Abstract
The effective Hamiltonian-molecular orbital and valence bond (EH-MOVB) method based on non-orthogonal block-localized fragment orbitals has been implemented into the program CHARMM for molecular dynamics simulations of chemical and enzymatic reactions, making use of semiempirical quantum mechanical models. Building upon ab initio MOVB theory, we make use of two parameters in the EH-MOVB method to fit the barrier height and the relative energy between the reactant and product state for a given chemical reaction to be in agreement with experiment or high-level ab initio or density functional results. Consequently, the EH-MOVB method provides a highly accurate and computationally efficient QM/MM model for dynamics simulation of chemical reactions in solution. The EH-MOVB method is illustrated by examination of the potential energy surface of the hydride transfer reaction from trimethylamine to a flavin cofactor model in the gas phase. In the present study, we employed the semiempirical AM1 model, which yields a reaction barrier that is more than 5 kcal/mol too high. We use a parameter calibration procedure for the EH-MOVB method similar to that employed to adjust the results of semiempirical and empirical models. Thus, the relative energy of these two diabatic states can be shifted to reproduce the experimental energy of reaction, and the barrier height is optimized to reproduce the desired (accurate) value by adding a constant to the off-diagonal matrix element. The present EH-MOVB method offers a viable approach to characterizing solvent and protein-reorganization effects in the realm of combined QM/MM simulations.
1. Introduction
Combined quantum mechanical and molecular mechanical (QM/MM) methods offer an excellent opportunity for studying chemical and electron transfer reactions in solution and in biological systems.1–3 In principle, the accuracy of combined QM/MM potentials can be systematically improved; however, it is still time demanding to carry out QM/MM simulations using ab initio wave function theory (WFT) or density functional theory (DFT) for subsystems consisting of more than 100 atoms in the QM region. Consequently, it is useful to develop efficient QM/MM techniques that can be made accurate for specific chemical and biomolecular applications, yet sufficiently fast for extensive conformational sampling. Aside from the brute force approach by increasing the level of theory and the size of basis set, there are two other ways to achieve this goal. The first is to parameterize purely empirical energy functions to model a specific process,4–5 and the second is to parameterize quantum mechanical models against experimental data with specific reaction parameters (SRP) for a given class of reactions.6–9 In this article, we describe an effective Hamiltonian approach based on the molecular orbital-valence bond (MOVB) theory developed in our laboratories for the treatment of reactive potential surfaces of reactions.10–12 In particular, we illustrate this novel QM/MM technique in the hydride transfer reaction from trimethyl ammonium ion to flavin cofactor.
The mixed molecular orbital and valence bond (MOVB) theory,10–11 initially developed at the ab initio level and recently extended to multistate density functional theory (called MSDFT or equivalently, VBDFT),13 is designed to treat reactive potential energy surfaces for chemical reactions and electron transfer processes. In this approach, the whole process is described with two or more resonating configurations, including the reactant and product states. In each specific state, molecular orbitals (MOs) are strictly localized within individual fragments of a molecular system. 14–20 However, the block-localized molecular orbitals (BLMOs) are still delocalized within each orbital block, making the MOVB method extremely efficient in comparison with ab initio valence bond theory.21–25 Key features of the MOVB and MSDFT theories include (1) that the BLMOs (or block-localized Kohn-Sham orbitals)13 within each fragment are orthogonal, which makes it computationally efficient, and (2) that the BLMOs between different fragments are nonorthogonal,11,13,20 which retains important characters of valence bond (VB) theory. In the limiting case in which there is one fragment, MOVB and MSDFT reduces exactly to the standard Hartree-Fock theory and Kohn-Sham DFT, respectively.
Recently, we introduced an effective Hamiltonian MOVB approach,26 in which the ab initio electronic matrix elements are adjusted to yield accurate barrier height and reaction enthalpy. This approach has the apparent similarity in the “calibration” process used to adjust the barrier height and the energy of reaction in semiempirical or empirical valence bond models,27–32 although the theory and algorithm of MOVB are based on ab initio WFT and DFT approaches to define VB electronic configuration states. Effective Hamiltonian approaches are widely used in many different areas.33–40 A major advantage of the EH-MOVB approach is that all VB matrix elements, including off-diagonal terms, are determined by an electronic structure method, which depends explicitly on all degrees of freedom in the system. In the empirical and semiempirical valence bond approaches, typically a simple function, depending on one degree of freedom, or a constant is used to treat the off-diagonal matrix elements in a VB-like Hamiltonian.27,30–31,41 Note that empirical multiconfigurational models have been described to fit the energy, gradient and Hessian of ab initio potential surfaces40,42–44 using Gaussian and polynomial functions45-46 or Shepard interpolation.47–49
In this paper, we show that the EH-MOVB method can be constructed using semiempirical QM models such as the Austin model 1 (AM1),50 parameterization model 3 (PM3)51 or Recife model 1 (RM1)52 to yield the barrier height for a chemical reaction in agreement with experiments or with ab initio results. In the following, we first present the theoretical background, followed by computational details. Results and discussions are presented next. Finally, the paper concludes with a summary of the major findings of this study and future perspectives.
2. Method
A. The Mixed Molecular Orbital and Valence Bond (MOVB) Theory
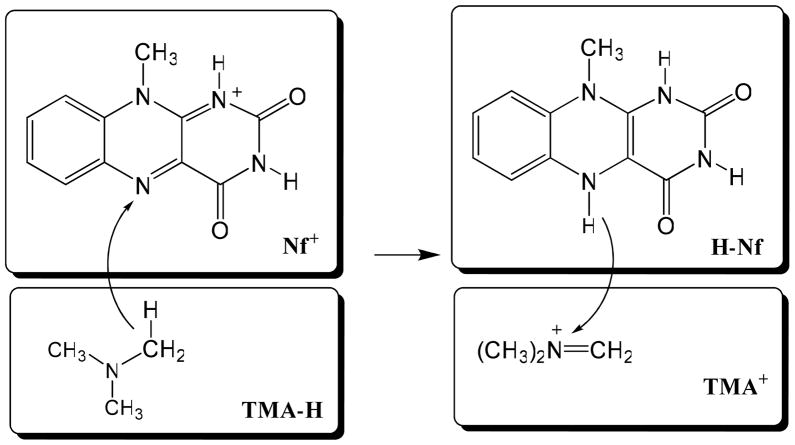
In MOVB,10–12,21 we use one Slater determinant wave function constructed using non-orthogonal block-localized molecular orbitals (BLMO) to define the reactant and product configurations. These electronic configurations are called diabatic states. The use of BLMO was explored by many groups in different applications such as reducing basis set superposition errors in weakly bound complexes,17–19,53–54 and it has been used in other contexts.14–16,55–61 For the hydride transfer reaction between trimethylamine, (CH3)3N (TMA-H), and a flavin cofactor (Nf+) model (hereafter simply called flavin), the wave function of the reactant diabatic state, Ψr(R), (see Scheme 1), is defined by a single Slater determinant wave function in which molecular orbitals are block-localized on the two subsystems:
| (1) |
where R specifies all Cartesian atomic coordinates of the system and  is an antisymmetrization operator. The notations and in eq 1 specify the products of occupied BLMOs that are defined as linear combinations of atomic orbitals located on atoms in fragments TMA-H and Nf+, respectively (Scheme 1). Similarly, the wave function of the product state (Scheme 1), Ψp(R), is expressed as
Scheme 1.
Schematic representation of the block-localization of molecular orbitals within individual molecular fragments for the reactant diabatic state (left) and the product diabatic state (right) for the hydride transfer reaction between trimethylamine (TMA-H) and a model for the flavin cofactor (Nf+). Atoms and charges in each rectangular specify the molecular block defined by the corresponding Lewis structure within which molecular molecular orbitals are localized. The antisymetric wave function constructed from the two blocks on the left-hand side of the arrow, TMA-H and Nf+, defines the reactant diabatic state, whereas that for the right-hand side blocks, TMA+ and H-Nf, define the product diabatic state.
| (2) |
where and denote the products of occupied BLMOs expanded over basis orbitals on atoms in fragments TMA+ and H-Nf, respectively (Scheme 1).
It is important to note that the MOs within each fragment for each state are constrained to be orthogonal, but they are nonorthogonal between different fragments.11 Consequently, the MOVB model retains key characteristic features of valence bond theory in the use of nonorthogonal orbitals. The structure of the transformation matrix for the reactant and product states is block-diagonal.
| (3) |
where Cr and Cp are the matrices of molecular orbital coefficients for the reactant and product state, respectively.11 Note that the dimensions of the matrix elements in eq 3 have different dimensions as the hydride atom is grouped in different blocks. The total number of electrons within each fragment of each diabatic state is also fixed according to the corresponding Lewis structure (Scheme 1), and there is no chemical bond between the two fragments in each state.
In the present EH-MOVB model employing a semiempirical method, a special situation must be considered because of the neglect diatomic differential overlap (NDDO) approximation.62. The two-center one-electron integral between two atoms that are located in different blocks is not included in either blocks, and it must be treated specifically. Note that these integrals are formally ignored in the NDDO approximation, but they are also treated as an exception in standard semiempirical methods because of the importance of these integrals in describing chemical bonding.50 In MOVB, when the two bonding atoms involved in bond formation are in different molecular blocks (fragments), the two-center one-electron integral is not treated by the standard semiempirical model, and the exclusion of this type of resonance integrals affects the bonding properties as the chemical bonds are partially formed and broken across different blocks (fragments) at the transition state. Consequently, we need to include these resonance integrals for the corresponding bonds in the diabatic energy term as follows. For the reactant state in the present hydride transfer reaction, the reactant state diabatic energy is given as follows:
| (4) |
and the energy for the product state is
| (5) |
where SHN and SCH are the s-type overlap integrals between the acceptor nitrogen atom and the transferring hydrogen atom, and between the donor carbon atom and the migrating hydrogen atom, specified by the subscripts respectively, and βsp(X) = [βs(X) + 3βp(X)]/4 at X = N or C and β’ s being the standard semiempirical parameters for these atoms.50–51 The use of s-type overlap integrals in eqs 4 and 5 is to preserve rotation invariance. In eqs 4 and 5, we treat αHN and αCH as semiempirical parameters, adjusted to yield the corresponding bond distances in agreement with DFT energies at the transition state. These two parameters associated with bonding interactions may also be considered as EH-MOVB parameters, in addition to the two parameters to adjust diabatic coupling results.
The MOVB wave function for the reactive system is written as a linear combination of the diabatic states.
| (6) |
where ar and ap are the configurational coefficients for the reactant and product diabatic state, respectively.15–16,20,26 The potential energy of the adiabatic ground state, Vg(R), is the lower energy root of the secular equation.
| (7) |
where V(R) is the adiabatic potential energy, Hrr(R) and Hpp(R) are the Hamiltonian matrix elements for the reactant and product diabatic states, respectively, Hrp(R) = Hpr(R) is the exchange integral (off-diagonal matrix element), and Srp(R) = Spr(R) is the overlap integral between the two diabatic states.
The Hamiltonian matrix elements in eq 7 are given as follows:11,13
| (8) |
where the subscripts a and b specify either the reactant (r) or the product (p) state or both, Enuc is the nuclear Coulomb energy, Sab and Dab are the overlap integral and density matrix over nonorthogonal determinant wave functions, and h, J, and K are the standard one-electron, Coulomb and exchange matrices. It is important to note that eq 8 is a general formula that is valid for ab initio and semiempirical WFT as well as for standard Kohn-Sham DFT.13 In the latter case, the exchange integral K is replaced by the exchange-correlation potential.13
In reference 20, we described two special situations to optimize the wave function of eq 6. In the first case, which is called the consistent diabatic configurations (CDC) MOVB, both the orbital coefficients (eq 3) and configurational coefficients are optimized as in the multiconfiguration self-consistent field method. An alternative approach is to variationally optimize the reactant and product state separately, followed by optimizing the configuraitonal coefficient in eq 6 with the orbital coefficients kept fixed. The latter configuration interaction procedure is called the variational diabatic configuration (VDC) MOVB to emphasize that the diabatic states are individually optimized. Both CDC and VDC states are useful in condensed phase simulations, although their applications will be addressed in future publications.
B. Effective Hamiltonian MOVB
We aim to develop an efficient (e.g., capable of carrying out ns to μs dynamics simulations using the current computer architecture) and accurate (within 1 to 2 kcal/mol of experimental barrier height) QM/MM method for simulation of enzymatic reactions and chemical processes in solution using MOVB. Although ab initio MOVB and multistate VBDFT, provide a natural choice, and the former has indeed been applied to a number of condensed phase reactions,10–12,59,63 it is still very time-demanding to carry out routine free energy simulations, in which a large number of atoms are treated quantum-mechanically. To this end, we have implemented the MOVB method into the CHARMM package,64 based on the NDDO approximations.62,65 The present implementation represents a significant advance in combined QM/MM methodology because (a) semiempirical methods are computationally efficient, allowing for statistical mechanical sampling in molecular dynamics simulations, and (b) the computational accuracy can be conveniently achieved using the nonorthogonal block-localized orbital approach described here.26
Experience shows that the qualitative features of the potential surface for chemical processes can be adequately represented by semiempirical models, such as AM1,50 PM351 or the self-consistent charge tight-bonding density functional algorithm (SCC-DFTB).66 Consequently, we define and describe the reactant and product diabatic states using a semiempirical Hamiltonian. The quantitative errors in the computed barrier height and the energy of reaction inherited in the semiempirical method are eliminated by adjusting the EH-MOVB matrix elements26 in a similar way as that in empirical or semiempirical models.27,30–31,41 It should be realized that all combined QM/MM methods are empirical models in that one has to employ empirical potential functions such as the Lennard-Jones terms to approximate the quantum mechanical exchange repulsion and dispersion interactions between the QM and MM regions. Thus, the adjustment of the EH-MOVB matrix elements in no stranger to combined QM/MM methodologies.
Specifically, we introduce a parameter in the off-diagonal Hamiltonian matrix element Hrp, which is optimized in order to reproduce the barrier height for a given chemical reaction:
| (9) |
In eq 9, Hrp is the MOVB off-diagonal matrix element that is determined directly (eq 8) using a given semiempirical model, γrp is a parameter that affects dominantly the computed barrier height, and is the total effective Hamiltonian (EH) resonance integral. Another formalism that we have explored is to scale the off-diagonal matrix element as follows:26
| (10) |
Both options can be useful, depending on the performance of the semiempirical model and the specific reaction considered, and both are available options in our implementation in CHARMM. In eq 9, the resonance integral is shifted by a constant value, whereas the scaling procedure in eq 10 affects the dependence of the resonance integral on the overlap between the reactant and product diabatic states. For the hydride transfer reaction between trimethylamine and flavin, we found that eq 9 yields the best results and it is employed in the present study.
The second parameter that we introduce in the EH-MOVB model is the adjustment of the relative energy between the reactant and product diabatic states. Thus, if necessary, the diagonal MOVB matrix element for the product state, Hpp, is shifted by an amount of Δε to yield the desired energy of reaction for the process of interest:
| (11) |
The value of the parameter Δε is readily estimated as follows:
| (12) |
where ΔEMOVB = Hpp(Rp) − Hrr(Rr), which is the relative energy of the un-shifted reactant and product diabatic state at their corresponding equilibrium geometries Rr and Rp, and ΔEexpt is the experimental energy of reaction.
The procedure outlined above (eqs 9–12) is identical to that used in the parameter “calibration” of empirical valence bond models, such as that in refs 32 and 41, or more generally, of semiempirical valence bond,27–31 which allows the energies (barrier height and reaction energy) to be readily fitted to their targets exactly. In general, however, it is much more challenging to “calibrate” the variation of molecular structure along the entire reaction path, especially the precise geometry of the transition state. The sophistication of the mathematical algorithm used by Schlegel and coworkers is a remarkable reflection of the difficulty in constructing accurate potential energy surface employing empirical models.45–46 The changes of the structural properties, including bond order and force constant, are critically important if one is interested in computing kinetic isotope effects, particularly the error-sensitive secondary effects (2° KIEs), for enzymatic reactions. Inaccuracy can easily be hidden in the large primary KIEs because they typically involve a significant loss of zero-point effects. Thus, agreement with experiment in primary KIEs, which could be simply due to the loss of the reactant state stretching mode, is not necessarily an indication of good geometry at the transition state. In fact, it is essential to examine both the optimized structure and energy at the transition state to validate the quality of a two-state (or multistate) model against high-level electronic structural data.20,26,45–46
To this end, the off-diagonal matrix element in EH-MOVB (eq 9) is an explicit function of all degrees of freedom of the system, i.e., Hrp(R) = < Ψr(R) | H | Ψp(R) >.10–13,20,26 Consequently, the full-dimensional potential surface can be adequately represented as accurately as the accuracy of the level of the electronic structure method permits, and the transition structure for a reaction can be obtained in accord with that optimized from WFT or DFT calculations. Note that the approach outlined in eqs 9–12 is in principle analogous to that used in effective Hamiltonian valence bond methods to parameterically model the ab initio matrix elements to reproduce the exact high-level results.33–40,42–49
3. Computational Details
All calculations are carried out using CHARMM c34a2,67 modified with the implementation of the present EH-MOVB. The current QM/MM module in CHARMM at the semiempirical level, called SQUANTUM,68 was implemented by Kwangho Nam and Ross Walker in 2004, based on a Fortran90 code.69 SQUANTUM has been incorporated into the standard distribution and has become the default QM/MM module of CHARMM since version c33a1. The EH-MOVB method was implemented by Lingchun Song in collaboration with Wangshen Xie and it has become a part of the SQUANTUM module with additional options to define the number of states and the number of blocks in each state as well as the associated options. The EH-MOVB method can provide a rigorous valence bond-like model for studying chemical reactions such that the users can conveniently calibrate the model to yield the desired barrier height and reaction energy as well as optimized geometry at the transition state.
The EH-MOVB module at the semiempirical level is computationally fast; for large systems, the computational bottleneck using our QM/MM potential is in the treatment of the classical long-range electrostatic effects with particle-mesh Ewald (PME) rather than the QM calculation itself. In addition, two options are available for determining the diabatic and adiabatic ground state energies: (1) the consistent diabatic state (CDC) method and (2) the variational diabatic state (VDC) model.20 For those who are interested in using the energy gap between the product and reactant diabatic state as the reaction coordinate,70 the VDC diabatic states should be used,10–12 which also provides the basis states to yield the adiabatic ground state potential energy surface. On the other hand, if geometrical parameters are used to define the reaction coordinate on the adiabatic ground state potential surface, the CDC model is appropriate since analytical gradients can be computed. Note that the CDC method is analogous to multiconfiguration self-consistent field (MCSCF) theory,20,26 whereas the VDC approach is akin to a configuration interaction (CI) method.10–12
DFT calculations are carried out using Gaussian0371 modified to include the M06-2X functionals.72–73 The 6–31+G(d) basis set is used throughout for all calculations. Geometries for the hydride transfer reaction between trimethylamine and flavin cofactor along the reaction coordinate defined below are optimized using the 6–31+G(d) basis set at each level of theory. The recently developed M06-2X functional are used to calibrate the EH-MOVB model.
To describe the change in energy and wave function of the two Lewis bond states as the reaction takes place, we define the reaction coordinate here as the difference between the bond lengths of the central hydrogen atom that is transferred to the donor atom (C) of TMA-H, and the acceptor atom (N) of Nf+:
| (13) |
Of course, one can use other definitions to monitor the progress of the reaction, including the difference between the corresponding bond orders or energies of the two Lewis bond states. The geometrical variable, corresponding to the asymmetric bond stretch coordinate, is a good choice and of chemical intuition.
4. Results and Discussion
The main goal of this study is to develop an effective Hamiltonian within MOVB theory to study chemical reactions in solution and in enzymes using CHARMM as a combined QM/MM potential. We hope to illustrate that the procedure can be conveniently used by biochemists as a research tool to help interpret experimental findings, with a straightforward calibration of the EH-MOVB model. We use the hydride transfer reaction from trimethylamine to a flavin cofactor model, designed as a chemical model to mimic the catalytic transformation by the enzyme lysine-specific demethylase in the histone tail modification process. The discussion of the chemical and biological simulations is beyond the scope of this report and will be reported separately. We first carry out ab initio electronic structural calculations using DFT to yield the structures and energies along the hydride transfer reaction pathway. Then, we optimize the EH-MOVB Hamiltonian to reproduce the “high-level” data. The qualitative features and quantitative results of the diabatic configurations and the adiabatic potential surface will be discussed.
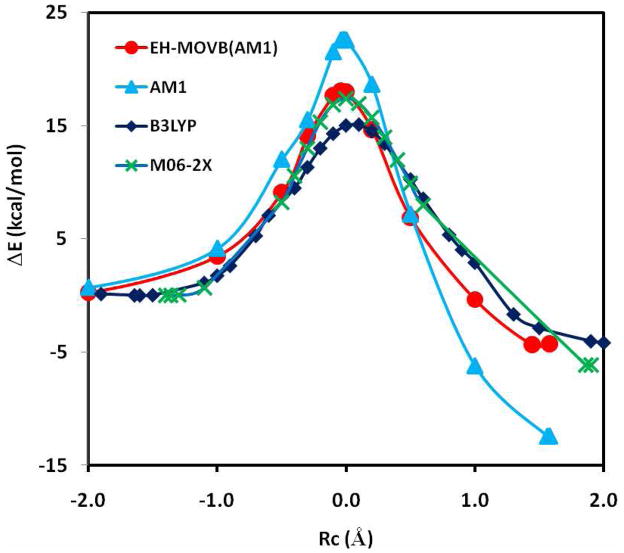
The adiabatic ground state potential energy surfaces determined using DFT with the B3LYP and M06-2X functional are compared with the standard semiempirical AM1 model and the EH-MOVB method in Figure 1 as a function of the reaction coordinate Rc (eq 13) for the hydride transfer reaction between trimethylamine and a flavin cofactor. Optimized structures at the reactant state and product state complex and the transition state are illustrated in Figure 2 along with key structural parameters. The M06-2X density functional calculations yield an estimated barrier height of 17.4 kcal/mol and a relative energy of −6.1 kcal/mol between the product and reactant state. The popular hybrid B3LYP method underestimates the hydride transfer barrier at 15.1 kcal/mol. The semiempirical AM1 energy profile is qualitatively correct, but it contains two main problems; the computed energy of activation is 22.6 kcal/mol, about 5 kcal/mol too high, and the predicted energy of reaction is too endothermic by 6.4 kcal/mol. The latter error is completely transferred into the MOVB relative energies of the reactant and product diabatic states, which can be easily corrected by shifting the product state up by an equal amount (Table 1). With an increase in the strength of diabatic coupling between the reactant and product states at the transition state, the barrier height can be lowered, and using the parameters listed in Table 1, we obtained an activation energy of 18.1 kcal/mol for the hydride transfer between trimethylamine and flavin and an energy of reaction of −4.4 kcal/mol. We note that the AM1 model finds another configuration in which the donor N-C-H unit is roughly coplanar with the flavin ring and it is slightly lower in energy (by about 2 kcal/mol) than the configuration in which TMA is under the plane of the flavin ring. The latter configuration is more closely aligned with the structure found in the active site in the human histone lysine-specific demethylase (LSD1) structure,74 which is most relevant to the hydride transfer reaction pathway.
Figure 1.
Computed potential energy profile along the minimum energy path (Rc = R[C – H ] − R[H – N] ) for the hydride transfer reaction between trimethylamine and flavin model using EH-MOVB(AM1) in red, AM1 in light blue, B3LYP/6-31G(d) in navy blue, and M06-2X/6-31G(d) in green.
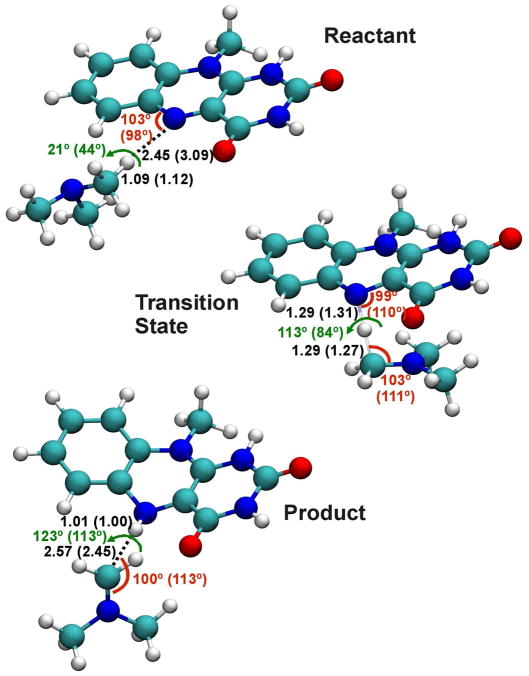
Figure 2.
Optimized geometries for the reactant and product complexes and the transition state for the hydride transfer reaction depicted in Scheme 1.
Table 1.
EH-MOVB parameters used in this study. The AM1 model is used to define the diabatic reactant and product states for the hydride transfer reaction between trimehylamine and a model flavin cofactor.
| αCH | αHN | Δε (kcal/mol) | γrp (eV) |
|---|---|---|---|
| 0.9 | 1.0 | 8.0 | 1.5 |
The optimized geometries at the reactant, product and transition states from EH-MOVB(AM1) calculations are in accord with those obtained using M06-2X density functional theory. In particular, the donor (C-H) and acceptor (H-N) distances from the hydride atom transferred are 1.29 and 1.29 Å, respectively, which may be compared with the M06-2X value of 1.27 and 1.31 Å. The potential energy surface about bond angles and torsional angles is relatively flat, and the accord between EH-MOVB(AM1) and M06-2X is reasonable (Figure 2).
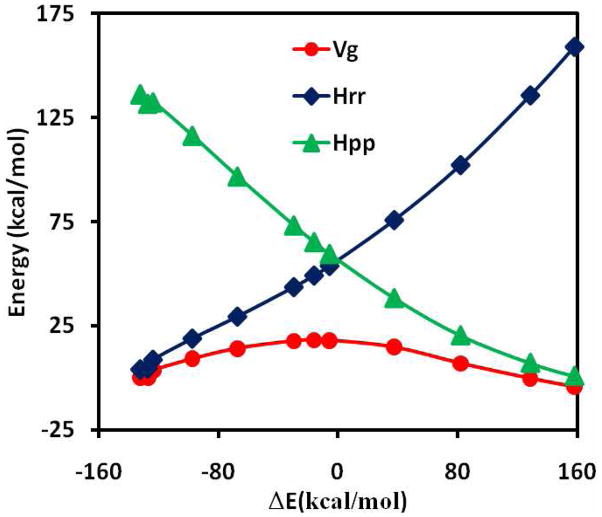
The minimum energy path (MEP) for the hydride transfer from trimethylamine to flavin has been optimized as a function of the reaction coordinate defined by eq 13. In the present study, we have constrained the hydride migration to be collinear with the donor (C) and acceptor (N) atoms, whereas all other degrees of freedom are fully minimized using the ABNR algorithm in CHARMM.64 The potential energy curves for the reactant and product diabatic states are shown in Figure 3 along with that for the adiabatic ground state. The reactant state potential shows a steady increasing as the reaction coordinate changes from the reactant to the product side. On the other hand, the potential energy surface is somewhat leveled off for the product state when the molecular geometry is in the reactant state configuration. The trend of the two diabatic potential energy curves is consistent with heterolytic bond cleavages of the reactant (C-H) and the product (N-H) species. At the diabatic state crossing point, which corresponds roughly to the location of the transition state of the hydride transfer reaction, the diabatic state is ca. 40 kcal/mol in energy above the adiabatic ground state, suggesting that there is significant electronic coupling between the reactant and product states. The coupling energy is similar to values determined for proton transfer and nucleophilic substitution reactions using ab initio WFT and DFT.10–13,20,26
Figure 3.
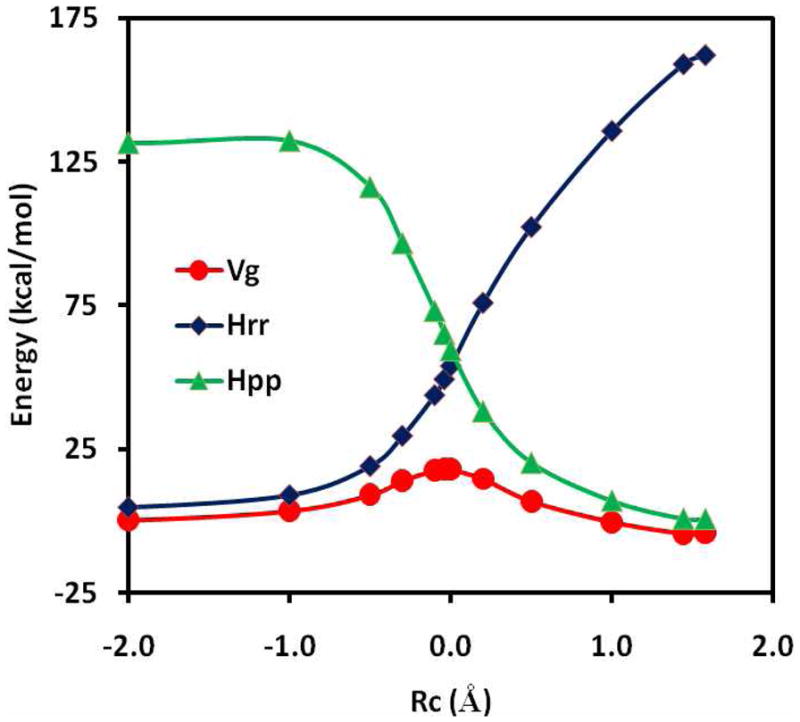
Computed potential energy surfaces for the diabatic reactant state (blue), the diabatic product state (green), and the adiabatic ground state (red) along the minimum energy path.
Figure 4 exhibits the same potential curves shown in Figure 3, but they are plotted against the diabatic energy difference, or the energy-gap reaction coordinate.
Figure 4.
Computed potential energy surfaces in Figure 3 for the diabatic reactant state (blue), the diabatic product state (green), and the adiabatic ground state (red) represented as a function of the energy difference between the reactant and product diabatic states (i.e., the energy-gap reaction coordinate).
| (14) |
Figure 4 shows that the minimum energy potential surface for the adiabatic ground state and those for the diabatic states can be fully represented with the use of either a geometrical or an energy-gap reaction coordinate when the reaction profile is determined by optimizing the geometrical reaction coordinate.
For reactions in solutions or in enzymes, it is of interest to consider the effects of solvent or protein reorganization and this is often presented using the energy-gap reaction coordinate (eq 14). Although this is easily modeled using an empirical force field to represent the diabatic states, it is far from straightforward if a combined QM/MM potential is employed. The MOVB theory is the first and only QM/MM approach at this time to provide well-defined diabatic states for condensed phase simulations, and ab initio MOVB-QM/MM methods have been utilized in the study of solvent effects and reorganization energies for several reactions in solution. Of course, empirical potential functions have been used extensively to describe the energy-gap coordinate.41,75–76 The present EH-MOVB approach in the context of a QM electronic structure theory can be conveniently calibrated to yield accurate results and applied to enzymatic catalysis. The free energy reaction profile as a function of the energy-gap reaction coordinate is typically obtained through a coupled free energy perturbation simulation,10–11,41 which drives the solvent and protein configurations from the reactant state to the product state using a reference potential (which is also called a mapping potential),41VRP(R), and umbrella sampling that transforms the biased simulations with VRP(R) into the true adiabatic ground-state potential surface, Vg(R).
The reference potential is typically expressed as a mixture of the diabatic reactant and product energy through a coupling parameter λ:
| (15) |
where λ is a parameter that varies from 0 (reactant) to 1 (product), and R specifies the instantaneous geometry of the system. In the present study of the model hydride transfer from TMA-H to flavin (Nf+), we optimized the reference minimum-energy path (RMEP) defined by eq 15. Then, using the geometries along this reference minimum-energy path, we carried out single-point energy calculations to determine the adiabatic ground state energy. Note that this “RMEP” is not the true adiabatic ground-state MEP (Figures 3 and 4) determined using the EH-MOVB potential, Vg(R), because the structures are optimized using different potential energy surfaces.
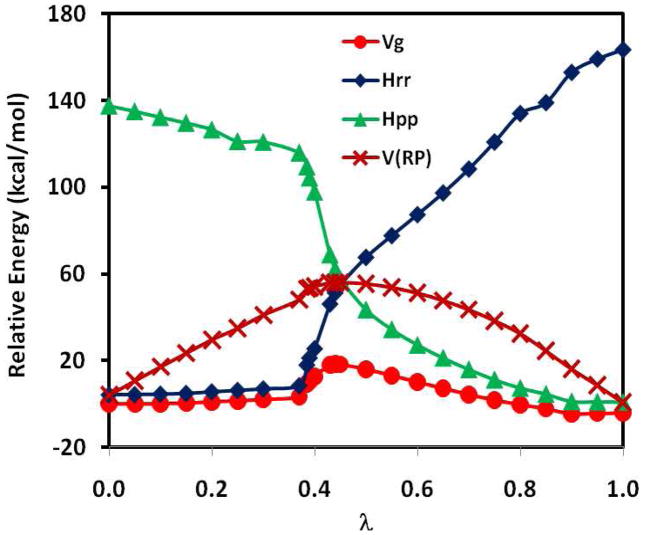
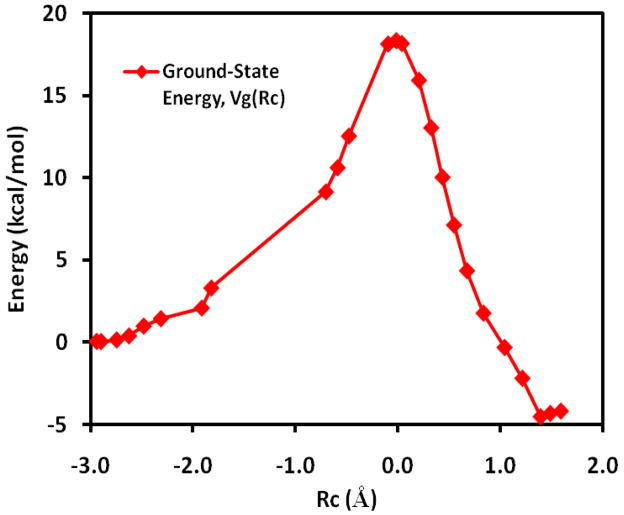
Figure 5 depicts the diabatic potential energies and the adiabatic ground state energy, along with the reference potential (eq 15), as a function of the coupling parameter. Since the reference potential is dominantly determined by the reactant diabatic state when λ is less than 0.5, there is a rapid geometry change in the hydride transfer coordinate, which is not explicitly specified by the coupling parameter λ and cannot be effectively restrained to yield a smooth variation. Consequently, there is a sudden change in the molecular geometry as the hydride is fully transferred to the carbon atom, corresponding to a geometrical description of Rc = −0.7 Å to Rc = −1.8 Å. The ground-state potential is shown as a function of the geometrical reaction coordinate in Figure 6. This is accompanied by a rather steep increase in the reactant diabatic state and the adiabatic ground state potential in the region of λ = 0.4 and 0.5 (Figure 5). Interestingly, the overall reference potential shows smoother variations (curve in maroon) due to the compensating contributions from the product diabatic state. The computed barrier height is 18.3 kcal/mol along the RMEP, similar to that of the MEP for the hydride transfer.
Figure 5.
Computed potential energy profiles for the reactant (blue) and product (green) diabatic states along with the adiabatic ground state (red) and the reference potential as a function of the coupling parameter linearly connecting the reactant and product potentials. This reaction path is called the reference minimum energy path, which has a different meaning from that of Figure 1.
Figure 6.
The potential energy profile for the hydride transfer reaction between methylamine and the model flavin cofactor plotted against the geometrical reaction coordinate (eq 14) following the reference minimum energy path in Figure 5.
Figure 7 recasts the data illustrated in Figure 5, but the adiabatic ground state potential energy surface Vg(R) is plotted against the energy-gap reaction coordinate Δ E. In contrast to Figures 5 and 6, the potential Vg(R) appears to be surprisingly smooth, despite the fact that part of the geometrical variations along the reaction path in fact is discontinuous in Figure 5. Figure 7 shows that a non-smooth geometrical transition that gives rise to an abrupt energy change can be hidden behind the seeming smooth energy curve when the adiabatic ground-state potential is given as a function of a geometry-implicit coordinate such as the energy-gap representation. This observation suggests that it is critically important to report and show both energy results and the corresponding geometries of the reactive molecule in calculations that employ the reference potential of eq 15.
Figure 7.
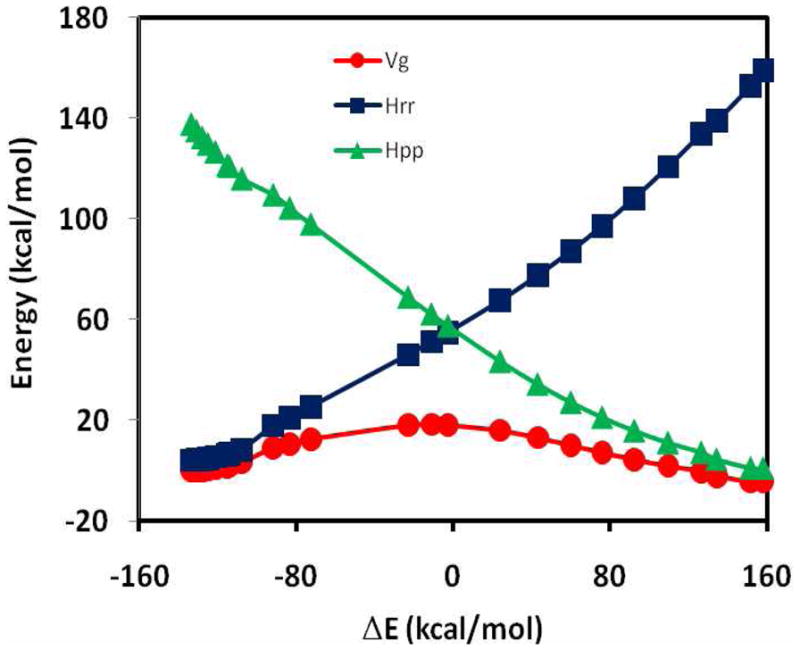
Computed potential energy profiles for the reactant (blue) and product (green) diabatic states and the adiabatic ground state (red) as a function of the energy gap-reaction coordinate for structures obtained along the reference minimum energy path in Figure 5.
Before we leave this section, we consider the procedure used in condensed phase and enzyme calculations.10–11,41 In this case, the reference potential of eq 15 will be used in a series of discrete free energy perturbation simulations with fixed values of λi to yield the free energy differences as λi changes from 0 to 1. Thus, the free energy at λ relative to the reactant state (λ0 = 0) is determined as follows:
| (16) |
where <···>i specifies an ensemble average over the potential VRP(λi), the summation runs to a value λ = λi+1. Here, the use of the arbitrary reference potential is purely for the purpose of moving the system to go from molecular configurations corresponding to the reactant state ensemble into the product state. To obtain the free energy of the true ground state potential surface, governed by the distribution e−Vg(Δ;E)/RT, an umbrella sampling-like procedure is applied to the configurations sampled on the basis of the distribution of e−VRP(λi)/RT. Thus,
| (17) |
where the quantity is the normalized distribution of configurations that have a value of ΔE in the ensemble sampled by the reference potential VRP(λi).
An important distinction should be made is that the procedure outlined in eqs 16 and 17 yields the free energy profile, or the potential of mean force, as a function of an ensemble of configurations, all having the energy-gap ΔE. Obviously, it is not and should not be compared with the potential energy surface. Furthermore, the “reaction path” mapped by eq 16 is not the minimum energy path of the adiabatic ground state, nor the reference minimum energy path. Thus, the energy computed, either by averaging over all configurations sampled on the basis of eq 16 or by selecting a single structure of its ensemble is not directly comparable to results rigorously defined by the MEP.
5. Conclusions
The effective Hamiltonian-molecular orbital and valence bond (EH-MOVB) method based on nonorthogonal block-localized molecular orbitals has been implemented into the program CHARMM for molecular dynamics simulations of chemical and enzymatic reactions, making use of semiempirical quantum mechanical methods. Building upon previous results using ab initio MOVB theory, we introduce two parameters in the EH-MOVB method, along with the addition of the two-center one-electron integrals across different molecular blocks which may be considered as parameters, such that the barrier height and the relative energy between the reactant and product state for a given chemical reaction can be fitted in good agreement with experiment or high-level ab initio and DFT results. The EH-MOVB method provides a highly accurate and computationally efficient QM/MM model for dynamics simulation of chemical reactions in solution. The MOVB theory is the first and only QM/MM method that allows the potential of mean force to be mapped into an energy-gap reaction coordinate for characterization of solvent reorganization effects.
The EH-MOVB method is illustrated by examination of the potential energy surface of the hydride transfer reaction from trimethylamine to a flavin cofactor model in the gas phase. In the present study, we employ the semiempirical AM1 model, which yields a qualitatively correct energy profile along the minimum energy path (Figure 1). However, as in most practical applications using semiempirical Hamiltonians, the quantitative results are not satisfactory. Traditionally, there is no systematic way of improving the semiempirical model, even though the qualitative features of structure and energy are reasonable. In EH-MOVB, the barrier height is optimized to reproduce the desired (accurate) value either by scaling or by adding a constant to the off-diagonal matrix element. The present EH-MOVB method offers an alternative approach to characterization of solvent and protein-reorganization effects in the realm of truly combined QM/MM simulations.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM46736) for support of this work. AP is a recipient of the Thailand Research Fund, under the Royal Golden Jublee Ph.D. Graduate Program (PHD/0211/2547).
References
- 1.Senn HM, Thiel W. Angew Chem, Int Ed. 2009;48:1198. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 2.Gao J, Xia X. Science. 1992;258:631. doi: 10.1126/science.1411573. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Ma S, Major DT, Nam K, Pu J, Truhlar DG. Chem Rev. 2006;106:3188. doi: 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandrasekhar J, Smith SF, Jorgensen WL. J Am Chem Soc. 1985;107:154. [Google Scholar]
- 5.Gao J. J Am Chem Soc. 1991;113:7796. [Google Scholar]
- 6.Rossi I, Truhlar DG. Chem Phys Lett. 1995;233:231. [Google Scholar]
- 7.Marti S, Moliner V, Tunon I. J Chem Theory Comput. 2005;1:1008. doi: 10.1021/ct058001a. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Viloca M, Truhlar DG, Gao J. Biochemistry. 2003;42:13558. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 9.Nam K, Cui Q, Gao J, York DM. J Chem Theory Comput. 2007;3:486. doi: 10.1021/ct6002466. [DOI] [PubMed] [Google Scholar]
- 10.Mo Y, Gao J. J Comput Chem. 2000;21:1458. [Google Scholar]
- 11.Mo Y, Gao J. J Phys Chem A. 2000;104:3012. [Google Scholar]
- 12.Gao J, Garcia-Viloca M, Poulsen TD, Mo Y. Adv Phys Org Chem. 2003;38:161. [Google Scholar]
- 13.Cembran A, Song L, Mo Y, Gao J. J Chem Theory Comput. 2009;5:2702. doi: 10.1021/ct9002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo Y, Peyerimhoff SD. J Chem Phys. 1998;109:1687. [Google Scholar]
- 15.Mo Y, Zhang Y, Gao J. J Am Chem Soc. 1999;121:5737. [Google Scholar]
- 16.Mo Y, Gao J, Peyerimhoff SD. J Chem Phys. 2000;112:5530. [Google Scholar]
- 17.Stoll H, Preuss H. Theor Chem Acc. 1977;46:12. [Google Scholar]
- 18.Gianinetti E, Raimondi M, Tornaghi E. Int J Quantum Chem. 1996;60:157. [Google Scholar]
- 19.Gianinetti E, Vandoni I, Famulari A, Raimondi M. Adv Quantum Chem. 1998;31:251. [Google Scholar]
- 20.Song L, Gao J. J Phys Chem A. 2008;112:12925. doi: 10.1021/jp803050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Mo Y. Prog Theor Chem Phys. 2000;5:247. [Google Scholar]
- 22.Cooper DL, Gerratt J, Raimondi M. Adv Chem Phys. 1987;69:319. [Google Scholar]
- 23.Hiberty PC, Flament JP, Noizet E. Chem Phys Lett. 1992;189:259. [Google Scholar]
- 24.Wu W, Song L, Cao Z, Zhang Q, Shaik S. J Phys Chem A. 2002;106:2721. [Google Scholar]
- 25.Song L, Mo Y, Zhang Q, Wu W. J Comput Chem. 2005;26:514. doi: 10.1002/jcc.20187. [DOI] [PubMed] [Google Scholar]
- 26.Song L, Mo Y, Gao J. J Chem Theory Comput. 2009;5:174. doi: 10.1021/ct800421y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato S. J Chem Phys. 1955;23:592. [Google Scholar]
- 28.Kuntz PJ, Nemeth EM, Polanyi JC, Rosner SD, Young CE. J Chem Phys. 1966;44:1168. [Google Scholar]
- 29.Raff LM, Stivers L, Porter RN, Thompson DL, Sims LH. J Chem Phys. 1970;52:3449. [Google Scholar]
- 30.Raff LM. J Chem Phys. 1974;60:2220. [Google Scholar]
- 31.Silver DM, Brown NJ. J Chem Phys. 1980;72:3859. [Google Scholar]
- 32.Warshel A, Weiss RM. J Am Chem Soc. 1980;102:6218. [Google Scholar]
- 33.Sheppard MG, Freed KF. J Chem Phys. 1981;75:4507. [Google Scholar]
- 34.Hurtubise V, Freed KF. Adv Chem Phys. 1993;83:465. [Google Scholar]
- 35.Martin CH, Graham RL, Freed KF. J Phys Chem. 1994;98:3467. [Google Scholar]
- 36.Bernardi F, Olivucci M, Robb MA. J Am Chem Soc. 1992;114:1606. doi: 10.1021/ja00226a011. [DOI] [PubMed] [Google Scholar]
- 37.Bearpark MJ, Robb MA, Bernardi F, Olivucci M. Chem Phys Lett. 1994;217:513. [Google Scholar]
- 38.Bearpark MJ, Bernardi F, Olivucci M, Robb MA. J Phys Chem A. 1997;101:8395. [Google Scholar]
- 39.Bearpark MJ, Smith BR, Bernardi F, Olivucci M, Robb MA. ACS Symp Ser. 1998;712:148. [Google Scholar]
- 40.Chang YT, Miller WH. J Phys Chem. 1990;94:5884. [Google Scholar]
- 41.Aqvist J, Warshel A. Chem Rev. 1993;93:2523. [Google Scholar]
- 42.Schmitt UW, Voth GA. J Phys Chem B. 1998;102:5547. [Google Scholar]
- 43.Day TJF, Soudackov AV, Cuma M, Schmitt UW, Voth GA. J Chem Phys. 2002;117:5839. [Google Scholar]
- 44.Maupin CM, Wong KF, Soudackov AV, Kim S, Voth GA. J Phys Chem A. 2006;110:631. doi: 10.1021/jp053596r. [DOI] [PubMed] [Google Scholar]
- 45.Schlegel HB, Sonnenberg JL. J Chem Theory Comput. 2006;2:905. doi: 10.1021/ct600084p. [DOI] [PubMed] [Google Scholar]
- 46.Sonnenberg JL, Schlegel HB. Mol Phys. 2007;105:2719. [Google Scholar]
- 47.Kim Y, Corchado JC, Villa J, Xing J, Truhlar DG. J Chem Phys. 2000;112:2718. [Google Scholar]
- 48.Tishchenko O, Truhlar DG. J Phys Chem A. 2006;110:13530. doi: 10.1021/jp0640833. [DOI] [PubMed] [Google Scholar]
- 49.Lin H, Zhao Y, Tishchenko O, Truhlar DG. J Chem Theory Comput. 2006;2:1237. doi: 10.1021/ct600171u. [DOI] [PubMed] [Google Scholar]
- 50.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. J Am Chem Soc. 1985;107:3902. [Google Scholar]
- 51.Stewart JJP. J Comp Chem. 1989;10:209. [Google Scholar]
- 52.Rocha GB, Freire RO, Simas AM, Stewart JJP. J Comput Chem. 2006;27:1101. doi: 10.1002/jcc.20425. [DOI] [PubMed] [Google Scholar]
- 53.Stoll H, Wagenblast G, Preuss H. Theor Chim Acta. 1980;57:169. [Google Scholar]
- 54.Raimondi M, Famulari A, Specchio R, Sironi M, Moroni F, Gianinetti E. Theochem. 2001;573:25. [Google Scholar]
- 55.Mo Y, Gao J. J Phys Chem A. 2001;105:6530. [Google Scholar]
- 56.Mo Y, Subramanian G, Gao J, Ferguson DM. J Am Chem Soc. 2002;124:4832. doi: 10.1021/ja0174433. [DOI] [PubMed] [Google Scholar]
- 57.Mo Y, Schleyer PvR, Wu W, Lin M, Zhang Q, Gao J. J Phys Chem A. 2003;107:10011. [Google Scholar]
- 58.Mo Y, Wu W, Song L, Lin M, Zhang Q, Gao J. Angew Chem, Int Ed. 2004;43:1986. doi: 10.1002/anie.200352931. [DOI] [PubMed] [Google Scholar]
- 59.Mo Y, Gao J. J Phys Chem B. 2006;110:2976. doi: 10.1021/jp057017u. [DOI] [PubMed] [Google Scholar]
- 60.Khaliullin RZ, Head-Gordon M, Bell AT. J Chem Phys. 2006;124:204105/1. doi: 10.1063/1.2191500. [DOI] [PubMed] [Google Scholar]
- 61.Mo Y, Gao J. Acc Chem Res. 2007;40:113. doi: 10.1021/ar068073w. [DOI] [PubMed] [Google Scholar]
- 62.Pople JA, Santry DP, Segal GA. J Chem Phys. 1965;43:S129. [Google Scholar]
- 63.Mo Y-r, Alhambra C, Gao J-l. Huaxue Xuebao. 2000;58:1504. [Google Scholar]
- 64.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187. [Google Scholar]
- 65.Pople JA, Segal GA. J Chem Phys. 1965;43:S136. [Google Scholar]
- 66.Elstner M, Porezag D, Juugnickel G, Elsner J, Haugk M, Frauenheim T, Sukai S, Seifect G. Phys Rev B. 1998;58:7260. [Google Scholar]
- 67.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nam K, Prat-Resina X, Garcia-Viloca M, Devi-Kesavan LS, Gao J. J Am Chem Soc. 2004;126:1369. doi: 10.1021/ja039093l. [DOI] [PubMed] [Google Scholar]
- 69.Walker RC, Crowley MF, Case DA. J Comput Chem. 2008;29:1019. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 70.Marcus RA. Angew Chem, Int Ed Engl. 1993;32:1111. [Google Scholar]
- 71.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian03, Revision D01. Gaussian, Inc; 2004. [Google Scholar]
- 72.Zhao Y, Truhlar DG. J Chem Theory Comput. 2006;2:1009. doi: 10.1021/ct060044j. [DOI] [PubMed] [Google Scholar]
- 73.Zheng J, Zhao Y, Truhlar DG. J Phys Chem A. 2007;111:4632. doi: 10.1021/jp070252n. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Proc Nat Acad Sc USA. 2006;103:13956. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Billeter SR, Webb SP, Agarwal PK, Iordanov T, Hammes-Schiffer S. J Am Chem Soc. 2001;123:11262. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- 76.Hatcher E, Soudackov AV, Hammes-Schiffer S. J Am Chem Soc. 2007;129:187. doi: 10.1021/ja0667211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.