1. INTRODUCTION
The traditional role of synthetic chemistry in the study of biological systems has been the preparation and employment of small-molecule ligands to perturb biological pathways through highly specific, non-covalent interactions with their biological targets. While this ligand-based approach will undoubtedly continue to help the elucidation of a large fraction of biomolecular functions in many biological processes,1 alternative strategies are needed for the study of biomolecules that lack the obvious ligand-binding pockets. To this end, bioorthogonal chemistry offers an exciting new strategy for the study of biomolecular dynamics and function in living systems.2–4 Compared to ligand-based approach, bioorthogonal chemistry relies upon a specific, covalent attachment of probe molecules to the biomolecule of interest. As a result, it offers several unique advantages: (1) it is applicable to all classes of biomolecules in living systems, including proteins, nucleic acids, carbohydrates, and lipids; (2) it is extremely versatile, with the choice of probe molecules limited only by the imagination of a researcher; (3) it is highly scalable, suitable for functional annotation of a single biomolecular target in living cells as well as a class of biomolecules during the genome-wide functional profiling. Because of these unique features, bioorthogonal chemistry has been successfully employed in protein functional studies to visualize protein expression, track protein localization, measure protein activity, identify protein interaction partners, and study protein turnover in living systems.
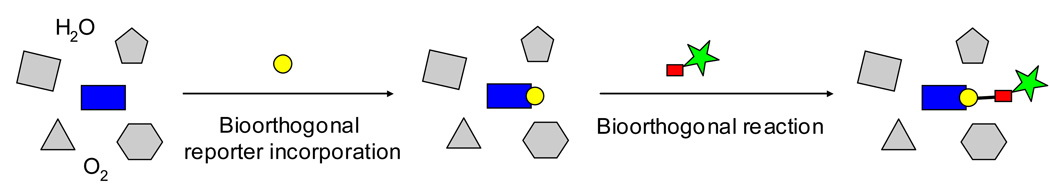
Two sequential steps are typically involved during the implementation of bioorthogonal chemistry: (1) incorporation of a bioorthogonal reporter into the biomolecule of interest by either native or the engineered biosynthetic pathway; (2) bioorthogonal reaction between the bioorthogonal reporter and the cognate, externally introduced chemical probe (Figure 1). While a plethora of methods have been developed to address the first step, e.g., amber codon suppression mutagenesis,5–8 expressed protein ligation,9–14 metabolic engineering,15–17 and tagging-via-substrate,18–23 very few bioorthogonal reactions are known to date that permit the covalent bond formations between the bioorthogonal reporter and the small-molecule probe. This can be explained by numerous constraints encountered during the development of bioorthogonal reactions, some of which are: (1) reactions need to be robust, with high yields and fast rates at relatively low concentrations; (2) reactions need to take place in the physiological conditions at neutral pH; (3) reactants must not cross-react with the abundant biological electrophiles and nucleophiles inside cells, and should only react with the externally introduced reaction partners—a property referred to as bioorthogonality; (4) reactants need to be stable both thermally and metabolically inside cells before the reaction, and non-toxic to living systems; and (5) reaction products need to be stable over a broad range of physiological conditions so that the functional measurement can be carried out.
Figure 1.
Schematic representation of bioorthogonal chemistry approach for the attachment of small-molecule probes onto a biomolecule: Blue rectangle: biomolecular target; yellow circle, bioorthogonal reporter; red rectangle, cognate reagent for the reporter; green star, small-molecule probe.
Despite these barriers, a number of bioorthogonal reactions have been developed to date that show excellent biocompatibility and selectivity in living systems. Since each of these reactions is unique, in this review, we will discuss the key attributes of each reaction as well as their representative applications in biological systems. Since the incorporation of bioorthogonal reporters —the first step in bioorthogonal chemistry has been extensively reviewed in the literature, we will not discuss the merits of each incorporation methods even though it inevitably dictates the reporter structures available for the bioorthogonal reaction studies.
2. BIOORTHOGONAL REACTIONS
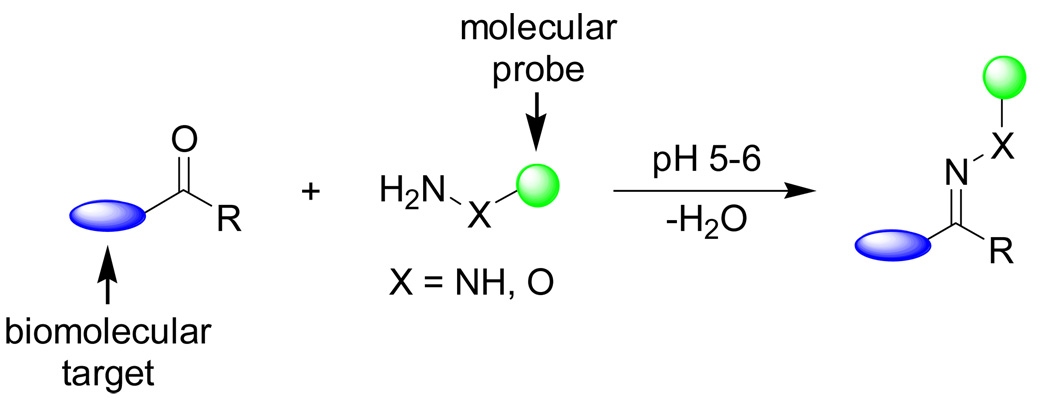
2.1 Aldehyde/ketone-based condensation
Aldehyde and ketone functionalities are attractive bioorthogonal reporters because: (1) they are small in size; (2) they can be readily incorporated into biomolecules via the biosynthetic machineries; 15, 24 and (3) they are virtually inert towards other endogenous functional groups at neutral pH. Under the acidic condition (pH = 5 ∼ 6), carbonyl group reacts with a primary amine to form a reversible Schiff base where equilibrium typically favors the free carbonyl form. However, when hydrazines (or hydrazides) and hydroxyamines are used, the equilibrium favors the imine forms, giving rise to the stable hydrazone and oxime adducts, respectively (Scheme 1).25–27
Scheme 1.
The mutual reactivity between aldehyde and hydrazine in living cells was first exploited by Rideout in 1986 for the in situ drug assembly inside cancer cells.28 In this seminal study, human erythrocytes were exposed to a mixture of decanal and N-amino-N’-1-octylguanidine (AOG) at 28 µM each in phosphate buffered saline (PBS) at 37 °C, leading to the formation of cytotoxic hydrozones which killed the cells after 80 min. The pseudo first-order rate constants between decanal and AOG were measured to be in the range of 5.6 ± 1.6 × 10−4 s−1 to 9.8 ± 1.0 × 10−4 s−1, depending on the medium conditions. More than 10 years later, Bertozzi and co-workers elegantly demonstrated the capacity of this chemistry on cell surface remodeling by treating Jurkat cells displaying a ketone reporter in the form of N-levulinoylmannosamine (ManLev) modified sialic acids on their surfaces with a biotin-hydrazide reagent.15 In general, this aldehyde/ketone-based bioorthogonal reaction is best suitable for extracellular applications15,22,29,30 because: (1) the reaction requires an optimum pH of 5 ∼ 6, incompatible with normal intracellular environment; and (2) the presence of abundant biological electrophiles inside the cells, e.g, sugars, pyruvate, and oxaloacetate, interferes with this reaction. To make the reaction feasible at neutral pH, Dawson and co-workers recently discovered that by the use of an aniline catalyst,31 the oxime adducts can be obtained in high yields at neutral pH on cell surfaces after the cells were treated with periodate under a mild condition to generate the aldehyde functionality.32
2.2 Staudinger ligation
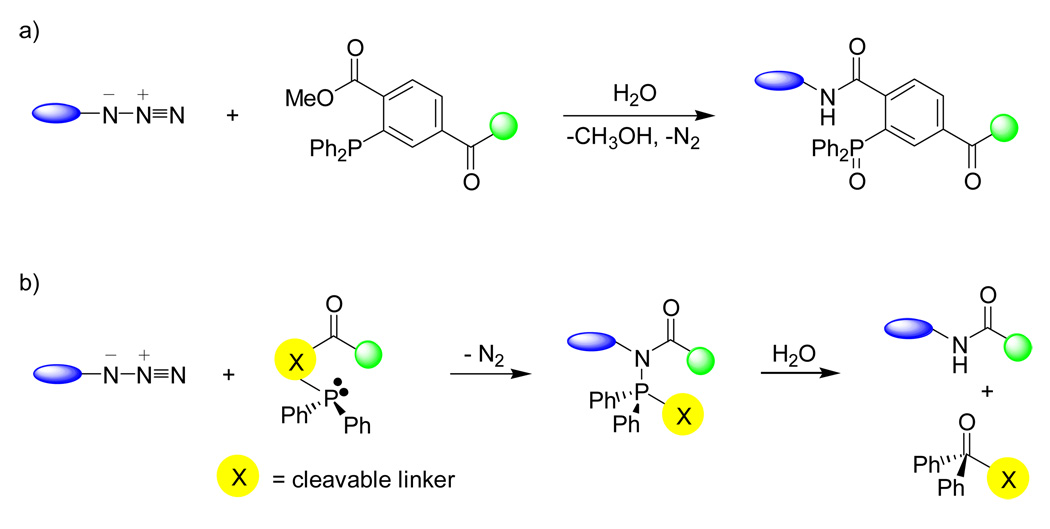
Azide group is absent in biomolecules and is stable at physiological conditions. Similar to aldehyde and ketone, azide is small in size and therefore it is relatively straightforward to be incorporated into biomolecules.33,34 One of the classic reactions involving organic azides is Staudinger reaction in which azides are converted to primary amines upon treatment with phosphines.35 A reactive aza-ylide intermediate is formed initially which, in the presence of water, hydrolyzes spontaneously to yield primary amine and phosphine oxide.36 Recognizing the mutual reactivity of azide and phosphine, Bertozzi and co-workers cleverly designed a phosphine reagent such that the aza-ylide intermediate reacts with an adjacent electrophilic carbonyl group to form a stable amide bond (Scheme 2a).37 This modified Staudinger reaction is now known as Staudinger ligation because of its exquisite ability in covalently linking two molecules together.38
Scheme 2.
Shortly after the original report, modifications termed as “traceless” Staudinger ligation were reported in which the products were devoid of phosphine oxide motif.39,40 Taking cues from the native chemical ligation, Raines and co-workers described a ligation method between a thioester and azide using phosphinothiol where the ligated product was linked by an amide bond.39 In a parallel study, Bertozzi and co-workers designed several phosphine reagents with a cleavable linker connecting the acyl group and the phosphine such that once the aza-ylide intermediate is formed, attack of the aza-ylide nitrogen on the carbonyl displaces the linker and the attached phosphonium group. Hydrolysis of the rearranged adduct produces an amide bonded ligation product and liberates a phosphine oxide (Scheme 2b).40
Staudinger ligation has been widely utilized in various biological systems. In the pioneer work, Bertozzi and co-workers engineered the surface of Jurkat cells to carry an azide reporter, and subsequently rendered the cell surfaces with biotin by employing Staudinger ligation reaction with a water-soluble phosphine reagent.37 Other applications include the enrichment of glycoprotein subtypes in the cell lysates,41,42 the addition of new functionality to recombinant proteins,43 the site-specific fluorescent labeling of proteins,34 the detection of active proteasome in living cells,44 and the live-cell imaging using a FRET-based fluorogenic phosphine.45 Oxidation of phosphine in air or by metabolic enzymes is by far the major disadvantage of this reaction, which can be overcome through the use of a large excess of phosphine reagents. Another drawback is its slow reaction rate (see more discussion below). In spite of this, Staudinger ligation has proved to be a very practical tool for biomolecular manipulation with a very mild reaction condition and high chemoselectivity.
2.3 CuI-catalyzed azide-acetylene 1,3-dipolar cycloaddition (“click chemistry”)
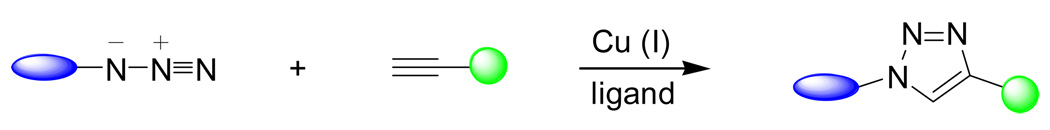
The 1,3-dipolar cycloaddition reaction between azides and acetylenes to yield triazoles was first described by Huisgen more than four decades ago.46 Since the reaction requires heating in order to achieve appreciable rates, it was not feasible for use in the biological system. However, a dramatic rate acceleration was uncovered when catalytic amount of CuI salt was used in the reaction (Scheme 3).47, 48 This CuI-catalyzed azide-alkyne1,3-dipolar cycloaddition reaction, now known as “click chemistry”, proceeds extremely efficiently at the physiological conditions at neutral pH.
Scheme 3.
Because of its fast reaction kinetics and excellent functional group tolerability, click chemistry has been widely employed in many biological studies, e.g., tagging of a variety of biomolecules, 49–51 virus surface remodeling,49 nucleic acid immobilization,52 selective protein modification,53 and the activity-based proteome profiling.54 The main advantage of click chemistry is its fast rate; the reaction between azides and alkynes proceeds at least 25 times faster than Staudinger ligation between azides and triarylphosphines in cell lysates.55 As a result, click chemistry has become an extremely valuable tool in situations that require manipulation of a very small quantity of biomolecules.56 However, the need for a copper catalyst, which is known to be toxic to cells,50 precludes its potential applications in live cells.
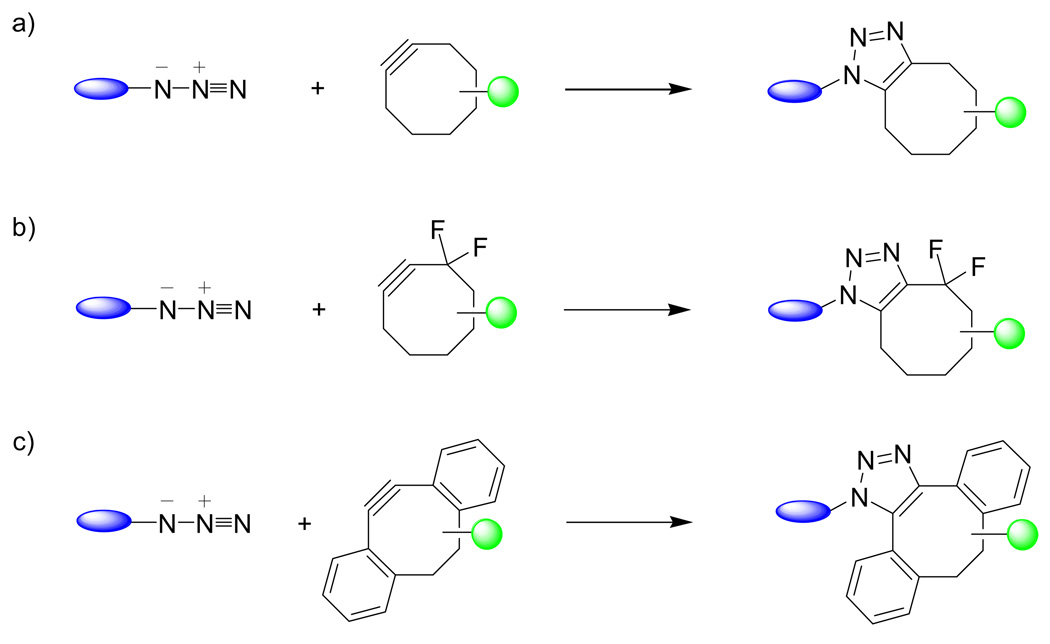
2.4 Strain-promoted azide-alkyne 1,3-dipolar cycloaddition
To circumvent the cytotoxicity associated with the copper salt, several metal-free azide-alkyne cycloaddition reactions have been developed.57 One approach was reported by Bertozzi and co-workers whereby they harnessed the ring strain present in cyclooctyne (ca. 18 kcal/mol)58 to accelerate the reaction with azide at room temperature (Scheme 4a).59 The second-order rate constant for the cycloaddition reaction between a cyclooctyne derivative and benzyl azide in aqueous CD3CN was determined to be 0.0012 M−1s−1, lower than that of a typical Staudinger ligation (0.0025 M−1s−1).55,60 Despite its slower kinetics, this earlier version of the strain-promoted cycloaddition reaction was successfully employed to label biomolecules both in vitro and on cell surfaces without observable cytotoxicity.59 Further optimization of this reaction led to the development of difluorocyclooctynes (DIFO) reagents (Scheme 4b) that showed an increased reaction rate (k2 = 0.076 M−1s−1). Because of this improved kinetics, the DIFO reagents allow the dynamic in vivo imaging of both CHO cells61 and the developing zebrafish.62 An improved synthesis of the second-generation DIFO reagents that showed similar reaction kinetics was also reported recently.63 The main disadvantage of the DIFO reagents is their synthetic demands; it takes 12 steps to prepare the first-generation DIFO reagents with an overall yield of ∼1%61 and 8 steps to synthesize the second-generation DIFO reagents with a total yield of 28%.63
Scheme 4.
An alternative cyclooctyne derivative was introduced by Boons and co-workers wherein two benzene rings were fused to the cyclooctyne to impose additional strain, thereby increasing greatly its reactivity with azides (Scheme 4c).64 The second-order rate constant between dibenzocyclooctyne and benzyl azide in acetonitrile–H2O (4:1) was determined to be 2.3 M−1s−1, approximately three orders of magnitude greater than that of simple cyclooctyne. This alkyne-based reagent was then used to visualize the metabolically labeled, azide-containing glycoconjugates on the surfaces of living cells. Compared to the DIFO reagents, the major advantages of the dibenzocyclooctyne-based reagents appear to be their synthetic accessibility and the possibility of further rate enhancement through substituent effect.
2.5 Cross-metathesis of allyl sulfides
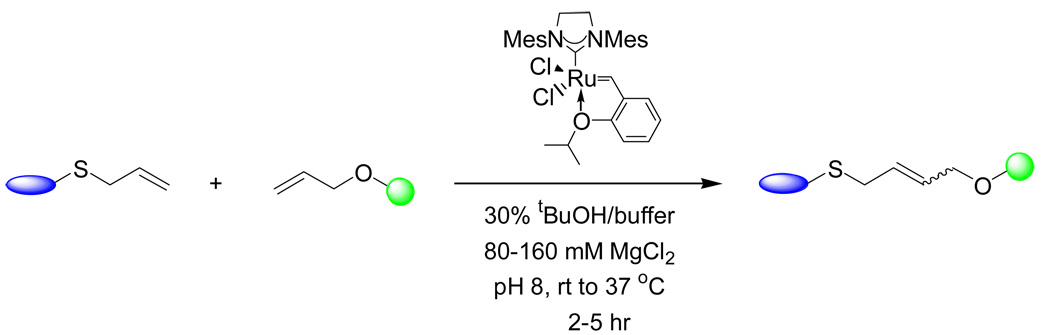
Olefin metathesis is one of the most powerful organic reactions for the construction of new carbon-carbon bonds. The suitability of cross metathesis in modifying proteins containing an allyl sulfide group in biological buffer was recently demonstrated by Davis and co-workers.65 In their studies, they screened a small panel of alkenes and found S-allylcysteine (Sac) to be the most efficient substrate for the cross metathesis reaction with allyl alcohol using the Hoveyda-Grubbs second-generation catalyst.66 Enhanced reactivity of allyl sulfides was believed to be the result of sulfur coordination to the ruthenium center. By introducing Sac to the surface of a single cysteine mutant of serine protease subtilisin Bacillus lentus (SBL), cross metathesis reaction between allyl alcohol and allyl sulfide-containing SBL can be carried out with 50–90% conversion in the presence of MgCl2 and 30% tert-butanol (Scheme 5). A multitude of site-specific SBL modifications such as glycosylation and PEGylation were subsequently demonstrated with this reaction. Importantly, there was no loss of enzymatic activity after the modifications, indicating that the reaction conditions were indeed very mild. In the same paper, they also demonstrated that Sac can be metabolically incorporated into a methionine mutant of Sulfolobus solfataricus β-glycosidase, raising an interesting possibility for future studies to use Sac as an alkene tag for cross metathesis reactions in vivo. For cellular applications, however, it remains to be investigated whether the ruthenium catalyst is toxic to cells and whether it can permeate the cell membrane necessary for the intracellular reactions.
Scheme 5.
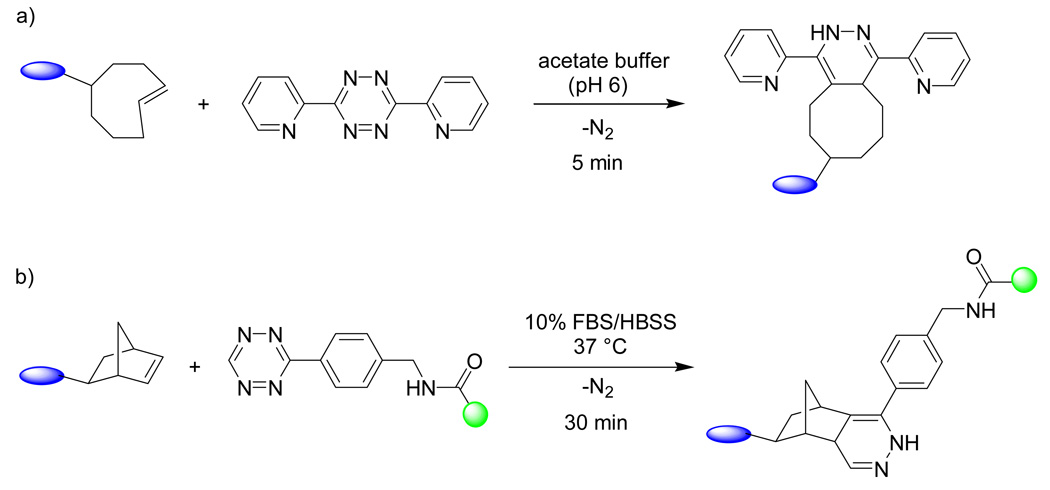
2.6 Tetrazine ligation
Diels-Alder reaction is a highly selective transformation that is known to proceed faster in water than in organic solvents due to the hydrophobic effect.67, 68 As a result, it is not surprising that Diels-Alder reaction has been employed previously in bioconjugation.69–72 Recently, Fox and co-workers reported an unusually fast bioorthogonal reaction based on the inverse-electron-demand Diels-Alder reaction between s-tetrazine and trans-cyclooctene.73 During this reaction, tetrazine acts as a voracious diene to produce the dihydropyrazine cycloadduct along with N2 as the only byproduct upon a retro-[4 + 2] cycloaddition (Scheme 6a).74 In their studies, they found that the reaction reached completion within 40 min at 25 °C and the apparent second-order rate constant for the cycloaddition between trans-cyclooctene and 3,6-di-(2-pyridyl)-s-tetrazine was determined to be 2000 ± 400 M−1s−1 in a 9:1 methanol-water mixture, faster than all other bioorthogonal reactions known to date. The suitability of this reaction in modifying proteins was illustrated with thioredoxin (Trx) in which a trans-cyclooctene derivative was first conjugated to Trx (15 µM) in an acetate buffer (pH = 6.0) through Michael addition followed by a ligation reaction with 30 µM 3,6-di-(2-pyridyl)-s-tetrazine. A 100% conversion was observed in just 5 min as analyzed by ESI-MS. At about the same time, Hilderbrand and co-workers also independently demonstrated the bioorthogonality of this reaction by using a tetrazine-derived fluorescent probe to modify a norbornene-conjugated antibody both in serum and in live cells (Scheme 6b).75
Scheme 6.
Because of its fast kinetics, the tetrazine ligation may be particularly useful in cases where rapid reactions are essential for tracking of fast biological events as well as for labeling of biomolecules of low abundance. While this reaction is clearly suitable for in vitro experiments, it is not clear how tetrazines or other strained alkenes can be genetically encoded in living system so that this ligation can be further developed for in vivo applications.
2.7 Photoinduced 1,3-dipolar cycloaddition (“photoclick chemistry”)
For a bioorthogonal reaction to be broadly useful in addressing complex and dynamic biological problems in living system, we believe three attributes are essential: 1) true bioorthogonality; 2) fast reaction rate; and 3) rapid inducibility. While the importance of excellent bioorthogonality and fast reaction rate is well recognized in the field, the importance of inducibility has not been widely appreciated. By analogy, many biological processes such as gene transcription and protein posttranslational modifications show a high level of temporality or inducibility, in order to achieve rapid response to environmental cues. In studying complex and dynamic biological processes, it would be advantageous if we can control the initiation as well as the extent of the bioorthogonal reactions such that a kinetic perturbation can be applied to a biological system. To this end, our group has developed a tetrazole-based photoinducible bioorthogonal reaction (“photoclick chemistry”) that in principle should provide a time-resolved chemical tool for the study of biological temporality.
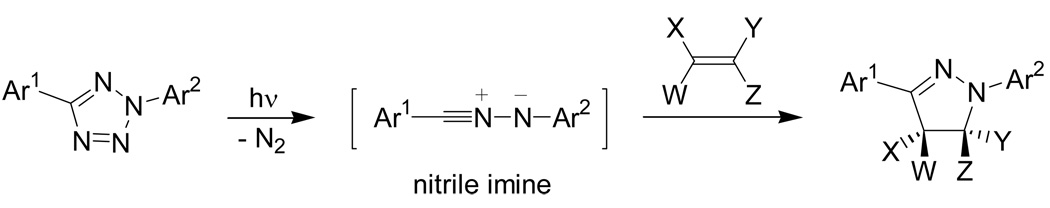
About four decades ago, Huisgen and co-workers reported a photoactivated 1,3-dipolar cycloaddition reaction between 2,5-diphenyltetrazole and methyl crotonate.76 A concerted reaction mechanism was proposed whereby the diaryltetrazole undergoes a facile cycloreversion reaction upon photoirradiation to release N2 and generate in situ a nitrile imine dipole which then cyclizes spontaneously with an alkene dipolarophile to afford a pyrazoline cycloadduct (Scheme 7). Attracted by this novel mode of substrate activation, our group initially identified an extremely mild photoactivation procedure with the use of a handheld low-powered UV lamp that allows the cycloaddition to proceed with excellent solvent compatibility including water, functional group tolerance, regioselectivity, and yield.77
Scheme 7.
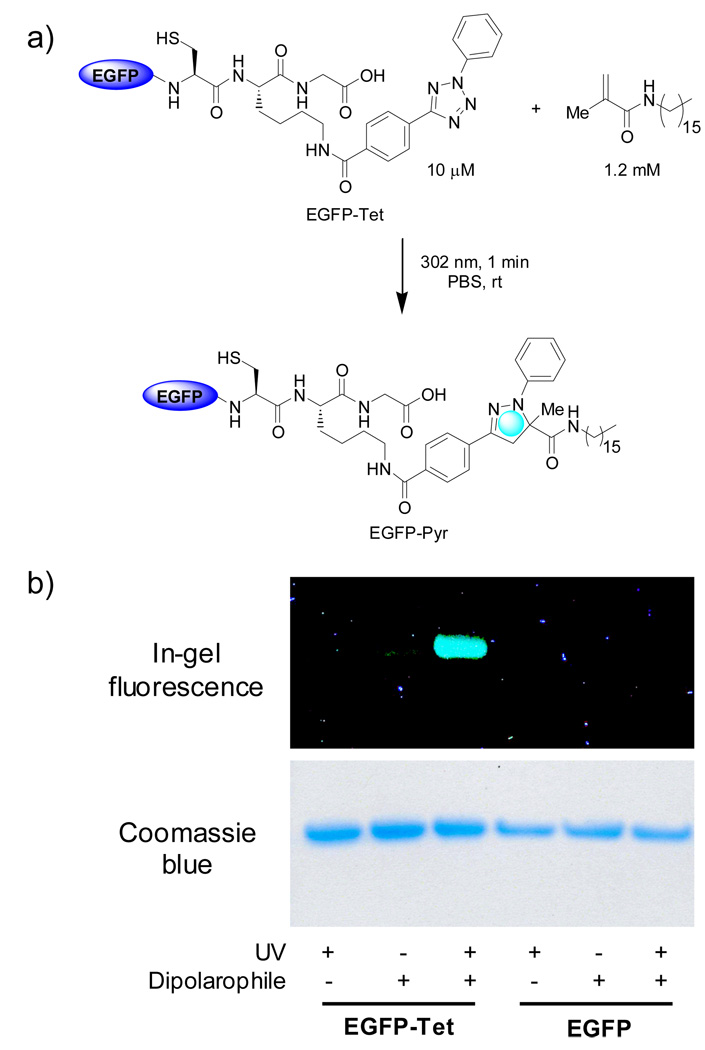
Our subsequent study demonstrated that this photoinduced cycloaddition reaction can be employed to modify the tetrazole-containing proteins in biological media.78 In a kinetic study between a tetrazole peptide and acrylamide in PBS buffer at pH 7.5, we found that under 302-nm photoirradiation the nitrile imine intermediate was generated rapidly (k1 = 0.14 s−1) and that the subsequent cycloaddition with acrylamide proceeded very efficiently with the second-order rate constant equal to 11.0 M−1s−1, significantly faster than Staudinger ligation and the strain-promoted azide-alkyne cycloaddition. Following this kinetic study, the reaction bioorthogonality was evaluated in both the residue-specific and the site-specific protein modifications. In the former case, diphenyltetrazole was introduced into lysozyme via acylation of the protein surface lysine sidechains. The resulting tetrazole-lysozyme conjugate was irradiated with a 302-nm handheld UV light for 2 min in the presence of 50 equiv of acrylamide in PBS buffer. The pyrazoline adduct was formed with greater than 90% conversion based on the LC-MS analysis. It is noteworthy that the pyrazoline cycloadducts were fluorescent, allowing us to use in-gel fluorescence analysis to confirm specific cycloadduct formation. In the latter case, diphenyltetrazole was introduced into the C-terminus of enhanced green fluorescent protein (EGFP) via the intein-based chemical ligation method. The tetrazole-containing EGFP (EGFP-Tet; 10 µM in 20 mM Tris, 500 mM NaCl, 1 mM EDTA, pH 7.5) was then reacted with 1.2 mM of a lipid dipolarophile via irradiation under 302 nm for 1 min. The in-gel fluorescence analysis revealed the presence of a fluorescent band, consistent with the formation of the fluorescent pyrazoline cycloadduct (Figure 2). The conversion from the tetrazole-EGFP to pyrazoline-EGFP was estimated to be 52% based on the LC-MS analysis. It is noteworthy that the cycloadduct showed an emission band at 460 nm along with a twofold increase in EGFP fluorescence intensity at 509 nm which was attributed to fluorescence resonance energy transfer (FRET) from the pyrazoline fluorophore to EGFP fluorophore.
Figure 2.
Photoinduced lipidation of EGFP carrying a tetrazole motif at its C-terminus: (a) Scheme for a photoinduced lipidation by a lipid dipolarophile. (b) Fluorescence imaging (top panel, λex = 365 nm) and Coomassie Blue staining (bottom panel) of EGFP and EGFP-Tet upon photoirradiation in the presence or absence of lipid dipolarophile. Duration of 1 min under 302 nm UV light was used for photoirradiation.
To avoid potential photo damage associated with short-wavelength UV light, additional effort was directed at the design of long-wavelength photoactivatable diaryltetrazoles that undergo ring-opening at 365 nm to generate the reactive nitrile imine dipoles. It was found that by placing an auxochromic or conjugative substituent on the N-phenyl ring, diaryltetrazoles exhibited good photoreactivity at 365 nm and the resulting dipoles showed excellent reactivity towards the electron-deficient and conjugated alkenes as well as the alkene-modified lysozymes.79
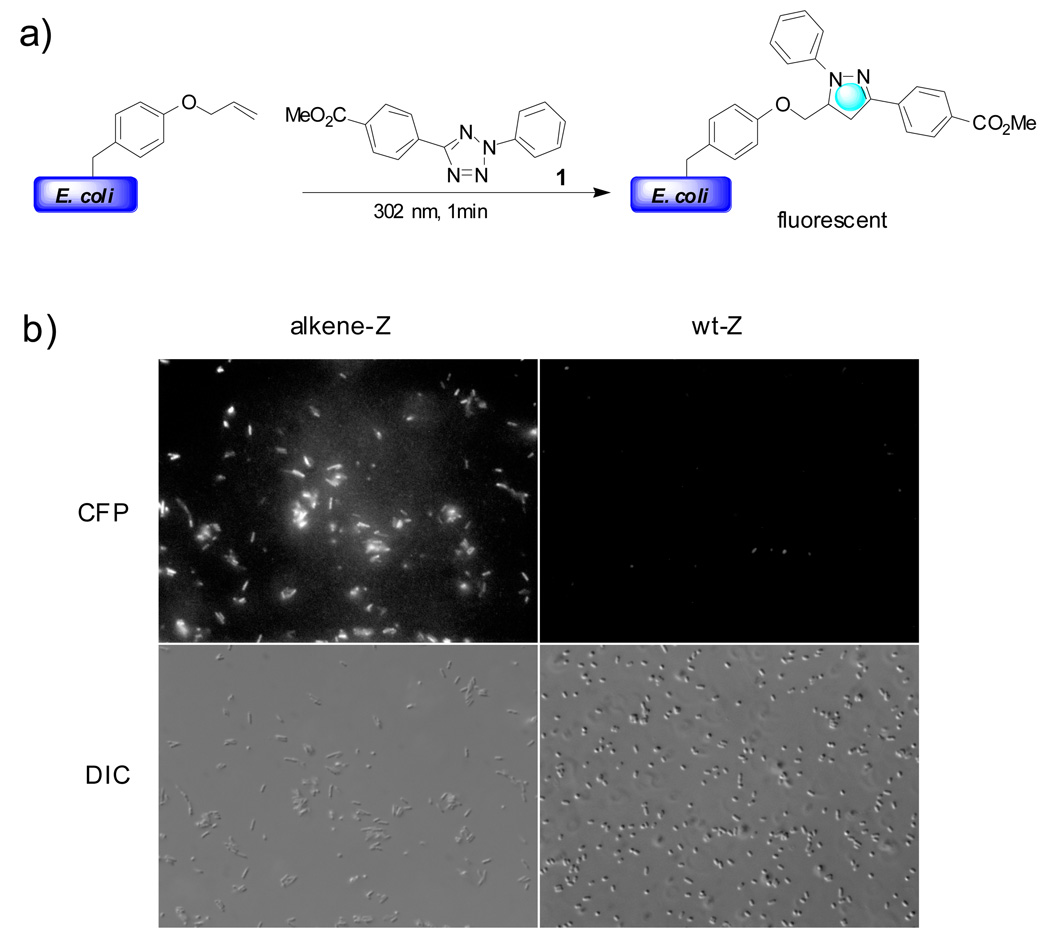
The power of this photoclick chemistry in selectively functionalizing proteins in vivo was demonstrated with E. coli cells overexpressing an alkene-containing Z-domain protein (alkene-Z).80 By employing a previously reported E. coli strain that charges O-allyltyrosine site-specifically into the Z-domain protein,81 BL21(DE3) cells expressing either wild-type Z (wt-Z) or alkene-Z protein were suspended in PBS buffer containing 5% glycerol and treated with 100 µM of tetrazole 1. After incubation at 37 °C for 30 min, the cell suspension was photoirradiated at 302 nm for 4 min followed by overnight incubation at 4 °C. In the cyan fluorescent protein (CFP) channel (which captures the fluorescent light emitted by the pyrazoline cycloadduct), only alkene-Z expressing bacterial cells showed strong fluorescence, indicative of selectivity toward the alkene functionality in intact bacterial cells (Figure 3).
Figure 3.
Selective functionalization of alkene-Z by tetrazole in E. coli cells: (1) reaction scheme; (2) CFP channel (top row) and DIC (Differential Interference Contrast) channel (bottom row) images of bacterial cells expressing either alkene-Z (left) or wt-Z (right) proteins after treatment with 100 µM tetrazole 1.
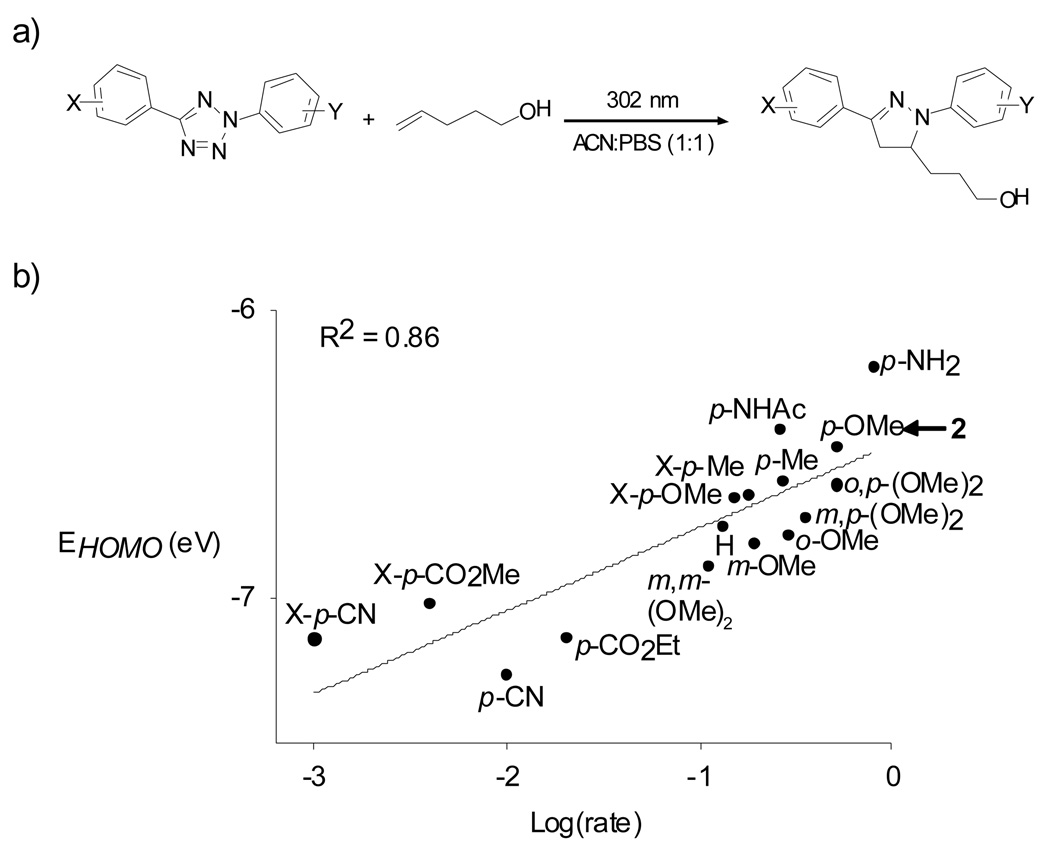
A significant observation was that the cycloaddition reaction between tetrazole 1 and O-allyltyrosine proceeded much slower than the corresponding reaction with acrylamide (k2 = 0.00202 ± 0.00007 M−1s−1 vs. 0.15 M−1s−1), presumably due to the significantly higher LUMO (Lowest Unoccupied Molecular Orbital) energy of O-allyltyrosine relative to acrylamide. This result led us to investigate the effect of systematically tuning the HOMO (Highest Occupied Molecular Orbital) energies of the nitrile imine dipoles by introducing specific substituents onto the aryl rings (Figure 4a). We found that the electron-donating groups in general lead to higher HOMO energies, which gave rise to accelerated cycloaddition rates (Figure 4b).82 One of the optimized reagents, 2-(p-methoxyphenyl)-5-phenyltetrazole (2), showed a robust reaction with O-allyltyrosine with k2 value of 0.95 M−1s−1, 475-fold faster than that of tetrazole 1. Gratifyingly, with the improved kinetics it has now become possible to use this reaction to functionalize the alkene-containing proteins in bacteria in less than one minute.82
Figure 4.
Cycloaddition rates are dependent on the nitrile imine HOMO energies. (a) Reaction scheme; (b) Plot of HOMO energies vs. Log(rate). The HOMO energies of the nitrile imines were calculated using Hartree-Fock 3–21G model based on the AM1 optimized geometries.
Since activated terminal alkenes are absent in the biological system (internal alkenes are prevalent in cellular membrane structures), this nitrile imine-mediated photoclick chemistry offered a high degree of bioorthogonality as demonstrated in our various applications. Through the use of the orbital-matched alkene dipolarophiles and HOMO-lifted tetrazole reagents, fast reaction rates can also be obtained. Additional advantages of this bioorthogonal reaction include mild reaction conditions (with incident photons to be the only reagent), easy access of tetrazole reagents (only 2 steps are required for their syntheses),83 and convenient fluorescent monitoring due to the formation of the pyrazoline adducts.
Compared to other bioorthogonal reactions, the main advantage of photoclick chemistry is its light inducibility. Since tetrazoles become activated only upon UV irradiation, the use of light allows a temporal and spatial control over the reaction initiation. While the applications in live cells so far have focused on the functionalization of the genetically encoded alkenes because of their pre-existence, it is also possible to contemplate the genetic incorporation of the tetrazole-derived building blocks. Indeed, we have recently reported the synthesis of a series of tetrazole amino acids and found that some of them showed excellent reactivity toward alkenes upon photoactivation.84 While efforts in incorporating these tetrazole amino acids are still ongoing, it is clearly possible that the reactions with the tetrazole-encoded biomolecules can be employed to regulate biomolecular function with spatial and temporal resolution. In this regard, the development of tetrazole reagents with two-photon activatability will be also warranted.
3. CONCLUSION & PERSPECTIVE
Bioorthogonal chemistry has made tremendous strides in recent years. A number of water-compatible organic transformations such as nucleophilic carbonyl addition, 1,3-dipolar cycloaddition reactions, Diels-Alder reactions, and olefin cross-metathesis have been optimized for use with biomolecular substrates in biological systems. Unlike conventional organic reaction development, a different set of parameters need to considered, e.g., bioorthogonality, reaction rate, inducibility, and substrate genetic encodability in addition to reaction yields and selectivity. However, in optimizing these parameters, the same physical organic principles that are applied to small-molecule chemistry work equally well in macromolecular chemistry, e.g., the ring-strain effect, the fluorine effect, and the HOMO-lifting effect. While each reaction has its own strengths and drawbacks, the dexterous use of these reactions in combination in functionalizing biomolecules in vivo could further enhance the utility above and beyond what is possible with their own.85
In a manner similar to the ubiquitous use of green fluorescent proteins (GFP) in cell biology, the major use of bioorthogonal chemistry has been in labeling biomolecules with fluorescent dyes in vivo. Bioorthogonal chemistry is particularly advantageous in situations where the GFP encoding is not feasible, e.g., the study of protein posttranslational modification dynamics. Because a diverse set of molecular probes can be appended onto the bioorthogonal reagents, it should be possible to engineer other detection modality such as magnetic resonance imaging (MRI) contrast agents into the reaction partners for whole animal studies. Moreover, by employing reactants with unique properties, bioorthogonal chemistry can be used to either mimic86 or expand the native biomolecular functions beyond what is known in nature—an important goal of synthetic biology.87 Evidently, the future of bioorthogonal chemistry—the use of organic reactions to directly manipulate biomolecular function—in chemical biology is bright indeed.
Acknowledgments
Our work is supported by U. S. National Institutes of Health (R01GM085092) and New York State Center of Excellence in Bioinformatics and Life Sciences.
REFERENCES
- 1.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 3.van Swieten PF, Leeuwenburgh MA, Kessler BM, Overkleeft HS. Bioorthogonal organic chemistry in living cells: novel strategies for labeling biomolecules. Org. Biomol. Chem. 2005;3:20–27. doi: 10.1039/b412558d. [DOI] [PubMed] [Google Scholar]
- 4.Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 6.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. A new strategy for the synthesis of glycoproteins. Science. 2004;303:371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Schultz PG. Adding amino acids to the genetic repertoire. Curr. Opin. Chem. Biol. 2005;9:548–954. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Schnolzer M, Kent SB. Constructing proteins by dovetailing unprotected synthetic peptides: backbone-engineered HIV protease. Science. 1992;256:221–225. doi: 10.1126/science.1566069. [DOI] [PubMed] [Google Scholar]
- 10.Low DW, Hill MG. Rational fine-tuning of the redox potentials in chemically synthesized rubredoxins. J. Am. Chem. Soc. 1998;120:11536–11537. [Google Scholar]
- 11.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotton GJ, Ayers B, Xu R, Muir TW. Insertion of a synthetic peptide into a recombinant protein framework: A protein biosensor. J. Am. Chem. Soc. 1999;121:1100–1101. [Google Scholar]
- 13.Arnold U, Hinderaker MP, Nilsson BL, Huck BR, Gellman SH, Raines RT. Protein prosthesis: A semisynthetic enzyme with a beta-peptide reverse turn. J. Am. Chem. Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]
- 14.Pellois JP, Muir TW. Semisynthetic proteins in mechanistic studies: using chemistry to go where nature can't. Curr. Opin. Chem. Biol. 2006;10:487–491. doi: 10.1016/j.cbpa.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 16.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 17.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 18.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 20.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
- 22.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 23.Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 25.Rose K. Facile synthesis of homogeneous artificial proteins. J. Am. Chem. Soc. 1994;116:30–33. [Google Scholar]
- 26.Canne LE, Ferre- D'Amare AR, Burley SK, Kent SBH. Total chemical synthesis of a unique transcription factor-related protein: cMyc-Max. J. Am. Chem. Soc. 1995;117:2998–3007. [Google Scholar]
- 27.Gaertner HF, Rose K, Cotton R, Timms D, Camble R, Offord RE. Construction of protein analogs by site-specific condensation of unprotected fragments. Bioconjugate Chem. 2002;3:262–268. doi: 10.1021/bc00015a010. [DOI] [PubMed] [Google Scholar]
- 28.Rideout D. Self-assembling cytotoxins. Science. 1986;233:561–563. doi: 10.1126/science.3523757. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Smith BAC, Wang L, Brock A, Cho C, Schultz PG. A new strategy for the site-specific modification of proteins in vivo. Biochemistry. 2003;42:6735–6746. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- 30.Sadamoto R, Niikura K, Ueda T, Monde K, Fukuhara N, Nishimura S. Control of bacteria adhesion by cell-wall engineering. J. Am. Chem. Soc. 2004;126:3755–3761. doi: 10.1021/ja039391i. [DOI] [PubMed] [Google Scholar]
- 31.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic catalysis of oxime ligation. Angew. Chem. Int. Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 32.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemieux GA, De Graffenried CL, Bertozzi CR. A fluorogenic dye activated by the Staudinger ligation. J. Am. Chem. Soc. 2003;125:4708–4709. doi: 10.1021/ja029013y. [DOI] [PubMed] [Google Scholar]
- 35.Staudinger H, Meyer J. Uber neue organische phosphoverbindungen III. Phosphinmethlenderivate und phosphiniimine. Helv. Chim. Acta. 1919;2:635–646. [Google Scholar]
- 36.Gololobov YG, Kasukhin LF. Recent advances in the Staudinger reaction. Tetrahedron. 1992;48:1353–1406. [Google Scholar]
- 37.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 38.Kohn M, Breinbauer R. The Staudinger ligation-a gift to chemical biology. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson BL, Kiessling LL, Raines RT. Staudinger ligation: A peptide from a thioester and azide. Org. Lett. 2000;2:1939–1941. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]
- 40.Saxon E, Armstrong JI, Bertozzi CR. A "traceless" Staudinger ligation for the chemoselective synthesis of amide bonds. Org. Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 41.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Metabolic functionalization of recombinant glycoproteins. Biochemistry. 2004;43:12358–12366. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- 44.Ovaa H, van Swieten PF, Kessler BM, Leeuwenburgh MA, Fiebiger E, van den Nieuwendijk AM, Galardy PJ, van der Marel GA, Ploegh HL, Overkleeft HS. Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew. Chem. Int. Ed. 2003;42:3626–3629. doi: 10.1002/anie.200351314. [DOI] [PubMed] [Google Scholar]
- 45.Hangauer MJ, Bertozzi CR. A FRET-based fluorogenic phosphine for live-cell imaging with the Staudinger ligation. Angew. Chem. Int. Ed. 2008;47:2394–2397. doi: 10.1002/anie.200704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huisgen R. 1,3-Dipolar cycloaddition. Angew. Chem. Int. Ed. 1963;2:565–598. [Google Scholar]
- 47.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by copper(I)-catalyzed azide-alkyne [3+2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 50.Link AJ, Tirrell DA. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2] cycloaddition. J. Am. Chem. Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 51.Gierlich J, Burley GA, Gramlich PME, Hammond DM, Carell T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
- 52.Seo TS, Bai X, Ruparel H, Li Z, Turro NJ, Ju J. Photocleavable fluorescent nucleotides for DNA sequencing on a chip constructed by site-specific coupling chemistry. Proc. Natl. Acad. Sci. USA. 2004;101:5488–5493. doi: 10.1073/pnas.0401138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson C, Schultz PG. Adding amino acids with novel reactivity to the genetic code of saccharomyces cerevisiae. J. Am. Chem. Soc. 2003;125:11782–11783. doi: 10.1021/ja0370037. [DOI] [PubMed] [Google Scholar]
- 54.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 55.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 56.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 57.Becer CR, Hoogenboom R, Schubert US. Click chemistry beyond metal-catalyzed cycloaddition. Angew. Chem. Int. Ed. 2009;48:4900–4908. doi: 10.1002/anie.200900755. [DOI] [PubMed] [Google Scholar]
- 58.Turner RB, Jarrett AD, Goebel P, Mallon BJ. Heat of hydrogenation. IX. Cyclic acetylenes andsome micellaneous olefins. J. Am. Chem. Soc. 1973;95:790–792. [Google Scholar]
- 59.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 60.Lin FL, Hoyt HM, van Halbeek H, Bergman RG, Bertozzi CR. Mechanistic investigation of the Staudinger ligation. J. Am. Chem. Soc. 2005;127:2686–2695. doi: 10.1021/ja044461m. [DOI] [PubMed] [Google Scholar]
- 61.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ning X, Guo J, Wolfert MA, Boons G-J. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast Huisgen cycloadditions. Angew. Chem. Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin YA, Chalker JM, Floyd N, Bernardes GJ, Davis BG. Allyl sulfides are privileged substrates in aqueous cross-metathesis: Application to site-selective protein modification. J. Am. Chem. Soc. 2008;130:9642–9643. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- 66.Garber SB, Kingsbury JS, Gray BL, Hoveyda AH. Efficient and recyclable monomeric and dendritic Ru-based metathesis catalysts. J. Am. Chem. Soc. 2000;122:8168–8179. [Google Scholar]
- 67.Breslow R, Rideout DC. Hydrophobic acceleration of Diels-Alder reactions. J. Am. Chem. Soc. 1980;102:7816–7817. [Google Scholar]
- 68.Otto S, Engberts JBFN. Diels-Alder reactions in water. Pure Appl. Chem. 2000;72:1365–1372. [Google Scholar]
- 69.Seelig B, Jäschke A. Site-specific modification of enzymatically synthesized RNA: Transcription initiation and Diels-Alder reaction. Tet. Lett. 1997;38:7729–7732. [Google Scholar]
- 70.Yousaf MN, Mrksich M. Diels-Alder reaction for the selective immobilization of protein to electroactive self-assembled monolayers. J. Am. Chem. Soc. 1999;121:4286–4287. [Google Scholar]
- 71.Latham-Timmons HA, Wolter A, Roach JS, Giare R, Leuck M. Novel method for the covalent immobilization of oligonucleotides via Diels-Alder bioconjugation. Nucleosides, Nucleotides & Nucleic Acids. 2003;22:1495–1497. doi: 10.1081/NCN-120023019. [DOI] [PubMed] [Google Scholar]
- 72.de Araújo AD, Palomo JM, Cramer J, Köhn M, Schröder H, Wacker R, Niemeyer C, Alexandrov K, Waldmann H. Diels-Alder ligation and surface immobilization of proteins. Angew. Chem. Int. Ed. 2006;45:296–301. doi: 10.1002/anie.200502266. [DOI] [PubMed] [Google Scholar]
- 73.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boger DL. Diels-Alder reactions of heterocyclic aza dienes. Scope and applications. Chem. Rev. 1986;86:781–793. [Google Scholar]
- 75.Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clovis JS, Eckell A, Huisgen R, Sustmann R. 1.3-Dipolare Cycloadditionen, XXV. Der Nachweis des freien Diphenylnitrilimins als Zwischenstufe bei Cycloadditionen. Chem. Ber. 1967;100:60–70. [Google Scholar]
- 77.Wang Y, Rivera Vera CI, Lin Q. Convenient synthesis of highly functionalized pyrazolines via mild, photoactivated 1,3-dipolar cycloaddition. Org. Lett. 2007;9:4155–4158. doi: 10.1021/ol7017328. [DOI] [PubMed] [Google Scholar]
- 78.Song W, Wang Y, Qu J, Madden MM, Lin Q. A photoinducible 1,3-dipolar cycloaddition reaction for rapid, selective modification of tetrazole-containing proteins. Angew. Chem. Int. Ed. 2008;47:2832–2835. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, Hu WJ, Song W, Lim RKV, Lin Q. Discovery of long-wavelength photoactivatable diaryltetrazoles for bioorthogonal 1,3-dipolar cycloaddition reactions. Org. Lett. 2008;10:3725–3728. doi: 10.1021/ol801350r. [DOI] [PubMed] [Google Scholar]
- 80.Song W, Wang Y, Qu J, Lin Q. Selective functionalization of a genetically encoded alkene-containing protein via "photoclick chemistry" in bacterial cells. J. Am. Chem. Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z, Wang L, Brock A, Schultz PG. The selective incorporation of alkenes into proteins in Escherichia coli. Angew. Chem. Int. Ed. 2002;41:2840–2842. doi: 10.1002/1521-3773(20020802)41:15<2840::AID-ANIE2840>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Song W, Hu WJ, Lin Q. Fast alkene functionalization in vivo by photoclick chemistry: HOMO lifting of nitrile imine dipoles. Angew. Chem. Int. Ed. 2009;48:5330–5333. doi: 10.1002/anie.200901220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ito S, Tanaka Y, Kakehi A, Kondo K. Facile Synthesis of 2,5-disubstituted tetrazoles by reaction of phenylsulfonylhydrazones with arenediazonium salts. Bull. Chem. Soc. Jpn. 1976;49:1920–1923. [Google Scholar]
- 84.Wang Y, Lin Q. Synthesis and evaluation of photoreactive tetrazole amino acids. Org. Lett. 2009;10 doi: 10.1021/ol901300h. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Kasteren SI, Kramer HB, Jensen HH, Campbell SJ, Kirkpatrick J, Oldham NJ, Anthony DC, Davis BG. Expanding the diversity of chemical protein modification allows post-translational mimicry. Nature. 2007;446:1105–1109. doi: 10.1038/nature05757. [DOI] [PubMed] [Google Scholar]
- 86.Serwa R, Wilkening I, Del Signore G, Muhlberg M, Claussnitzer I, Weise C, Gerrits M, Hackenberger CP. Chemoselective staudinger-phosphite reaction of azides for the phosphorylation of proteins. Angew. Chem. Int. Ed.l. 2009 doi: 10.1002/anie.200902118. in press. [DOI] [PubMed] [Google Scholar]
- 87.Benner SA, Sismour AM. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]