129/Sv region shared by all mutant mice covers approximately 14 Mb (from 67 to 81 Mb, Ensembl m3.3) (Fig. 4b), whereas all Rab3a+/+ mice were homozygous for C57BL/6J at the region surrounding Rab3a. In Ebd mutant and wild-type mice, almost all tested chromosome 8 loci (10/12) were homozygous for C57BL/6J alleles (data not shown), indicating that remnants of the C3H/HeJ chromosome are dispersed across the genome in Ebd mutant and wild-type mice. This result indicates that the differentially expressed genes in the congenic region were regulated in a strain-specific manner. Therefore, we conclude that the Rab3a-null mutation per se has subtle effect on gene expression, as does the Ebd mutation.
Figure 4. Differentially expressed genes in Rab3a−/− mice map to a congenic chromosomal region derived from the 129 strain.
(a) Distribution of differentially expressed genes on chromosome 8. The map position of the differential genes in the cortex and the hippocampus of Rab3a−/− mice showed a distribution similar to normal distribution as indicated by the curve. The distribution centered at the Rab3a locus. (b) Genotypes of the Rab3a+/+ and Rab3a−/− mice at 9 loci around Rab3a gene. The positions of the markers are indicated by the arrows along chromosome 8. To the right of the markers are numbers of the animals that were homozygous for C57BL/6J (B6), homozygous for 129/Sv (129) or heterozygous (B6/129) at markers indicated.
Behavioral assessments revealed the role of Rab3A in learning and memory and in anxiety-related behavior
As a part of the overall characterization and comparison of the two Rab3a alleles, we examined behavior of mutant mice and their wild-type littermates in a battery of behavioral paradigms. These paradigms were selected with a goal of assessing several aspects of brain function, from motor abilities (rotarod), emotional behavior (zero maze), sensorimotor gating (PPI), stress-induced behavior (FST) to learning and memory (FC and Morris water maze).
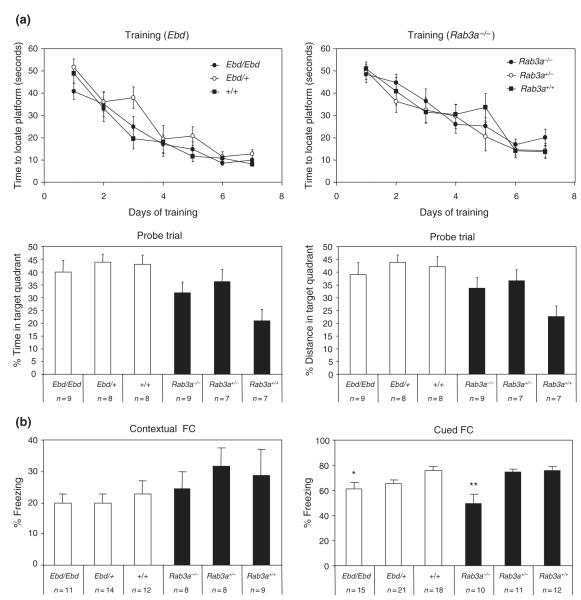
Ebd and Rab3a-null mice exhibit intact spatial learning but impaired cued FC
Spatial memory in Ebd and Rab3a knockout mice was assayed in a hidden platform version of the Morris water maze. This paradigm involves training mice for 7 days to locate a submerged platform, followed by a probe trial in which the platform is removed and the preference for the quadrant of the pool in which the platform was located is determined by measuring percent time spent and swim distance in the platform quadrant. Both Ebd and Rab3a knockout mice exhibited a rapid decline in latency as the training period progressed (Fig. 5a). In the Ebd line, no statistical difference was observed for either the percent total distance traveled in the platform quadrant (F2,24 = 0.396, P = 0.677) or percent time spent in the platform quadrant (F2,24 = 0.290, P = 0.751) during the probe trial. Similarly, Rab3a knockout mice showed no significant difference in percent distance (F2,22 = 2.911, P = 0.078) and percent time (F2,22 = 2.829, P = 0.083) spent in the platform quadrant during the probe trial. The results indicate that neither the Ebd nor the Rab3a−/− allele affects spatial learning and memory performance. Nonetheless, two-way anova detected significant effect of genetic background between Ebd and Rab3a knockout lines in both percent distance (F1,47 9.71, P = = 0.003) and percent time (F1,47 = 12.72, P = 0.0009) in the platform quadrant, although no interaction between background and genotypes was observed (F2,47 = 1.79, P = 0.180 for percentage distance and F2,47 = 1.84, P = 0.171 for percentage time). Our results support previous reports on differences between the 129/Sv and the C57BL/6J lines in the Morris water-maze test (Owen et al. 1997b).
Figure 5. Learning and memory deficits in earlybird (Ebd) and Rab3a−/− mice.
(a) Spatial learning and memory in the hidden platform version of the Morris water-maze test. The upper panels show the performance of Ebd (left) and Rab3a−/− (right) mice in the training trials. The lower panels show the performance in the probe trial. (b) Fear conditioning (FC). Hippocampal-dependent learning was examined by measuring freezing behavior upon presentation of the chamber (contextual FC), and amygdala-dependent learning was assessed by presentation of the white noise stimulus (cued FC). Data shown are means ± SEM. *P < 0.05; **P < 0.01, Tukey HSD test after one-way analysis of variance.
The FC test is another measure of learning and memory in mice. This paradigm tests a mouse’s ability to associate a mild foot shock (US) with a specific environment (contextual FC, environment CS) and a tone (cued FC, tone CS); memory of the US is signified by freezing behavior observed in the mouse. Ebd and Rab3a knockout mice were subjected to both FC tests and were compared in baseline (without US), immediate (following US), contextual (environment CS, no US), altered context (no US) and cued (tone CS, no US) freezing. Mice from both lines exhibited similar baseline and immediate freezing (data not shown). All mice performed similarly in the contextual FC [Ebd lines: F2,36 = 0.23, P = 0.796; Rab3a−/− lines: F2,24 = 0.265, P = 0.77; genetic backgrounds: F1,61 = 3.54, P = 0.065], thus neither mutations of Rab3A nor genetic background affected this type of hippocampal-dependent learning (Fig. 5b). In contrast, both Ebd/Ebd mice (F2,53 = 3.853, P < 0.05) and Rab3a−/− mice (F2,32 = 8.943, P < 0.001) displayed impaired cued FC learning, as compared to their wildtype controls (Fig. 5b). This potentially implicates Rab3A in some aspect of amygala-dependant learning.
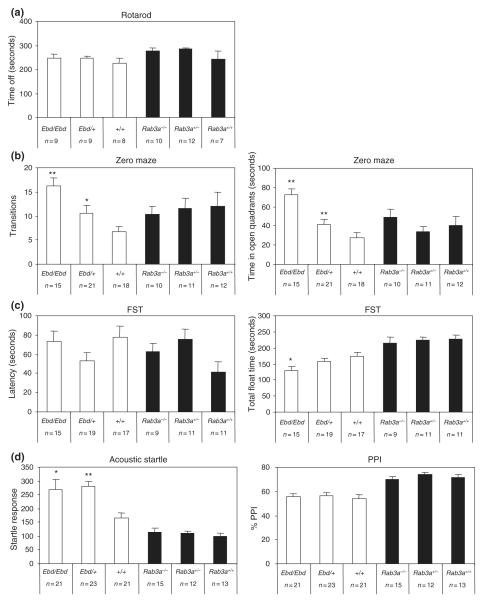
Ebd mice, but not Rab3a−/− mice, show reduced anxiety-like, decreased behavioral despair behavior as well as acoustic startle-induced hyperekplexia
To further evaluate the observed anomalies in Rab3A mutant mice, we assessed other behavioral phenotypes in these mice. Neuromuscular coordination and strength in Ebd and Rab3a−/− mice were examined using an accelerating rotarod (Fig. 6a). No difference was observed among the three genotypes of Ebd lines (Ebd/Ebd, Ebd/+, and +/+) (F2,25 = 0.533, P = 0.594) and the three genotypes of Rab3a−/− lines (Rab3a−/−, Rab3a+/– and Rab3a+/+)(F2,28 = 1.385, P = 0.268). This result indicates that neither the loss-of-function nor the Ebd mutation affect neuromuscular coordination or motor ability and affirmed that the anomalies observed in other behavioral tests were not consequences of mutant Rab3A-induced neuromuscular dysfunction. However, two-way anova did reveal an effect of genetic background between Ebd and Rab3a−/− lines (F1,54 = 4.52, P = 0.039).
Figure 6. Earlybird (Ebd) mice but not Rab3a−/− mice showed abnormal emotional status.
(a) Both the Ebd mice and the Rab3a-null mice have normal motor co-ordination on the accelerated rotarod. (b) Ebd mice displayed reduced anxiety-like behavior in the zero maze test. (c) Ebd mice showed reduced immobility in the Forced swim test. (d) Ebd mice had increased startle response compared to wild-type controls but showed normal prepulse Inhibition. Data shown are means ± SEM. *P < 0.05; **P < 0.01, Tukey HSD test after one-way analysis of variance.
We used the elevated zero maze to test anxiety-like behavior in Ebd and Rab3a−/− mice. The time that a mouse spends in the open areas and the number of transitions between open and closed quadrants are indicative of anxiety-like behavior, such that increased time in the open quadrants and increased transitions are predictive of low anxiety (Borsini & Meli 1988). The Ebd/Ebd and Ebd/+ mice showed an increased number of transitions (F2,53 = 8.878, P < 0.001) and spent longer time in open quadrants (F2,53 = 15.613, P < 0.001), indicating reduced anxiety-like behavior in these mice (Fig. 6b). Among the Rab3a−/− lines, no difference was observed in either the number of transitions (F2,32 = 0.124, P = 0.884) or the time spent in the open quadrants (F2,32 = 0.903, P = 0.416) (Fig. 6b). Two-way anova revealed significant interaction between genetic background and genotypes in both transitions (F2,86 = 4.08, P = 0.021) and time in open quadrants (F2,86 = 3.73, p = 0.028), suggesting that the genetic backgrounds of Ebd and Rab3a−/− lines affect anxiety-like behavior differently.
Mice from the Ebd and Rab3a−/− lines were subjected to the FST to evaluate response to stress. In this test, increased float time or shorter latency (swim time before first float) is indicative of increased ‘behavioral despair’. No difference in the latency before first float was observed in either the Ebd line (F2,50 = 1.736, P = 0.187) or the Rab3a−/− line (F2,30 = 2.996, P = 0.066) (Fig. 6c). The genetic background of the Ebd and Rab3a−/− lines have negligible effect on the latency in FST (F1,81 = 0.76, P = 0.386). In contrast, a significant strain effect of total float time was observed in the Ebd lines (F2,50 = 3.592, P = 0.035), with decreased total float time in Ebd/Ebd mice relative to wild-type controls (P < 0.05, Tukey HSD test) (Fig. 6c). Rab3a−/− mice, however, showed no difference in total float time (F2,30 = 0.148, P = 0.863). Genetic background had a significant effect on this behavior (F1,81 = 38.53, P < 0.001). These observations suggest that the Ebd mutation leads to decreased behavioral despair in the FST and that the different performance of Ebd and Rab3a−/− lines in this test is due to their different genetic backgrounds.
The startle test assesses sensorimotor processing (Koch & Schnitzler 1997), while PPI of the acoustic startle reflex (ASR) is a measure of sensorimotor gating in humans and rodents (for a review, see Swerdlow et al. 2001). The magnitude of the ASR was significantly increased in Ebd/Ebd and Ebd/+mice (F2,64 = 5.482, P = 0.01) compared to wild-type controls, although no difference was observed in Rab3a−/− lines (F2,39 = 0.313, P = 0.734) (Fig. 6d). Genetic background had a significant effect on acoustic startle (F1,104 = 42.32, P < 0.001) (Fig. 6d). For PPI, there were no differences within either Ebd lines (F2,64 = 0.297, P = 0.74) or Rab3a−/− = lines (F2,39 = 0.622, P = 0.542); however, genetic background affects PPI significantly (F1,104 = 52.75, P < 0.001), with the Rab3a−/− lines showing higher percentage PPI than the Ebd lines (Fig. 6d). This result suggests that the Ebd mutation affects sensorimotor processing without affecting sensorimotor gating and that differences observed between Ebd and Rab3a−/− lines in this test are due to their different genetic backgrounds.
Discussion
Previous genetic studies of Rab3A, a small GTP-binding protein, focussed on studies of the null mutation in the mouse and its role in vesicle trafficking and neurotransmitter release (Geppert et al. 1994; Geppert et al. 1997; Schluter et al. 2004). We have described the identification of Ebd, an ENU-induced point mutation in the Rab3a gene, which results in the replacement of aspartate with glycine in the highly conserved GTP-binding domain of Rab3A (Kapfhamer et al. 2002). Here, we compare two Rab3A mutations, null and Ebd, at a molecular and biochemical level, with the goal of relating their effects to observed behavioral anomalies.
D77G mutation in Rab3A causes reduced binding of effectors and reduced stability of Rab3A and rabphilin3A proteins
Previous in vitro studies of Rab3A involved analysis of a series of mutations in rat Rab3A and a yeast ortholog Ypt1, including their effects on intrinsic GTPase activity, GDP dissociation rate and GDP or GTP binding (Becker et al. 1991; Brondyk et al. 1993; Burstein et al. 1992; Wagner et al. 1987). Mutations in the aspartic acid of the Asp-X-X-Gly (DXXG) motif common to all GTP-binding proteins have not been reported in any Rab3A ortholog. The crystal structures of the GppNp-bound Rab3A and Ypt1-GDI complexes implicate the role of D77 in Rab3A and D63 in Ypt1 in Mg2+ binding, which is important for nucleotide binding and hydrolysis (Ostermeier & Brunger 1999; Rak et al. 2003). Functional consequences are known for mutations in the corresponding amino acid in human H-Ras; H-RasD57Y shows higher affinity for GDP than for GTP (Jung et al. 1994), and H-RasD57A has increased GDP binding and reduced GTPase activity (John et al. 1993). However, replacement of the same aminoacid by asparagine (H-RasD57N) did not alter the biochemical properties of H-Ras (Farnsworth & Feig 1991). Our studies of mouse Rab3A showed that replacement of aspartic acid by glycine in Rab3AD77G lowers the affinity for both GTP and GDP but leaves GTPase activity intact. On the basis of these findings, we postulate that under physiological conditions, the Ebd mutation shifts Rab3A away from its active form.
In the Rab3A-null mutant, the absence of Rab3A protein leads to a decreased level of rabphilin3A protein (Li et al. 1994). We have also observed reduced levels of rabphilin3A in Ebd mutants, and the reduction seems to be comparable to that caused by the null mutation. Co-immunoprecipitation revealed reduced binding between rabphilin3A and Rab3AD77G, suggesting that the binding of rabphilin3A to functional Rab3A protein is required for its stability. This, in accord with previous studies of synapsin I and synapsin II (Rosahl et al. 1995), illustrates that in a presynaptic neuron, loss of specific protein–protein interactions can lead to protein instability and degradation. Though the absence of rabphilin3A does not affect neurotransmission in hippocampal neurons in mice (Schluter et al. 1999), rabphilin3A does potentiate SNARE function independently of Rab3A (Staunton et al. 2001). Furthermore, it has been shown that phosphorylated rabphilin3A is region-specifically distributed in rat brain, with the highest levels in the cerebellum, midbrain and medulla (Foletti & Scheller 2001). It is possible therefore that the reduction of rabphilin3A protein perturbs the function of a subset of synapses in some specific brain regions. Rab3AD77G also has reduced binding affinity for Rab GDI and Rab GAP, though neither the Ebd nor the null mutations of Rab3A affected the protein levels of these Rab3A effectors. Rab GDI retrieves GDP bound form of Rab proteins from the membrane by forming a heterodimer that serves as a cytosolic reservoir for inactive Rabs (Sasaki et al. 1990). Systematic expression profiling indicates that Rab3A is the most abundantly expressed Rab family member in the brain (Gurkan et al. 2005) and thus represents the major substrate for Rab GDI. The reduced affinity between Rab3AD77G and Rab GDI may cause an increase of inactive Rab3AD77G bound to the membrane, which may interfere with normal vesicle trafficking. It is important to note that mutations in GDI1 have been implicated in X-linked non-specific mental retardation (D’Adamo et al. 1998), and Gdi1-deficient mice show altered associative memory and social behavior (D’Adamo et al. 2002). On the basis of the genetic evidence, we suggested that the Ebd mutation represents a dominant negative allele of Rab3a (Kapfhamer et al. 2002). Although our results describe biochemical changes in several Rab3A effectors caused by the D77G substitution; further studies are required to show which, among a large number of Rab3A interacting proteins, are responsible for the robust behavioral consequences of the mutation.
Mutations of Rab3A do not cause robust changes at the transcriptome level
Expression profiling of Ebd and Rab3a−/− mice and their wildtype littermates revealed subtle, if any, changes caused by the presence of the Rab3A mutations. Therefore, we conclude that disruption of Rab3A function, either by its absence or a mutation in the GTP-binding domain, leads to molecular changes and behavioral anomalies mainly through changes at the post-transcriptional level. For example, our microarray experiment, as well as Q-PCR, showed that the reduced level of rabphilin3A is not caused by a reduced level of mRNA but rather by the stability of rabphilin3A protein. We do not attribute the absence of global changes in gene expression in Rab3A mutants to technical problems or our inability to detect subtle changes in gene expression by microrrray technology because of the successful detection of 18 genes that are differentially expressed between the 129/Sv and C57BL/6J in the congenic line. We also considered that the lack of transcriptional changes may be explained by functional redundancy between the Rab3a, Rab3b, Rab3c and Rab3d genes; however, in our microarray experiment, we did not detect changes in the expression levels of these paralogs in Rab3a−/− or Ebd mice.
It is remarkable that the only robust changes in gene expression between mutant Rab3a−/− and wild-type mice can be attributed to differences in genetic background. Two mutant lines were originally generated on two different chromosomes, Ebd on C57BL/6J and Rab3a−/− on 129/Sv. Both lines were backcrossed to C57BL/6J for three generations; however, in the case of Ebd, the series of backcrosses followed an original outcross to C3H/HeJ. Therefore, during backcrossing and selection for a mutant Rab3a−/− locus, remnants of the 129/Sv (donor strain) were inherited as so-called passenger loci. In Ebd mice, differential chromosomal segments are scattered throughout the genome. On the basis of the calculations (Flaherty 1981; Silver 1995) as well as empirical data, we expected that a three-generation backcross of the Rab3a−/− line on a 129/Sv chromosome to a C57BL/6J background would leave a large segment (approximately 50 cM) of the donor chromosome (129/Sv) around the Rab3a locus that serves as a target in selection. Through genotyping of all lines used for microarray analysis, we showed that the differential chromosomal segment in Rab3a−/− covers > 14 Mb and contains 178 genes according to ENSEMBL (mouse m33 version). This high gene number supports reports that this segment of mouse chromosome 8, and the orthologous region on human chromosome 19, represents the most gene-dense region in the mammalian genome (Dehal et al. 2001). Furthermore, our finding that 18 among 178 genes show differential expression between 129/Sv and C5BL/6J chromosomes may be explained by an exceptionally high degree of nucleotide variation, including non-synonymous coding SNPs and stop codons, between these two genomes (Adams et al. 2005).
Rab3A is involved in learning and memory as well as in emotional regulation
In our original publication, we reported that Ebd and Rab3a−/− mice have shortened circadian period and decreased response to sleep loss (Kapfhamer et al. 2002). Mutants with sleep and circadian anomalies provide useful models for the analysis of a link between these neurobiological systems and mood disorders in humans. For example, Clock mice have sex-specific changes in exploratory and escape-seeking behavior, with no changes in anxiety and depression-like behavior (Easton et al. 2003). Moreover, studies of circadian mutants in Drosophila showed that the period gene plays a key role in long-term memory formation (Sakai et al. 2004). Therefore, we conducted a comparison of two alleles, Ebd and Rab3a−/−, using a comprehensive battery of behavioral tests, including those for anxiety, sensorimotor gating, response to stress and learning and memory.
Electrophysiological experiments in Rab3a knockout mice established a role for Rab3A in hippocampal mossy fiber LTP and LTD (Castillo et al. 1997; Geppert et al. 1997), two forms of synaptic plasticity generally accepted as necessary for the consolidation of certain types of long-term memory. On the basis of these LTP/LTD phenotypes, one might predict that Rab3a mutant mice would exhibit altered performance in hippocampal-dependent learning paradigms. However, both contextual FC and performance in the Morris water maze were normal in Ebd and Rab3a knockout mice, indicating that hippocampal-dependent learning/memory does not require Rab3A. Both contextual and cued FC depend on the amygdala, whereas contextual FC also involves the hippocampal system, depending on the exact nature of the FC protocol followed (Kim & Fanselow 1992; Matus-Amat et al. 2004; Phillips & LeDoux 1992). The observed deficit in cued FC in both Ebd/Ebd and Rab3a−/− mice suggests that alterations in Rab3A-meditated neurotransmission may underlie this deficit and provides evidence for a role of Rab3A in memory processes outside the hippocampus. Our results to some extent support a role for Rab3A in LTP in the cortical pathway to the amygdala as observed by Huang et al. (Huang et al. 2005), but how this explains the selective deficits in cued but not contextual conditioning remains unclear. Other studies failed to detect an effect on cued FC in the Rab3a knockout line (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004), probably due to differences in genetic background of the mice used in the studies, differences in age of tested mice and/or experimental environments (see below). Although one may question the validity of our findings for the Rab3a knockout mice due to differential expression of >18 genes in the 129/Sv chromosomal segment surrounding the Rab3a gene, anomalies in cued FC detected in the Ebd allele are more likely to be due to the disruption of the Rab3a gene because in Ebd mutants a small contributions from the C3H-derived genome (6%) is randomly distributed in both mutant and wild-type progeny. It is interesting that our studies reveal impairments in cued FC but not in contextual conditioning. This suggests that different neuronal circuits (perhaps within the amygdala) may mediate these distinct forms of FC, an idea that is supported by several other studies in which a similar phenotype of impaired cued FC but normal contextual conditioning was observed (Barnes & Good 2005; Gould & Feiro 2005) as well as the finding of distinct QTLs for cued and contextual conditioning (Owen et al. 1997a).
In contrast to the Rab3a knockout line, Ebd mice exhibited altered performance in the zero maze, forced swim and startle tests, which may be due either to a dominant-negative effect of the Ebd mutation or a background effect that masks the phenotypes in Rab3a knockout mice. The reduced anxiety-like behavior is consistent with impaired performance in cued FC, given the fact that both fear and anxiety involve the amygdala (Charney & Deutch 1996; Davis 1998) and the same mesolimbic dopaminergic pathways (Pezze & Feldon 2004). However, the enhanced acoustic startle response seems to be inconsistent with decreased anxiety in Ebd mice, because anxiety enhances the acoustic startle response in both rodents and humans (Koch 1999). A possible resolution lies in the fact that the neuronal circuits mediating acoustic startle response and anxiety are different. The acoustic startle response circuit is located in the lower brainstem with the caudal pontine reticular nucleus playing a key role in the primary acoustic startle response pathway, and both excitatory transmitter glutamate and inhibitory GABA are involved (Koch 1999). Anxiety, by contrast, is regulated by the limbic-hypothalamic-pituitary-adrenal axis (Lopez et al. 1999), and a pivotal role for the amygdala was suggested in the transmission and interpretation of fear and anxiety (Charney & Deutch 1996).
Although the main purpose of behavioral studies reported in this paper is to compare anomalies in two Rab3a alleles, Ebd and Rab3a−/−, we can compare or calibrate our findings to several previously reported behavioral studies of Rab3a−/− mice (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004). The most consistent finding observed by all four studies is normal hippocampus-dependent learning and memory in contextual FC assay (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004). Moreover, all reports are consistent with normal spatial learning performance of Rab3a−/− mutants in the conventional form of the Morris water maze (D’Adamo et al. 2004; Hensbroek et al. 2003; Powell et al. 2004), although D’Adamo et al. (D’Adamo et al. 2004) reported moderately impaired platform reversal learning in the water maze in reference memory and episodic-like memory tasks. Hensbroek et al. found reduced exploratory behavior in Rab3a knockout mice, while D’Adamo et al. reported the opposite result (D’Adamo et al. 2004; Hensbroek et al. 2003), and we detected no anomalies in exploratory behavior tested in the hole board (data not shown). We and D’Adamo et al. report reduced anxiety-like behavior, which was not detected by Powell et al. (D’Adamo et al. 2004; Powell et al. 2004). What makes the behavioral phenotypes in Rab3a−/− mutants so different? First, mutant lines examined in these studies have different genetic backgrounds. Hensbroek et al. used mice on a 129/Sv background, except for (C57BL/6JolaHsd X 129/SvImJ) F1 hybrids used in the Morris water-maze test (Hensbroek et al. 2003). The other three studies used hybrid 129/Sv/C57BL/6J mice with a different number of backcrossed generations, which may account for the discrepancies observed among these behavioral studies. These disparate results indicate that genetic background has a strong effect on behavioral phenotypes in Rab3a knockout mice. Another potential explanation for differences between the current study and those of D’Adamo et al. (D’Adamo et al. 2004), Powell et al. (Powell et al. 2004) and Hensbroek et al. (Hensbroek et al. 2003) is the age at which the mice were tested. D’Adamo and Powell et al. tested adults, beginning behavioral testing at or after age 12 weeks, whereas our study used younger mice (ages 6–10 weeks). Finally, as previously reported, experimental environments in different laboratories may also contribute to different behavioral observations (Crabbe et al. 1999).
Our studies show that the Ebd and loss-of-function mutations of Rab3A have subtle, if any, effects on the overall level of gene transcription. However, they have significant effects on the stability of Rab3A and rabphilin3A, as well as the binding of Rab3A to its effectors. We therefore conclude that the associated changes in behavior, including learning and memory, emotion, and the previously described anomalies in circadian rhythm and response to sleep-loss, are mediated by mutation-induced changes in the stability of and the interaction between Rab3A and its effectors. Our comparison of two Rab3a alleles illustrates the utility as well as challenges in linking different genetic lesions in the same gene to behavioral phenotypes.
Supplementary Material
Figure S1. Differentially expressed genes on chromosome 8: microarray results and validation by Q-PCR or RT-PCR. (a) The expression patterns of Lipoprotein lipase (Lpl) in the hippocampus were examined by microarray (left) and Q-PCR (right). In both experiments Lpl showed more than 2-fold higher expression in Rab3a−/− mice than in Rab3a+/+ mice. Q-PCR also showed that Lpl expressed more than 2-fold higher in 129SvJ strain than in C57BL/6J strain. (b) The expression pattern of the longer form of Peroxiredoxin II (Prdx2-L) in microarray experiment (left) was confirmed by RT-PCR (right). The Rab3a−/− and 129SvJ mice expressed less Prdx2-L than the Rab3a+/+ and C57BL/6J mice. (c) Decreased Slc5a5 expression in the Rab3a−/− mice relative to Rab3a+/+ mice. The expression level of Slc5a5 in Rab3a−/− mice is similar to that in 129SvJ mice, which was much lower than in Rab3a+/+ and C57BL/6J mice. Microarray data are fold change comparing mean expression values of Rab3a−/− mice to that of Rab3a+/+. Q-PCR showed representative results from two or three experiments in quadruplicate. RT-PCR showed mean ± SD of the quantified image data (using ImageJ) from two experiments.
Figure S2. The tissue specific expression of Lpl and strain specific splicing of Prdx2. (a) Lpl expression was increased in the hippocampus but not in the cortex of Rab3a−/− mice as shown in microarray (left) and Q-PCR (right) experiments. Similarly, Lpl showed increased expression only in the hippocampus of the 129SvJ strain compared to the C57BL/6J strain. (b) Prdx2 isoforms in the hippocampi of Rab3a+/+, Rab3a−/−, 129SvJ and C57BL/6J mice. Two Affymetrix probes targeting different regions of Prdx2 mRNA gave different results in microarray experiments (left). The probe 1430979_a_at targeting the 3′ region (therefore detects the longer version of Prdx2, Prdx2-L) showed decreased levels in Rab3a−/− mice. The other probe 1418506_a_at, which targets the 5′ region and detects both the long and the short version of Prdx2 (Prdx2-LS), didn’t show any difference in gene expression. RT-PCR (right) using primers specific for Prdx2-L and Prdx2-LS showed that only the Prdx2-L was decreased in Rab3a−/− and 129SvJ mice as compared to that in Rab3a+/+ and C57BL/6J mice, respectively.
Acknowledgments
We thank Yoshimi Takai for rabphilin3A construct and antibodies for rabphilin3A, Rab GDI and Rab GAP; Adam Crystal and Virginia Lee for providing HEK293 cells; Don Baldwin and Warren Ewens for advice on microarray experiments and statistical analysis; Margaret Chou and Jeff Field for advice on GTP/GDP binding experiments; Irwin Lucki for advice on the FST; Lois Maltais for help with the nomenclature and Bucan lab members for technical help and critical reading of the manuscript. This work was supported by the NIH grant (R01 MH604687) and, in part, by a grant with the Pennsylvania Department of Health.
References
- Adams DJ, Dermitzakis ET, Cox T, Smith J, Davies R, Banerjee R, Bonfield J, Mullikin JC, Chung YJ, Rogers J, Bradley A. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat Genet. 2005;37:532–536. doi: 10.1038/ng1551. [DOI] [PubMed] [Google Scholar]
- Barnes P, Good M. Impaired pavlovian cued fear conditioning in Tg2576 mice expressing a human mutant amyloid precursor protein gene. Behav Brain Res. 2005;157:107–117. doi: 10.1016/j.bbr.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Becker J, Tan TJ, Trepte HH, Gallwitz D. Mutational analysis of the putative effector domain of the GTP-binding Ypt1 protein in yeast suggests specific regulation by a novel GAP activity. EMBO J. 1991;10:785–792. doi: 10.1002/j.1460-2075.1991.tb08010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Thakur P, Junge HJ, Ashery U, Rhee JS, Scheuss V, Rosenmund C, Rettig J, Brose N. Functional interaction of the active zone proteins Munc13–1 and RIM1 in synaptic vesicle priming. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Borsini F, Meli A. Is the forced swimming test a suitable model for revealing antidepressant activity? Psychopharmacology (Berl) 1988;94:147–160. doi: 10.1007/BF00176837. [DOI] [PubMed] [Google Scholar]
- Brondyk WH, McKiernan CJ, Burstein ES, Macara IG. Mutants of Rab3A analogous to oncogenic Ras mutants. Sensitivity to Rab3A-GTPase activating protein and Rab3A-guanine nucleotide releasing factor. J Biol Chem. 1993;268:9410–9415. [PubMed] [Google Scholar]
- Burstein ES, Brondyk WH, Macara IG. Amino acid residues in the Ras-like GTPase Rab3A that specify sensitivity to factors that regulate the GTP/GDP cycling of Rab3A. J Biol Chem. 1992;267:22715–22718. [PubMed] [Google Scholar]
- Burton JL, Burns ME, Gatti E, Augustine GJ, De Camilli P. Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J. 1994;13:5547–5558. doi: 10.1002/j.1460-2075.1994.tb06892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Janz R, Sudhof TC, Tzounopoulos T, Malenka RC, Nicoll RA. Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature. 1997;388:590–593. doi: 10.1038/41574. [DOI] [PubMed] [Google Scholar]
- Charney DS, Deutch A. A functional neuroanatomy of anxiety and fear: implications for the pathophysiology and treatment of anxiety disorders. Crit Rev Neurobiol. 1996;10:419–446. doi: 10.1615/critrevneurobiol.v10.i3-4.70. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, Tandon A, Valtorta F, Balch WE, Chelly J, Toniolo D. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet. 1998;19:134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Welzl H, Papadimitriou S, Raffaele di Barletta M, Tiveron C, Tatangelo L, Pozzi L, Chapman PF, Knevett SG, Ramsay MF, Valtorta F, Leoni C, Menegon A, Wolfer DP, Lipp HP, Toniolo D. Deletion of the mental retardation gene Gdi1 impairs associative memory and alters social behavior in mice. Hum Mol Genet. 2002;11:2567–2580. doi: 10.1093/hmg/11.21.2567. [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Wolfer DP, Kopp C, Tobler I, Toniolo D, Lipp HP. Mice deficient for the synaptic vesicle protein Rab3a show impaired spatial reversal learning and increased explorative activity but none of the behavioral changes shown by mice deficient for the Rab3a regulator Gdi1. Eur J Neurosci. 2004;19:1895–1905. doi: 10.1111/j.1460-9568.2004.03270.x. [DOI] [PubMed] [Google Scholar]
- D’Hooge R, De Deyn PP. Applications of the Morris water maze in the study of learning and memory. Brain Res Brain Res Rev. 2001;36:60–90. doi: 10.1016/s0165-0173(01)00067-4. [DOI] [PubMed] [Google Scholar]
- Davis M. Are different parts of the extended amygdala involved in fear versus anxiety? Biol Psychiatry. 1998;44:1239–1247. doi: 10.1016/s0006-3223(98)00288-1. [DOI] [PubMed] [Google Scholar]
- Dehal P, Predki P, Olsen AS, et al. Human chromosome 19 and related regions in mouse: conservative and line-age-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Easton A, Arbuzova J, Turek FW. The circadian Clock mutation increases exploratory activity and escape-seeking behavior. Genes Brain Behav. 2003;2:11–19. doi: 10.1034/j.1601-183x.2003.00002.x. [DOI] [PubMed] [Google Scholar]
- Farnsworth CL, Feig LA. Dominant inhibitory mutations in the Mg(2+) -binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991;11:4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty L. Congenic strains. In: Foster HL, Small JD, Fox JG, editors. The Mouse in Biomedical Research. Vol. 1. Academic Press; New York: 1981. pp. 215–222. [Google Scholar]
- Foletti DL, Scheller RH. Developmental regulation and specific brain distribution of phosphorabphilin. J Neurosci. 2001;21:5461–5472. doi: 10.1523/JNEUROSCI.21-15-05461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- Geppert M, Bolshakov VY, Siegelbaum SA, Takei K, De Camilli P, Hammer RE, Sudhof TC. The role of Rab3A in neurotransmitter release. Nature. 1994;369:493–497. doi: 10.1038/369493a0. [DOI] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Stevens CF, Sudhof TC. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature. 1997;387:810–814. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- Giovedi S, Darchen F, Valtorta F, Greengard P, Benfenati F. Synapsin is a novel RAB3 effector protein on small synaaptic vesicles II. Functional effects of the Rab3A-synapsin 1 interaction. J Biol Chem. 2004;279:43769–43779. doi: 10.1074/jbc.M404168200. [DOI] [PubMed] [Google Scholar]
- Gould TJ, Feiro OR. Age-related deficits in the retention of memories for cued fear conditioning are reversed by galantamine treatment. Behav Brain Res. 2005;165:160–171. doi: 10.1016/j.bbr.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Grant GR, Liu J, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell. 2005;16:3847–3864. doi: 10.1091/mbc.E05-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensbroek RA, Kamal A, Baars AM, Verhage M, Spruijt BM. Spatial, contextual and working memory are not affected by the absence of mossy fiber long-term potentiation and depression. Behav Brain Res. 2003;138:215–223. doi: 10.1016/s0166-4328(02)00243-7. [DOI] [PubMed] [Google Scholar]
- Huang YY, Zakharenko SS, Schoch S, Kaeser PS, Janz R, Sudhof TC, Siegelbaum SA, Kandel ER. Genetic evidence for a protein-kinase-A-mediated presynaptic component in NMDA-receptor-dependent forms of longterm synaptic potentiation. Proc Natl Acad Sci USA. 2005;102:9365–9370. doi: 10.1073/pnas.0503777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+) -binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- Jung V, Wei W, Ballester R, Camonis J, Mi S, Van Aelst L, Wigler M, Broek D. Two types of RAS mutants that dominantly interfere with activators of RAS. Mol Cell Biol. 1994;14:3707–3718. doi: 10.1128/mcb.14.6.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D, Valladares O, Sun Y, Nolan PM, Rux JJ, Arnold SE, Veasey SC, Bucan M. Mutations in Rab3a alter circadian period and homeostatic response to sleep loss in the mouse. Nat Genet. 2002;32:290–295. doi: 10.1038/ng991. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Nakanishi H, Takai Y. Purification and properties of Rab3A. Methods Enzymol. 1995;257:57–70. doi: 10.1016/s0076-6879(95)57010-1. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science. 1992;256:675–677. doi: 10.1126/science.1585183. [DOI] [PubMed] [Google Scholar]
- Klinz FJ. GTP-blot analysis of small GTP-binding proteins. The C-terminus is involved in renaturation of blotted proteins. Eur J Biochem. 1994;225:99–105. doi: 10.1111/j.1432-1033.1994.00099.x. [DOI] [PubMed] [Google Scholar]
- Koch M. The neurobiology of startle. Prog Neurobiol. 1999;59:107–128. doi: 10.1016/s0301-0082(98)00098-7. [DOI] [PubMed] [Google Scholar]
- Koch M, Schnitzler HU. The acoustic startle response in rats – circuits mediating evocation, inhibition and potentiation. Behav Brain Res. 1997;89:35–49. doi: 10.1016/s0166-4328(97)02296-1. [DOI] [PubMed] [Google Scholar]
- Lattal KM, Abel T. Different requirements for protein synthesis in acquisition and extinction of spatial preferences and context-evoked fear. J Neurosci. 2001;21:5773–5780. doi: 10.1523/JNEUROSCI.21-15-05773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Takei K, Geppert M, Daniell L, Stenius K, Chapman ER, Jahn R, De Camilli P, Sudhof TC. Synaptic targeting of rabphilin-3A, a synaptic vesicle Ca2+/phospholipid-binding protein, depends on rab3A/3C. Neuron. 1994;13:885–898. doi: 10.1016/0896-6273(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Lopez JF, Akil H, Watson SJ. Neural circuits mediating stress. Biol Psychiatry. 1999;46:1461–1471. doi: 10.1016/s0006-3223(99)00266-8. [DOI] [PubMed] [Google Scholar]
- Lucki I, Dalvi A, Mayorga AJ. Sensitivity to the effects of pharmacologically selective antidepressants in different strains of mice. Psychopharmacology (Berl) 2001;155:315–322. doi: 10.1007/s002130100694. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Nagata E, Ye K, Yu H, Jung TS, Luo X, Jain S, Sawa A, Snyder SH. GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31:439–451. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Kikuchi A, Araki S, Hata Y, Kondo J, Teranishi Y, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for smg p25A, a ras p21-like GTP-binding protein. Mol Cell Biol. 1990;10:4116–4122. doi: 10.1128/mcb.10.8.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus-Amat P, Higgins EA, Barrientos RM, Rudy JW. The role of the dorsal hippocampus in the acquisition and retrieval of context memory representations. J Neurosci. 2004;24:2431–2439. doi: 10.1523/JNEUROSCI.1598-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano F, Sasaki T, Fukui K, Asakura T, Imazumi K, Takai Y. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J Biol Chem. 1998;273:24781–24785. doi: 10.1074/jbc.273.38.24781. [DOI] [PubMed] [Google Scholar]
- Nagano F, Kawabe H, Nakanishi H, Shinohara M, Deguchi-Tawarada M, Takeuchi M, Sasaki T, Takai Y. Rabconnectin-3, a novel protein that binds both GDP/GTP exchange protein and GTPase-activating protein for Rab3 small G protein family. J Biol Chem. 2002;277:9629–9632. doi: 10.1074/jbc.C100730200. [DOI] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Owen EH, Christensen SC, Paylor R, Wehner JM. Identification of quantitative trait loci involved in contextual and auditory-cued fear conditioning in BXD recombinant inbred strains. Behav Neurosci. 1997a;111:292–300. doi: 10.1037//0735-7044.111.2.292. [DOI] [PubMed] [Google Scholar]
- Owen EH, Logue SF, Rasmussen DL, Wehner JM. Assessment of learning by the Morris water task and fear conditioning in inbred mouse strains and F1 hybrids: implications of genetic background for single gene mutations and quantitative trait loci analyses. Neuroscience. 1997b;80:1087–1099. doi: 10.1016/s0306-4522(97)00165-6. [DOI] [PubMed] [Google Scholar]
- Pezze MA, Feldon J. Mesolimbic dopaminergic pathways in fear conditioning. Prog Neurobiol. 2004;74:301–320. doi: 10.1016/j.pneurobio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Sudhof TC, Nestler EJ. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/s0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- Rosahl TW, Spillane D, Missler M, Herz J, Selig DK, Wolff JR, Hammer RE, Malenka RC, Sudhof TC. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci USA. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Kikuchi A, Araki S, Hata Y, Isomura M, Kuroda S, Takai Y. Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:2333–2337. [PubMed] [Google Scholar]
- Schluter OM, Schnell E, Verhage M, Tzonopoulos T, Nicoll RA, Janz R, Malenka RC, Geppert M, Sudhof TC. Rabphilin knock-out mice reveal that rabphilin is not required for rab3 function in regulating neurotransmitter release. J Neurosci. 1999;19:5834–5846. doi: 10.1523/JNEUROSCI.19-14-05834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter OM, Khvotchev M, Jahn R, Sudhof TC. Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J Biol Chem. 2002;277:40919–40929. doi: 10.1074/jbc.M203704200. [DOI] [PubMed] [Google Scholar]
- Schluter OM, Schmitz F, Jahn R, Rosenmund C, Sudhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- Shirataki H, Kaibuchi K, Yamaguchi T, Wada K, Horiuchi H, Takai Y. A possible target protein for smg-25A/rab3A small GTP-binding protein. J Biol Chem. 1992;267:10946–10949. [PubMed] [Google Scholar]
- Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol Cell Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM. Mouse Genetics: Concepts and Applications. Oxford University Press; New York: 1995. [Google Scholar]
- Staunton J, Ganetzky B, Nonet ML. Rabphilin potentiates soluble N-ethylmaleimide sensitive factor attachment protein receptor function independently of rab3. J Neurosci. 2001;21:9255–9264. doi: 10.1523/JNEUROSCI.21-23-09255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler O, Moya KL, Zahraoui A, Tavitian B. Developmental changes in the localization of the synaptic vesicle protein rab3A in rat brain. Neuroscience. 1994;62:587–600. doi: 10.1016/0306-4522(94)90391-3. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: current knowledge and future challenges. Psychopharmacology (Berl) 2001;156:194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Shirataki H, Nakanishi H. Rab3A small GTP-binding protein in Ca(2 )-dependent exocytosis. Genes Cells. 1996;1:615–632. doi: 10.1046/j.1365-2443.1996.00257.x. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, Gould TJ, Druhan JP, Bucan M. Behavior and mutagenesis screens: the importance of baseline analysis of inbred strains. Mamm Genome. 2000;11:555–564. doi: 10.1007/s003350010107. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y. Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem. 1997;272:3875–3878. doi: 10.1074/jbc.272.7.3875. [DOI] [PubMed] [Google Scholar]
- Wagner P, Molenaar CM, Rauh AJ, Brokel R, Schmitt HD, Gallwitz D. Biochemical properties of the rasrelated YPT protein in yeast: a mutational analysis. EMBO J. 1987;6:2373–2379. doi: 10.1002/j.1460-2075.1987.tb02514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sudhof TC. Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins (small star, filled) Genomics. 2003;81:126–137. doi: 10.1016/s0888-7543(02)00024-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof TC. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Sudhof TC. The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem. 2000;275:20033–20044. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- Wieland S, Lucki I. Antidepressant-like activity of 5-HT1A agonists measured with the forced swim test. Psychopharmacology (Berl) 1990;101:497–504. doi: 10.1007/BF02244228. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Differentially expressed genes on chromosome 8: microarray results and validation by Q-PCR or RT-PCR. (a) The expression patterns of Lipoprotein lipase (Lpl) in the hippocampus were examined by microarray (left) and Q-PCR (right). In both experiments Lpl showed more than 2-fold higher expression in Rab3a−/− mice than in Rab3a+/+ mice. Q-PCR also showed that Lpl expressed more than 2-fold higher in 129SvJ strain than in C57BL/6J strain. (b) The expression pattern of the longer form of Peroxiredoxin II (Prdx2-L) in microarray experiment (left) was confirmed by RT-PCR (right). The Rab3a−/− and 129SvJ mice expressed less Prdx2-L than the Rab3a+/+ and C57BL/6J mice. (c) Decreased Slc5a5 expression in the Rab3a−/− mice relative to Rab3a+/+ mice. The expression level of Slc5a5 in Rab3a−/− mice is similar to that in 129SvJ mice, which was much lower than in Rab3a+/+ and C57BL/6J mice. Microarray data are fold change comparing mean expression values of Rab3a−/− mice to that of Rab3a+/+. Q-PCR showed representative results from two or three experiments in quadruplicate. RT-PCR showed mean ± SD of the quantified image data (using ImageJ) from two experiments.
Figure S2. The tissue specific expression of Lpl and strain specific splicing of Prdx2. (a) Lpl expression was increased in the hippocampus but not in the cortex of Rab3a−/− mice as shown in microarray (left) and Q-PCR (right) experiments. Similarly, Lpl showed increased expression only in the hippocampus of the 129SvJ strain compared to the C57BL/6J strain. (b) Prdx2 isoforms in the hippocampi of Rab3a+/+, Rab3a−/−, 129SvJ and C57BL/6J mice. Two Affymetrix probes targeting different regions of Prdx2 mRNA gave different results in microarray experiments (left). The probe 1430979_a_at targeting the 3′ region (therefore detects the longer version of Prdx2, Prdx2-L) showed decreased levels in Rab3a−/− mice. The other probe 1418506_a_at, which targets the 5′ region and detects both the long and the short version of Prdx2 (Prdx2-LS), didn’t show any difference in gene expression. RT-PCR (right) using primers specific for Prdx2-L and Prdx2-LS showed that only the Prdx2-L was decreased in Rab3a−/− and 129SvJ mice as compared to that in Rab3a+/+ and C57BL/6J mice, respectively.