Summary
Cellular therapies that either use modifications of a patient's own cells or allogeneic cell lines are becoming in vogue. Besides the technical issues of optimal isolation, cultivation and modification, quality control of the generated cellular products are increasingly being considered to be more important. This is not only relevant for the cell's therapeutic application but also for cell science in general. Recent changes in editorial policies of respected journals, which now require proof of authenticity when cell lines are used, demonstrate that the subject of the present paper is not a virtual problem at all. In this article we provide 2 examples of contaminated cell lines followed by a review of the recent developments used to verify cell lines, stem cells and modifications of autologous cells. With relative simple techniques one can now prove the authenticity and the quality of the cellular material of interest and therefore improve the scientific basis for the development of cells for therapeutic applications. The future of advanced cellular therapies will require production and characterization of cells under GMP and GLP conditions, which include proof of identity, safety and functionality and absence of contamination.
Key Words: Primary human cells, Cross-contamination, Cell identity, DNA typing, Karyotype
Zusammenfassung
Zelltherapien, die auf patienteneigenen Zellen oder allogenen Zellen basieren, werden zunehmend attraktiv und realisierbar. Neben technischen Fragen nach der optimalen Isolation und geeigneten Kultivierungsund Modifikationsverfahren tritt dabei auch die Bedeutung der Qualitätskontrolle für das Zellprodukt zunehmend ins Bewusstsein. Diese Entwicklung ist nicht nur bei Zellen für die therapeutische Anwendung wichtig, sondern auch für die Zellbiologie als Wissenschaft im Allgemeinen. Dass dies nicht nur ein virtuelles Problem ist, wird schon dadurch deutlich, dass angesehene Zeitschriften ihre Veröffentlichungspolitik geändert haben und nun einen Nachweis der Authentizität der verwendeten Zellen als Voraussetzung für die Veröffentlichung fordern. In diesem Beitrag stellen wir zwei Beispiele für die Kontamination von Zelllinien vor, gefolgt von einem Überblick über neuere Entwicklungen für die Verifikation von Zelllinien, Stammzellen und Zellmodifikationen. Mit relativ einfachen Techniken ist es heute möglich, die Authentizität der Zellen nachzuweisen, die Qualität des Zellmaterials zu beschreiben und damit die wissenschaftliche Basis für die Entwicklung von Zellen für die Therapie zu verbessern. Die Zukunft der sogenannten «Advanced Cellular Therapies» wird die Einhaltung von GMP- und GLP-Bedingungen erfordern. Dies schließt den Nachweis der Identität, Sicherheit und Funktionalität und der Freiheit von Kontamination für das Zellprodukt ein.
Introduction
Over the past decade, cell-based therapies have become a treatment option for a variety of disorders. In contrast to well-established cell-based therapies in transfusion medicine and organ transplantation, the new advanced therapies are based on prolonged cultivation and ex vivo manipulation, e.g. differentiation and genetic modification [1, 2]. Our improved knowledge of cellular functioning and cultivation of cells are key elements which have made these cellular therapies possible. Our understanding into the plasticity of precursor cells allows us to differentiate, mature or manipulate target cells ex vivo. However, the extended cultivation time and manipulation enhance the risk for mutation and contamination which can potentially lead to loss of function, infection-associated problems and even tumorigenesis. For transformation of research-based cell cultures and processes into cGMP procedures, which are required for therapeutic application, a number of hurdles have to be overcome. A strict standardization of the production is one of the most fundamental parameters [3,4,5]. Minimizing risks of contamination, especially microbial contamination during the cultivation process, is an important prerequisite for clinical application. Furthermore, the other chief and chronic problem with cellular products is the intra- or interspecies contamination with other cells.
Both issues of contamination and misidentification of eukaryotic cell lines have been documented from the 1950s where it is estimated that between 18–36% of all established cell lines are affected [6,7,8]. The situation of misidentification of cell lines has been recently described in an editorial in Nature as ‘identity crisis’. This wrong classification of cells is basis of erroneous results/interpretation in many scientific publications where it is very difficult to correct for. One example is the endothelial cell line ECV-304 [9] which was shown to be cross-contaminated with the T24 bladder carcinoma line [10]. Even after this disclosure, more than 500 publications based on this cell line can be accessed (W. Dirks, personal communication).
The following two examples presented in this review demonstrate how misinterpretation can be generated from cell lines which were not ensured for their authenticity.
Examples
Example 1: Generation of Human Neurons from Blood-Derived Precursor Cells
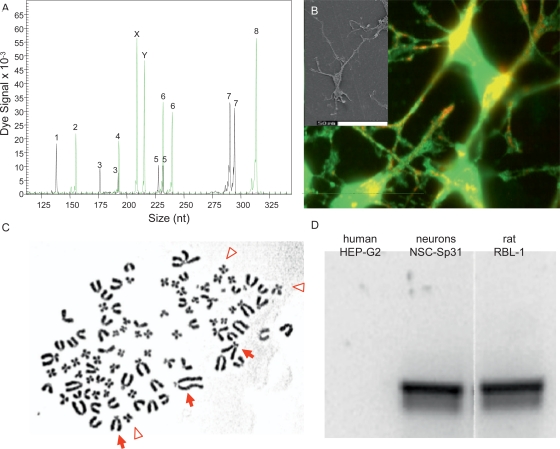
During a collaborative project, one institute received a human cell line from another institute and was able to generate neurons using a specific isolation and cultivation protocol. Two students worked on this project investing more than 2 years of dedicated time with the aim to present their work as part of their dissertations. Initial results looked promising, and a number of manuscripts were being prepared for publication. However, prior to submission one of the group leaders requested for the authenticity of the cell line and asked us to document the human origin of the cell line by performing species specific mitochondrial DNA (mtDNA) analysis. To everyone's astonishment the mtDNA analysis showed that the cell line was not of human origin but in fact originated from the rat. Further genetic analyses confirmed this to be the origin of the cell line and thus reaffirmed the mtDNA analysis; however, the primary isolates consisted of human cells as expected (fig. 1). A possible explanation for this phenomenon is simultaneous cultivation of different cell lines with the potential of contamination. If a certain cell line is not affected by culture conditions, it will overgrow and cause extinction to a cell line which is more sensitive. So although their manuscripts were yet to be submitted for publication, one can only imagine the personal devastation and disappointment of the students and project leaders in respect to both their dissertations and reputation. However, this situation could have been prevented by an initial verification of the cell line.
Fig. 1.
Analysis of ‘human’ immature neuronal cells. A The short tandem repeat (STR) patterns of uncultivated male human donor cells isolated by positive magnetic cell sorting. STR loci tested were D5S818 (1), vWA (2), D13S317 (3), THO1 (4), D7S820 (5), TPOX (6), D16S539 (7), CSF1 (8) and amelogenin (X and Y) [37]. A unique human STR pattern without indication of cross-contamination was obtained. B The cultivated cells reacted with antibodies against neuron specific proteins, anti-human bassoon, a large multidomain protein in the presynaptic active zone (green fluorescence) and anti-human CaC, a voltage-gated Ca2+ channel protein (P/Q-type, [.alpha]-1A subunit) – red fluorescence). The antibodies were known to cross-react to other mammalian species such as rats. Insert: scanning electron microscopy of cultured cells. C Chromosome analysis of the cultured cells revealed that the cells are not of human origin. The large acrocentric chromosomes (arrows) and the small metacentric chromosomes (arrowheads) suggest a rat karyotype [67], which D was confirmed by PCR analysis of the cultured neurons.
Example 2: Stem-1 Equals SAOS 2
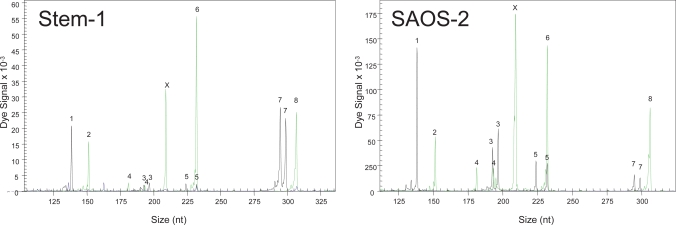
Alarmed by the first example, several cell lines were assessed for their authenticity. The first to be examined was Stem-1, donated from another collaborating institute as human mesenchymal stem cell (MSC) line able to produce differentiated cells depending on the differentiation cocktail. The specific Stem-1 cell line was in culture for more than 1 year before its authenticity analysis. No papers had yet been submitted or published using this specific cell line. Genetic profiling demonstrated that the Stem-1 line as well as the earliest samples available had an identical profile with SAOS-2 (ACC No. 243 DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; German Collection of Microorganisms and Cell Cultures) database) (fig. 2, table 1). This cell line was established in 1973 from primary osteosarcoma tissue from an 11-year-old Caucasian female.
Fig. 2.
Bone marrow-derived Stem-1 cell line identified as SAOS-2 cells (osteosarcoma cell line). Comparison of STR analysis with a cell line database (DSMZ) indicated that the STR pattern of Stem1 is identical to the pattern of SAOS-2 (see table 1). The same STR loci as in figure 1 were used. Further analysis of different passages and samples confirmed the mix-up/contamination (data not shown).
Table 1.
Summary of STR-analysis of mesenchymal cellsa
| Cell line | STR alíele |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D5S818 | D5S818' | D13S317 | D13S317' | D7S820 | D7S820' | D16S539 | D16S539' | vWA | vWA' | TH01 | TH01' | TPOX | TPOX' | CSF1 | CSF1' | Amel | Amel' | M | R | CH | SH | |
| MSC-G3 | 11 | 12 | 8 | 12 | 9 | 11 | 11 | 12 | 18 | 19 | 6 | 10 | 8 | 11 | 10 | 10 | X | Y | − | − | − | − |
| M7 | 11 | 11 | 12 | 13 | 9 | 12 | 9 | 11 | 16 | 18 | 9 | 9 | 11 | 12 | 9 | 11 | X | X | − | − | − | − |
| M10 | 11 | 12 | 8 | 11 | 12 | 12 | 11 | 11 | 16 | 17 | 7 | 10 | 8 | 8 | 11 | 12 | X | Y | − | − | − | − |
| Stem1 | 12 | 12 | 12 | 13 | 8 | 10 | 12 | 13 | 18 | 18 | 6 | 9 | 8 | 8 | 10 | 10 | X | X | − | − | − | − |
| SAOS-2 | 12 | 12 | 12 | 13 | 8 | 10 | 12 | 13 | 18 | 18 | 6 | 9 | 8 | 8 | 10 | 10 | X | X | − | − | − | − |
| HELA + animal | 11 | 12 | 12 | 14 | 9 | 12 | 9 | 10 | 16 | 18 | 7 | 7 | 8 | 12 | 9 | 10 | X | X | + | + | + | + |
MSC-G3, MSC-M7 and MSC-M10 cells (derived from human bone according to published methods [68]) and cell lines Stem-1 and SAOS-2 were analyzed with the STR panel indicated. The control reaction contains DNA of HELA (human), NIH3T3 (M: mouse), RGE (R: rat), CHO-K1 (CH: Chinese hamster) and BHK-21 (SH: Syrian hamster). DNA of MSC-G3, Stem1, MSC-M7 und MSC-M10 showed no cross-contamination with DNA derived from mouse, rat, Chinese or Syrian hamster. Designation of the STR loci used is given in row 1, numbers for both alleles are listed.
Possible Solutions and Discussion
Recent Approaches to Verify Cellular Material
The most important obstacle for the culture of cells is contamination caused from: i) cross-contamination with other cell lines; ii) chromosomal stability and/or ii) contamination with microorganisms (particularly Mycoplasma spp. in 20–30% of cases) [4, 5]. The cross-contamination problem does not appear to be sufficiently recognized even though it is estimated that more than one third of all cell cultures in use are cross-contaminated with either inter- or intraspecies [8, 11,12,13].
Traditional methods used for cell line authentication were: i) isoenzyme analysis which detects interspecies contamination of at least 10% [14], ii) DNA fingerprinting based on restriction fragment length polymorphism (RFLP), generating the specific banding pattern of each individual's DNA (exclusion rates of 99% or higher) [15], and iii) cytogenetic analysis (karyotyping), identifying chromosomal alterations. While karyotyping is currently considered to be the most sensitive method available, it is on the other hand labor-intensive, time-consuming and a rather expensive procedure [15]. These techniques are now supplemented or replaced by simpler, more informative and less time-consuming DNA-based approaches and are further extended by ‘omics’-technologies which allow describing the state of cell population more generally.
Species-Specific Mitochondrial DNA Analysis
mtDNA is present in every cell in multiple copies and has a low sequence complexity thus representing a simple and sensitive target for genetic analysis. First described as a quality control analytical method for gourmet meats [16], the possibility to analyze the sequence of the mitochondrially encoded gene for cytochrome b is regarded a relative simple measure for quality control. The stability of mtDNA is so good that these investigations could be extended to highly processed meat (e.g. cooked and canned tuna species in commercial preparations) [17]. Besides interspecies analysis, mtDNA analysis has also been applied in several biomedical investigations into human background. For instance, it has been used in studies tracing the origin of mankind [18,19,20] and certain human populations [21,22,23]. mtDNA analysis has also been extremely effective in forensic science for both the identification of felons and in identifying victims of crime, accidents or war [24,25,26]. Previously, mtDNA polymorphisms were applied in transfusion medicine and transplantation immunology to distinguish blood donors even in a relatively homogenous Caucasian population [27, 28], focusing on the hypervariable region of the control D-loop of the human mtDNA. This non-coding D-loop region contains regulatory elements for replication and transcription [29] and has high variability based on its high mutation rate (7–10 times faster) compared with genomic (nuclear) DNA because of the limited availability of repair mechanisms [30,31,32,33]. It has been shown [34, 35] that by comparing the mtDNA sequences of the hypervariable regions with the reference sequence it is possible to define cell line-specific polymorphisms. The highest degree of resolution of unique genotypes for the cell lines could be achieved from the sequence information from hypervariable region 1 (HRV1) or the combined sequence information from HRV2 and HRV3. Both approaches reliably discriminate cell lines by their specific polymorphisms of these regions. Although these regions are known to be highly variable, the cell line-specific polymorphisms remained unaltered after a high number of passages (75–80) of cell cultures (data not shown). We therefore conclude that sequence information from the hypervariable regions of the D-loop of the human mitochondrial DNA can be used for the discrimination and identification of human cell lines [36]. This simple, less labor-intensive method may enable the establishment of an in silico library of characterized polymorphisms as an additional method for the authentication of cell lines and the detection of cross-contamination.
By applying mitochondrial PCR analysis on 30 different human cell lines amplifying three hypervariable segments of the control region of mtDNA – HRV1, HRV2 and HRV3 – using previously published primers [28], we could determine mtDNA polymorphisms specific for each cell line [36].
We could also demonstrate that PCR with human-specific primers for HRV1, HRV2 and HRV3 and primers specific for the mouse mtDNA D-loop region reliably discriminated between human and mouse cell lines. With modern techniques like SNaPshot™ techniques or other automated SNP detection methods it will be possible to detect very low intraspecies contamination based on these polymorphisms. With increased application and the definition of polymorphisms in the HRV regions of the mtDNA of even more cell lines, this method will prove to be a simple and efficient approach for the authentication of cell lines and identification of cross-contamination.
Short Tandem Repeat Analysis
A more informative and complex technique for authentication of human cell lines is based on short tandem repeat (STR) DNA typing. The multi allelic variation of the sequences is so great that combination of a few loci allows distinguishing individuals only with the exception of identical twins. STR analysis is therefore becoming the principal method for DNA analyses in forensic cases.
Masters et al. [37] and Dirks et al. [38] have established STR-based typing systems for cell lines which exclude accidental identity with a probability of less than 1.14 × 108. It represents a robust and quick method for the generation of DNA profiles. In less than 8 h the uniqueness of the DNA profile can be examined and due to the international standardization comparisons to the cell data bases of the DSMZ, the American Type Culture Collection (ATCC) and the Japanese Colletion of Research Bioresources (JCRB) can easily be performed [37]. The STR method has been recommended as a possible solution to the identity crisis [11].
The test panel also includes primers for the detection of interspecies contamination. For this purpose, sequences from mouse, rat, Syrian and Chinese hamster – the species of the biotechnologically most relevant cell lines – are included. Interspecies contamination can be detected at a frequency of 1 contaminating genome in 104−105 target genomes.
In the examples described at the beginning of this review, STR analysis identified the starting material as human and unique but identified the deduced neuronal cells as rat-derived cells (fig. 1). In contrast, all available samples of the Stem-1 cell line had an STR pattern identical to the established SAOS-2 cell line (fig. 2, table 1). The identification of Stem-1 as osteogenic tumor cell line SAOS-2 was also supported by additional observations which discriminated it from the other MSC lines: faster growth, restricted (osteogenic but not adipogenic) differentiation potential and an expression profile with up-regulation of genes involved in cell growth and osteogenesis. However, the suspicion was finally confirmed by STR DNA profiling. Additional bone- and bone marrow-derived MSC (G3, M7, M10) had unique human STR pattern not present in the STR databases (table 1). The results of the X and Y chromosome-specific amelogenin probe corresponded to the gender of the donors. No cross-contamination with mouse, rat or hamster cells was detected in the mesenchymal cells.
Karyotyping
Karyotyping allows analyzing the total chromosomal content of individual cells. Specific numbers and forms of the chromosomes give information on species, gender and chromosomal integrity. Sophisticated isolation and staining techniques allow the identification of up to 900 bands, resulting in an optimal resolution of about 3 megabases for human cells [39].
Karyotypic analysis therefore allows determining aneuploidy (loss and gain of chromosomes), which is frequently observed in established cell lines and in genetic diseases, but also smaller and more subtle chromosomal anomalies like deletions, duplications, inversions and translocations. Loss of heterozygosity is also a hallmark of oncogenic transformation.
Karyotyping therefore is often used to analyze the genetic stability of stem cells [40,41,42,43,44,45]. It is regarded as a necessary control for the genetic integrity of stem cells by the International Stem Cell Initiation [46]. The resolution of GTG banding in stem cells karyotypes can be reduced [41, 47] which decreased the significance of this type of analysis. Resolution on the genomic scale can be improved by spectral karyotyping [48], a molecular cytogenetic technique which stains each chromosome with a different color. Therefore, smaller rear-rangements and translocations can be visualized. An even higher resolution of genetic gain or loss can be achieved by comparative genome hybridization (CGH), especially array CGH. However, only significant changes in the population but not at the individual cell level can be determined with this technology [49, 50].
In general, karyotyping is the method of choice for the analysis of genetic stability down to the single cell level. However, karyotyping fluorescence in situ hybridization (FISH), CGH and spectral karyotyping, are time-consuming, costly and require experienced skills for optimal results.
Microsatellite Instability and Chromosome-Specific PCR
For the analysis of specific problems of chromosomal instability technically simple PCR-based approaches have been developed. Instability of microsatellites is a consequence of defects in the DNA mismatch repair system [51]. Analysis of microsatellite instability (MSI) is an important tool for diagnosis and risk management of patients with Lynch syndromes and hereditary nonpolyposis colorectal cancer [52]. For diagnosis, at least 10% of the tissue has to be affected. Therefore, demanding isolation procedures are required for sensitive diagnosis. Defects in the DNA mismatch repair system could also be involved in chromosomal instability of cell lines, but in the cases tested (M10, Stem-1) no changes of markers used for MSI detection could be observed. The technique of chromosome-specific PCR of microsatellites on chromosome 13, 16, 18, 21 and X, which is used for diagnosis of aborts, allows to identify loss or gain of the specified chromosomal regions and identified a chromosome 18 monosomy in Stem-1 (SAOS-2) (data not shown).
Stability of Genotype and Phenotype
Stability and integrity of cell cultures is not only threatened by mix-ups and contamination in cell culture laboratories. Isolation and cultivation conditions actually may select alterations of cell phenotypes and genotypes. In general, cells are deprived from their normal context, and the requirements for maintenance of a specific phenotype are most probably only met partially by current cultivation techniques for most cell types. In addition, expansion and over-passaging of cells can select for cell variants with altered growth characteristics. Standardized controlled cultivation conditions and painstaking analysis of chromosomal integrity are therefore necessary for a defined cell product.
Human embryonal stem cells (hESC) apparently have normal karyotypes in most studies [53]. Nevertheless, long-term cultivation can lead to significant accumulation of genetic aberrations [54]. Aneuploidies and additions of chromosomes 12, 17, 20 or X are frequently observed [40, 45, 47, 55,56,57,58,59,60]. Genetic stability has also been shown to be dependent on specific cultivation conditions. Chemical and enzymatic passaging techniques may enhance the propensity to genetic alterations. Most reports of hESC with genetic alterations used chemical and enzymatic passaging [40, 45, 55,56,57], whereas genetically stable hESC lines were passaged by microdissection [41, 45, 53]. Mitalipova et al. [45] showed that manual dissection allowed passaging of hESC for more than 100 passages without alterations of the karyotype. These cells are considered to be very sensitive to small alterations of growth conditions.
Inconsistent results have been reported on the genetic stability of human MSC [43, 44, 61,62,63,64]. It is conventionally thought that cultivation conditions influence the appearance of genetic alterations during long-term expansion necessary to generate sufficient numbers of cells needed for therapeutic application. Most cells still showed a normal karyotype after expansion [43, 62, 64,65,66], but genetic stability could be influenced by cultivation in different sera (fetal calf serum (FCS) vs. autologous serum). Genetic alteration in early passages seemed to be more frequent in FCS-cultured MSC [61]. In some cases long-term cultivation seems to favor cells with correct karyotype [44, 61]. Since FCS is a complex mixture with great lot-to-lot variation, it is difficult to ascertain the relevant factors and to compare between different studies. This problem also applies to the use of autologous serum. Synthetic defined media for stem cell cultivation would avoid this variation problem but in turn may itself execute undefined selective pressure.
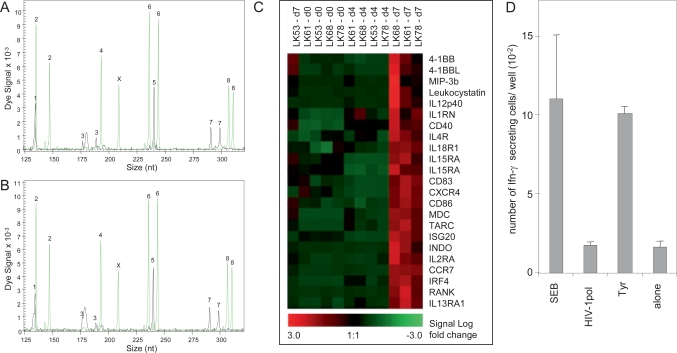
For the generation of genetically modified dendritic cells we developed a completely closed bag culture system based on defined media and standardized culture conditions. When STR analyses were performed on both CD14+ precursor cells and matured dendritic cells, no alterations could be detected during cultivation and differentiation (fig 3a, 3b). In addition, expression profiling was used to confirm the differentiation state of the cells (fig. 3c). Immunostimulatory capacity was tested in vitro to confirm functional capacity of the antigen presenting dendritic cells (fig. 3d).
Fig. 3.
Example for characterization of dendritic cell products. STR analyses of freshly isolated CD14+ monocytes (A) and of mature dendritic cells after 7 days in cell culture (B) confirm authenticity. STR loci are numbered as in figure 1. Detection of X-amelogenin and Y-amelogenin indicates a male donor. C) Expression profiling of dendritic cells derived from CD14+ monocytes (day 0) of leukapheresis samples (LK53, LK61, LK68 and LK78) during cultivation and maturation (day 4 and day 7) using Affymetrix HG-U95a DNA arrays. Heat map of selected genes indicates changes in gene expression during maturation. Up-regulation of maturation specific genes is evident LK61, LK68 and LK78 D) Immunostimulatory function of the mature dendritic cell product was confirmed by interferon [γ] – ELISPOT assays: Tyrosinase-specific T cells were stimulated by superantigen staphylococcal enterotoxin (SEB) (unspecific positive control) and specifically by tyrosinase peptide-presenting mature dendritic cells (tyr) but not by > dendritic cells alone or dendritic cells loaded with an unrelated peptide. The experiments were performed in triplicate; mean and standard deviation are shown.
Taken together, for safe and effective cellular therapies, characterization of the final cell products including karyotype genetic and epigenetic alterations, expression profiles, phenotype and differentiation potential as well as functional characterization depending on their intended use is fundamental.
Disclosure
The authors declared no conflict of interest.
Acknowledgements
We thank R. MacLeod, DSMZ, Braunschweig, for help with karyotyping, K. Steube for PCR analysis of neuronal cells, M. Rohde for scanning electron microscopy. We are very grateful to Melissa Wos for critical reading of the manuscript. We also thank Astrid Lootz for expert secretarial assistance.
This work was supported by grants from the Federal Ministry of Economics and Technology (Innonet project 16IN0546) and the EC (Clinigene-NoE, LSHB-CT-2006-018933).
References
- 1.Cheng L, Xiao L, Zeng F, Zhang YA. Stem cells shine in Shanghai. Cell Stem Cell. 2008;2:34–37. doi: 10.1016/j.stem.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Hay RJ. Cell Quantitation and characterization. In: Atala A, Lanza RP, editors. Methods of Tissue Engineering. San Diego: Academic Press; 2002. pp. 55–84. [Google Scholar]
- 3.EU Regulation (EC) No 1394/2007 of the European Parliament and of the Council of 13 November 2007 on Advanced Therapy Medicinal Products and Amending Directive 2001/83/EC and Regulation (EC) No 726/2004. Official Journal of the European Union. 2007:121–137. [Google Scholar]
- 4.Stacey GN. Cell contamination leads to inaccurate data: we must take action now. Nature. 2000;403:356. doi: 10.1038/35000394. [DOI] [PubMed] [Google Scholar]
- 5.Stacey GN. Standardisation of cell lines. Dev Biol (Basel) 2002;111:259–272. [PubMed] [Google Scholar]
- 6.Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007;43:575. doi: 10.2144/000112598. 577-778, 581-582. [DOI] [PubMed] [Google Scholar]
- 7.MacLeod RA, Dirks WG, Matsuo Y, Kaufmann M, Milch H, Drexler HG. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer. 1999;83:555–563. doi: 10.1002/(sici)1097-0215(19991112)83:4<555::aid-ijc19>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Markovic O, Markovic N. Cell cross-contamination in cell cultures: the silent and neglected danger. In Vitro Cell Dev Biol Anim. 1998;34:1–8. doi: 10.1007/s11626-998-0040-y. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Sawasaki Y, Hata J, Mukai K, Goto T. Spontaneous transformation and immortalization of human endothelial cells. In Vitro Cell Dev Biol. 1990;26:265–274. doi: 10.1007/BF02624456. [DOI] [PubMed] [Google Scholar]
- 10.Dirks WG, MacLeod RA, Drexler HG. ECV304 (endothelial) is really T24 (bladder carcinoma): cell line cross-contamination at source. In Vitro Cell Dev Biol Anim. 1999;35:558–559. doi: 10.1007/s11626-999-0091-8. [DOI] [PubMed] [Google Scholar]
- 11.Editorial Identity crisis. Nature. 2009;457:935–936. doi: 10.1038/457935b. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee R. Cell biology. Cases of mistaken identity. Science. 2007;315:928–931. doi: 10.1126/science.315.5814.928. [DOI] [PubMed] [Google Scholar]
- 13.Drexler HG, Dirks WG, MacLeod RA. False human hematopoietic cell lines: cross-contaminations and misinterpretations. Leukemia. 1999;13:1601–1607. doi: 10.1038/sj.leu.2401510. [DOI] [PubMed] [Google Scholar]
- 14.Nims RW, Shoemaker AP, Bauernschub MA, Rec LJ, Harbell JW. Sensitivity of isoenzyme analysis for the detection of interspecies cell line cross-contamination. In Vitro Cell Dev Biol Anim. 1998;34:35–39. doi: 10.1007/s11626-998-0050-9. [DOI] [PubMed] [Google Scholar]
- 15.Drexler HG, Matsuo AY, MacLeod RA. Continuous hematopoietic cell lines as model systems for leukemia-lymphoma research. Leuk Res. 2000;24:881–911. doi: 10.1016/s0145-2126(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 16.Bartlett SE, Davidson WS. FINS (forensically informative nucleotide sequencing): a procedure for identifying the animal origin of biological specimens. Biotechniques. 1992;12:408–411. [PubMed] [Google Scholar]
- 17.Unseld M, Beyermann B, Brandt P, Hiesel R. Identification of the species origin of highly processed meat products by mitochondrial DNA sequences. PCR Methods Appl. 1995;4:241–243. doi: 10.1101/gr.4.4.241. [DOI] [PubMed] [Google Scholar]
- 18.Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 19.Di Rienzo A, Wilson AC. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991;88:1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 21.Bonne-Tamir B, Johnson MJ, Natali A, Wallace DC, Cavalli-Sforza LL. Human mitochondrial DNA types in two Israeli populations - a comparative study at the DNA level. Am J Hum Genet. 1986;38:341–351. [PMC free article] [PubMed] [Google Scholar]
- 22.Harihara S, Hirai M, Suutou Y, Shimizu K, Omoto K. Frequency of a 9-bp deletion in the mitochondrial DNA among Asian populations. Hum Biol. 1992;64:161–166. [PubMed] [Google Scholar]
- 23.Torroni A, Brown MD, Lott MT, Newman NJ, Wallace DC. African, Native American, and European mitochondrial DNAs in Cubans from Pinar del Rio Province and implications for the recent epidemic neuropathy in Cuba. Cuba Neuropathy Field Investigation Team. Hum Mutat. 1995;5:310–317. doi: 10.1002/humu.1380050407. [DOI] [PubMed] [Google Scholar]
- 24.Gill P, Ivanov PL, Kimpton C, Piercy R, Benson N, Tully G, Evett I, Hagelberg E, Sullivan K. Identification of the remains of the Romanov family by DNA analysis. Nat Genet. 1994;6:130–135. doi: 10.1038/ng0294-130. [DOI] [PubMed] [Google Scholar]
- 25.Holland MM, Fisher DL, Mitchell LG, Rodriquez WC, Canik JJ, Merril CR, Weedn VW. Mitochondrial DNA sequence analysis of human skeletal remains: identification of remains from the Vietnam War. J Forensic Sci. 1993;38:542–553. [PubMed] [Google Scholar]
- 26.Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Berry DL, Holland KA, Weedn VW, Gill P, Holland MM. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 27.Garritsen HS, Szuflad P, Sibrowski W, Dzik WH. A sequence-specific polymerase chain reaction assay for mitochondrial DNA polymorphisms in human platelets and white cells. Transfusion. 1997;37:1012–1019. doi: 10.1046/j.1537-2995.1997.371098016438.x. [DOI] [PubMed] [Google Scholar]
- 28.Garritsen HS, Hoerning A, Hellenkamp F, Cassens U, Mittmann K, Sibrowski W. Polymorphisms in the non-coding region of the human mitochondrial genome in unrelated plateletapheresis donors. Br J Haematol. 2001;112:995–1003. doi: 10.1046/j.1365-2141.2001.02662.x. [DOI] [PubMed] [Google Scholar]
- 29.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;066:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 30.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci U S A. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enzmann H, Kuhlem C, Loser E, Bannasch P. Damage to mitochondrial DNA induced by the hepatocarcinogen diethylnitrosamine in ovo. Mutat Res. 1995;329:113–120. doi: 10.1016/0027-5107(95)00021-a. [DOI] [PubMed] [Google Scholar]
- 32.Shen CC, Wertelecki W, Driggers WJ, LeDoux SP, Wilson GL. Repair of mitochondrial DNA damage induced by bleomycin in human cells. Mutat Res. 1995;337:19–23. doi: 10.1016/0921-8777(95)00008-8. [DOI] [PubMed] [Google Scholar]
- 33.Wallace DC, Shoffner JM, Trounce I, Brown MD, Ballinger SW, Corral-Debrinski M, Horton T, Jun AS, Lott MT. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta. 1995;1271:141–151. doi: 10.1016/0925-4439(95)00021-u. [DOI] [PubMed] [Google Scholar]
- 34.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 35.Andrews RM, Kubacka I, Chinnery PF, Lightow-lers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 36.Hutter G, Nickenig C, Garritsen H, Hellenkamp F, Hoerning A, Hiddemann W, Dreyling M. Use of polymorphisms in the noncoding region of the human mitochondrial genome to identify potential contamination of human leukemia-lymphoma cell lines. Hematol J. 2004;5:61–68. doi: 10.1038/sj.thj.6200317. [DOI] [PubMed] [Google Scholar]
- 37.Dirks WG, Faehnrich S, Estella IA, Drexler HG. Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX. 2005;22:103–109. [PubMed] [Google Scholar]
- 38.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, Toji LH, Ohno T, Tanabe H, Arlett CF, Kelland LR, Harrison M, Virmani A, Ward TH, Ayres KL, Debenham PG. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98:8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaffer L, Tommerup N. ISCN 2005: An International System for Human Cytogenetic Nomenclature. Basel: Karger; 2009. [Google Scholar]
- 40.Brimble SN, Zeng X, Weiler DA, Luo Y, Liu Y, Lyons IG, Freed WJ, Robins AJ, Rao MS, Schulz TC. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 2004;13:585–597. doi: 10.1089/scd.2004.13.585. [DOI] [PubMed] [Google Scholar]
- 41.Caisander G, Park H, Frej K, Lindqvist J, Bergh C, Lundin K, Hanson C. Chromosomal integrity maintained in five human embryonic stem cell lines after prolonged in vitro culture. Chromosome Res. 2006;14:131–137. doi: 10.1007/s10577-006-1019-8. [DOI] [PubMed] [Google Scholar]
- 42.Grimes BR, Steiner CM, Merfeld-Clauss S, Traktuev DO, Smith D, Reese A, Breman AM, Thurston VC, Vance GH, Johnstone BH, Slee RB, March KL. Interphase FISH demonstrates that human adipose stromal cells maintain a high level of genomic stability in long-term culture. Stem Cells Dev. 2009;18:717–724. doi: 10.1089/scd.2008.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lange C, Cakiroglu F, Spiess AN, Cappallo-Obermann H, Dierlamm J, Zander AR. Accelerated and safe expansion of human mesenchymal stromal cells in animal serum-free medium for transplantation and regenerative medicine. J Cell Physiol. 2007;213:18–26. doi: 10.1002/jcp.21081. [DOI] [PubMed] [Google Scholar]
- 44.Meza-Zepeda LA, Noer A, Dahl JA, Micci F, Myklebost O, Collas P. High-resolution analysis of genetic stability of human adipose tissue stem cells cultured to senescence. J Cell Mol Med. 2008;12:553–563. doi: 10.1111/j.1582-4934.2007.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitalipova MM, Rao RR, Hoyer DM, Johnson JA, Meisner LF, Jones KL, Dalton S, Stice SL. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- 46.Loring JF, Rao MS. Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells. 2006;24:145–150. doi: 10.1634/stemcells.2005-0432. [DOI] [PubMed] [Google Scholar]
- 47.Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der EJ, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- 48.Schrock E, du MS, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 49.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 50.Urban AE, Korbel JO, Selzer R, Richmond T, Hacker A, Popescu GV, Cubells JF, Green R, Emanuel BS, Gerstein MB, Weissman SM, Snyder M. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci U S A. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peltomaki PT. Genetic basis of hereditary nonpoly-posis colorectal carcinoma (HNPCC) Ann Med. 1994;26:215–219. doi: 10.3109/07853899409147893. [DOI] [PubMed] [Google Scholar]
- 52.Soong R, Anuar D, Liu Y, Eu KW, Han HC, Salto-Tellez M, Iacopetta B. Denaturing high performance liquid chromatography for the detection of microsatellite instability using Bethesda and Pentaplex marker panels. Diagn Mol Pathol. 2008;17:127–133. doi: 10.1097/PDM.0b013e3181577daf. [DOI] [PubMed] [Google Scholar]
- 53.Darnfors C, Flodin A, Andersson K, Caisander G, Lindqvist J, Hyllner J, Wahlstrom J, Sartipy P. High-resolution analysis of the subtelomeric regions of human embryonic stem cells. Stem Cells. 2005;23:483–488. doi: 10.1634/stemcells.2004-0254. [DOI] [PubMed] [Google Scholar]
- 54.Gordeeva OF, Mitalipov S. Pluripotent stem cells: maintenance of genetic and epigenetic stability and prospects of cell technologies (in Russian) Ontogenez. 2008;39:405–419. [PubMed] [Google Scholar]
- 55.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 56.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 57.Inzunza J, Sahlen S, Holmberg K, Stromberg AM, Teerijoki H, Blennow E, Hovatta O, Malmgren H. Comparative genomic hybridization and karyotyping of human embryonic stem cells reveals the occurrence of an isodicentric X chromosome after long-term cultivation. Mol Hum Reprod. 2004;10:461–466. doi: 10.1093/molehr/gah051. [DOI] [PubMed] [Google Scholar]
- 58.Lefort N, Feyeux M, Bas C, Feraud O, NaceurGris-celli A, Tachdjian G, Peschanski M, Perrier AL. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- 59.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S, Carpenter M, Gazdar AF, Rao M, Chakravarti A. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 60.Rosler ES, Fisk GJ, Ares X, Irving J, Miura T, Rao MS, Carpenter MK. Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn. 2004;229:259–274. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- 61.Dahl JA, Duggal S, Coulston N, Millar D, Melki J, Shahdadfar A, Brinchmann JE, Collas P. Genetic and epigenetic instability of human bone marrow mesenchymal stem cells expanded in autologous serum or fetal bovine serum. Int J Dev Biol. 2008;52:1033–1042. doi: 10.1387/ijdb.082663jd. [DOI] [PubMed] [Google Scholar]
- 62.Rubio D, Garcia-Castro J, Martin MC, de la FR, Cigudosa JC, Lloyd AC, Bernad A. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035–3039. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 63.Rubio D, Garcia S, Paz MF, De la Cueva T, Lopez-Fernandez LA, Lloyd AC, Garcia-Castro J, Bernad A. Molecular characterization of spontaneous mesenchymal stem cell transformation. PLoS One. 2008;3:e1398. doi: 10.1371/journal.pone.0001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang ZX, Guan LX, Zhang K, Wang S, Cao PC, Wang YH, Wang Z, Dai LJ. Cytogenetic analysis of human bone marrow-derived mesenchymal stem cells passaged in vitro. Cell Biol Int. 2007;31:645–648. doi: 10.1016/j.cellbi.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 65.Bernardo ME, Zaffaroni N, Novara F, Cometa AM, Avanzini MA, Moretta A, Montagna D, Maccario R, Villa R, Daidone MG, Zuffardi O, Locatelli F. Human bone marrow derived mesen-chymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res. 2007;67:9142–9149. doi: 10.1158/0008-5472.CAN-06-4690. [DOI] [PubMed] [Google Scholar]
- 66.Miura Y, Gao Z, Miura M, Seo BM, Sonoyama W, Chen W, Gronthos S, Zhang L, Shi S. Mesenchymal stem cell-organized bone marrow elements: an alternative hematopoietic progenitor resource. Stem Cells. 2006;24:2428–2436. doi: 10.1634/stemcells.2006-0089. [DOI] [PubMed] [Google Scholar]
- 67.Buwe A, Steinlein C, Koehler MR, Bar-Am I, Katzin N, Schmid M. Multicolor spectral karyotyping of rat chromosomes. Cytogenet Genome Res. 2003;103:163–168. doi: 10.1159/000076306. [DOI] [PubMed] [Google Scholar]
- 68.Beresford JN. Osteogenic stem cells and the stromal system of bone and marrow. Clin Orthop Relat Res. 1989;240:270–280. [PubMed] [Google Scholar]