Abstract
The lifespan of a mammalian mRNA is determined, in part, by the binding of regulatory proteins and small RNA-guided complexes. The conserved endonuclease activity of Argonaute2 requires extensive complementarity between a small RNA and its target and is not used by animal microRNAs, which pair with their targets imperfectly. Here, we investigate the endonucleolytic function of Ago2 and other nucleases by transcriptome-wide profiling of mRNA cleavage products retaining 5′-phosphate groups in mouse ES. We detect a prominent signature of Ago2-dependent cleavage events and validate several such targets. Unexpectedly, a broader class of Ago2-independent cleavage sites is also observed, indicating participation of additional nucleases in site-specific mRNA cleavage. Within this class, we identify a cohort of Drosha-dependent mRNA cleavage events that functionally regulate mRNA levels in mES cells, including one in the Dgcr8 mRNA. Together, these results highlight the underappreciated role of endonucleolytic cleavage in controlling mRNA fates in mammals.
Introduction
The last two decades have provided a deep appreciation for the numerous roles of small RNAs in eukaryotic cells. Perhaps the best-characterized species are the subset that form components of RNAi-related pathways. These 21–30 nucleotide RNAs join Argonaute proteins and guide them to their targets via complementary base pairing. Once bound, effector complexes can elicit a variety of outcomes, with one of the most conserved being target RNA cleavage catalyzed by the Argonaute RNAse H-related nuclease domain.
Among the classes of Argonaute-associated RNAs, microRNAs form a conserved regulatory axis, which exerts post-transcriptional control over a broad set of cellular mRNAs (Carthew and Sontheimer, 2009). microRNAs are derived from partially duplexed precursors, called pri-miRNAs, via processing by RNAseIII family enzymes, Drosha and Dicer in animals or DCL1 in plants (Voinnet, 2009). Though superficially similar, plant and animal microRNAs differ not only in their mechanisms of biogenesis but also in the modes by which they regulate gene expression. In plants, extensive miRNA-mRNA pairing most often leads to cleavage of target mRNAs (Voinnet, 2009). In animals, target recognition is dominated by a short sequence comprising bases ~2–8 of the small RNA, termed the seed. The remainder of the small RNA pairs imperfectly, if at all, with its regulatory target. In these cases, a miRNA-target interaction commonly results in degradation of the mRNA by general cellular mRNA decay pathways rather than by Argonaute cleavage. Nevertheless, at least one mRNA, HOXB8, can be demonstrably cleaved in response to interaction with a highly complementary microRNA, miR-196 (Yekta et al., 2004). Additionally, cleavage products corresponding to miRNA target sites in the imprinted Rtl1/Peg11 locus have been observed (Davis et al., 2005).
The four mammalian Ago homologs evolved from a common ancestor early in the vertebrate lineage (unpublished observations). A significant expansion of novel miRNA families coincided with this Ago diversification, potentially explaining the emergent tissue complexity in vertebrates (Heimberg et al., 2008). Given the predominantly non-catalytic mode of miRNA action in mammals and the largely indiscriminate association of miRNAs with the Ago homologs (Azuma-Mukai et al., 2008; Ender et al., 2008), it is surprising that one of the Argonautes, Ago2, conserved its catalytic function throughout this expansion (Liu et al., 2004). Ago2 is the only mammalian Ago protein required for viability (Liu et al., 2004) (S.C. and G.J.H., unpublished), and the catalytic activity of Ago2 contributes to its essential role (Cheloufi et al., 2010). Ago2 catalysis is required for the unusual biogenesis of an erythroid microRNA, miR-451, but the degree of anemia caused by lack of this regulator probably does not fully explain the perinatal death of animals harboring only catalytically inactive Ago2 alleles (Cheloufi et al., 2010). Thus, there is a strong likelihood that cleavage by Ago2 has additional roles both during late embryogenesis and in adult animals.
The ends of cellular mRNAs are protected by a cap structure, and the major non-specific ribonucleases generally leave 5′ OH groups. Thus, 5′ phosphorylated RNA species are thought to be enriched in products of specific cellular RNA processing events. This property has been exploited to identify the mRNAs subject to microRNA-directed cleavage in plants by transcriptome-wide analysis of mRNA fragments bearing 5′ monophosphate termini (Addo-Quaye et al., 2008; German et al., 2008). Those studies confirmed many known microRNA targets and led to the identification of novel cleavage products of microRNA- and tasiRNA-primed RISC. The data also implied the existence of cleavage sites that could not be explained by known small RNAs.
Endonucleolytic cleavage likely plays a broader role in mRNA regulation than is currently appreciated, and evidence to this effect is beginning to emerge. In addition to its 3′-5′ exonuclease function, the exosome component Dis3/Rrp44 possesses a functionally relevant endonuclease activity via its PIN domain (Lebreton et al., 2008). Similarly, the PIN domain of SMG6 can cleave mRNAs near premature termination codons during NMD (Eberle et al., 2009; Huntzinger et al., 2008). Site-specific cleavage events in particular mRNAs have also been observed. For example, Drosha can regulate the expression of its DGCR8/Pasha cofactor by cleaving a site within its 5′ UTR that resembles a pri-miRNA, the canonical Drosha substrate (Han et al., 2009b). The stress-induced IRE1α enzyme promotes decay of a number of ER-localized mRNAs via endonucleolytic cleavage (Han et al., 2009a). Additional examples include cleavage of c-myc by G3BP and APE1 (Barnes et al., 2009; Tourriere et al., 2001) and cleavage of α-globin mRNA by an erythroid-enriched endonuclease (Wang and Kiledjian, 2000).
In this study, we examine transcriptome-wide mRNA cleavage patterns resulting in 5′-phosphorylated fragments in mammals. We identify a class of miRNA-guided, Ago2-dependent mRNA cleavage events that may contribute to the conservation of Ago2 catalytic potential. Surprisingly, we also discover a cohort of Drosha-dependent cleavage sites that indicate a broader extent of mRNA processing by this enzyme than was previously appreciated. We also noted a large class of evolutionarily conserved mRNA cleavage sites that depended neither on Ago2 nor Drosha. These highlight the participation of additional, yet to be described, nucleases in this mode of mRNA metabolism.
Results
Combined bioinformatic and experimental approaches reveal a set of miRNA cleavage targets
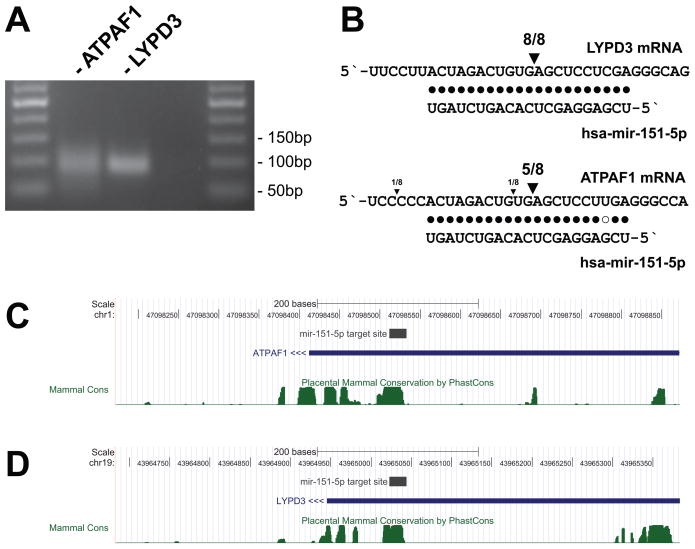
To define a set of potential targets of miRNA-guided cleavage, we performed a computational search of the transcriptome for extensive complementarity between miRNAs and mRNA transcripts. Based on known criteria for cleavage-competent pairing (Elbashir et al., 2001; Haley and Zamore, 2004; Martinez and Tuschl, 2004) we considered sites with perfect pairing at miRNA positions 9, 10, and 11, and ranked the sites by the number of non-GU mismatches and total mismatches. The resulting lists for human and mouse datasets showed a large number of targets with perfect or near-perfect complementarity (Supplementary Table S1). Cleavage of two predicted mir-151-5p targets, ATPAF1 and LYPD3, was confirmed by gene-specific 5′ RACE (Yekta et al., 2004) of 293S cellular RNA. Both reactions produced correctly sized amplicons, with 5/8 and 8/8 clones reflecting the predicted cleavage site upon sequencing (Figures 1A and 1B). The predicted mir-151-5p target sites in the 3′ UTRs of these genes showed significant conservation, suggesting a functionally relevant interaction (Figures 1C and 1D). Interestingly, mir-151-5p is derived from a LINE2 element (Smalheiser and Torvik, 2005) and complementarity to this microRNA can be found in a number of genes harboring LINE2 insertions, including the above transcripts. In these cases, however, the region of high conservation is confined to the miR-151-5p complementarity and does not extend throughout the LINE2 fragment.
Figure 1.
Validation of bioinformatically predicted miRNA cleavage targets by 5′ RACE in 293S cells. (A) Nested PCR of the 5′ RACE produces amplicons of the expected size for ATPAF1 and LYPD3, but not a negative control (lane 3). (B) Cloning and sequencing of the PCR amplicons indicates products corresponding to the expected miRNA cleavage site. (C) and (D) Conservation of the mir-151-5p target site in the 3′ UTRs of ATPAF1 and LYPD3. See also Supplementary Table S1.
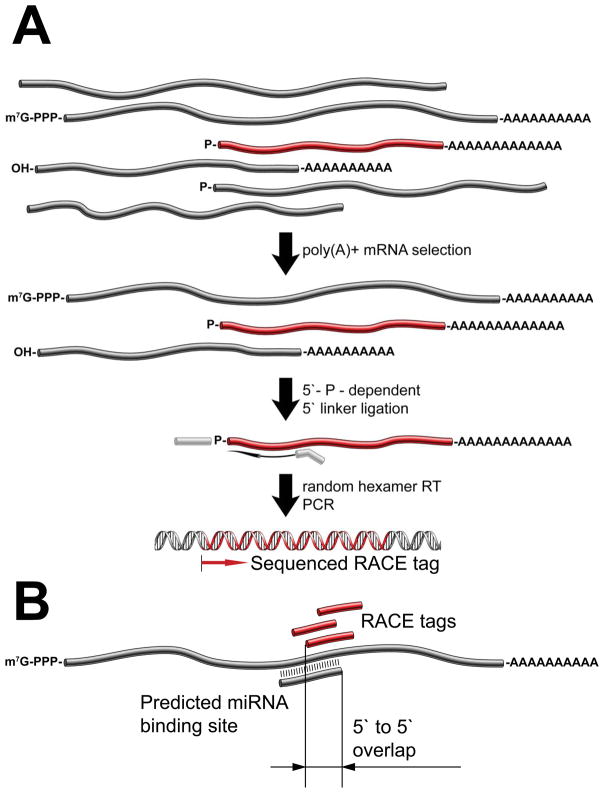
As an orthogonal approach, we undertook a purely experimental search for mRNA cleavage sites similar to that previously applied for the identification of Arabidopsis miRNA targets (Addo-Quaye et al., 2008; German et al., 2008). This procedure (Figure 2A) begins with the isolation of polyA+ RNA. Linkers are then ligated specifically to 5′ ends that bear monophosphate termini. After randomly primed reverse transcription and PCR amplification, libraries are analyzed by high throughput sequencing, generating a global set of RACE tags that reveal, based upon the presence of the linker, the precise 5′ ends of the 3′ fragments resulting from mRNA cleavage.
Figure 2.
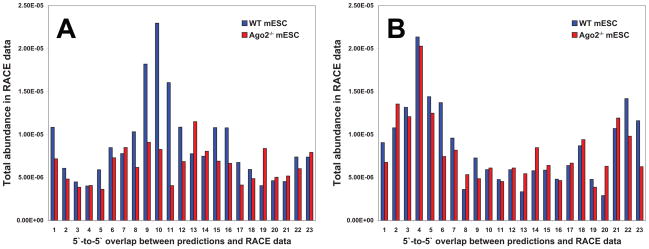
(A) Experimental scheme for transcriptome-wide 5′-phosphate-dependent RACE. PolyA+ RNAs are selected from total RNA and ligated to a 5′ adapter. The resulting RNAs are reverse-trancribed using random hexamer primers, PCR-amplified, and sequenced. (B) The experimental RACE tag data was combined with bioinformatic miRNA target site predictions by computing the 5′ to 5′ overlap between the tag and the miRNA at the binding site.
As a means to discriminate definitively microRNA-directed cleavage events from those produced by other cellular nucleases, we took advantage of the fact that Ago2 is the only catalytically active Argonaute family member. We established a series of ES cell lines using blastocysts collected from an intercross of animals heterozygous for Ago2 insertional mutations within the PIWI and the PAZ domains (Liu et al., 2004). From these, we identified Ago2−/− ES lines and used wild-type ES cells for comparison.
Global RACE tags from both cell lines were intersected with the computational predictions of miRNA cleavage sites. Each time an overlap was detected, we calculated the position of the 5′ end of the RACE tag with respect to the 5′ end of the miRNA predicted to pair at that site (Figure 2B). When considering all positions with up to 2 non-GU mismatches, libraries from wild-type cells showed a peak of tag abundance centered 10 nucleotides away from the miRNA 5′ end (Figure 3A, Supplementary Table S2). This is consistent with the known biochemical properties of Argonaute proteins, which cleave the phosphodiester bond opposite nucleotides 10–11 of the small RNA guide (Schwarz et al., 2004). The significance of this signal was indicated by three additional observations. First, libraries from Ago2−/− ES cells did not show a similar relationship between RACE tags and predicted sites of miRNA-mRNA pairing. Second, the peak disappeared if we performed similar analyses after randomizing the miRNA sequences (Figure 3B). Finally, if we included in our analysis sites with up to 5 non-GU mismatches, the enrichment at position 10 disappeared (Supplementary Figures S1A,B). These observations strongly support the ability of global RACE to identify targets of miRNA-directed Ago2 cleavage in mammalian cells.
Figure 3.
Combined experimental and bioinformatic approaches identify miRNA-guided mRNA cleavage sites in mouse embryonic stem cells. (A) Cumulative RACE tag abundances in WT (blue) and Ago2−/− (red) libraries as a function of overlap with predicted miRNA target sites with 2 or fewer non-GU mismatches. (B) Same as in (A), but for randomized miRNAs. See also Supplementary Table S3 and Figure S3.
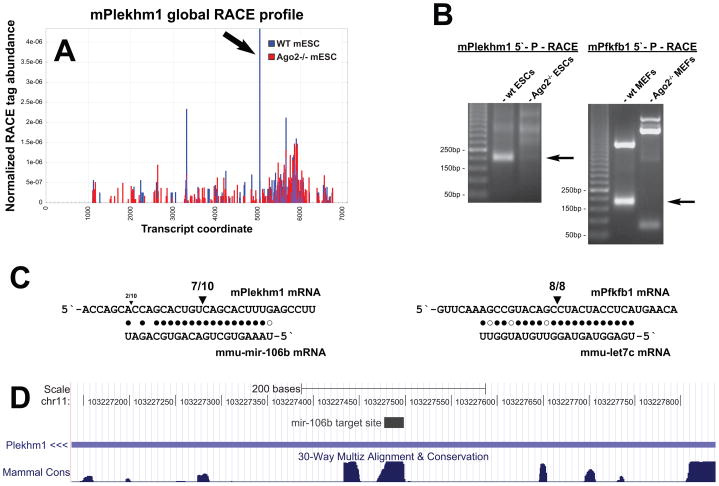
We chose two potential miRNA cleavage targets for further confirmation. Plekhm1 showed a strong Ago2-dependent RACE signal at a site with complementarity to miR-106b (Figure 4A). Consistent with this observation, conventional RACE amplified a product in wild-type, but not Ago2−/−-mESCs (Figure 4B), and the cleavage site was confirmed by clone sequencing (Figure 4C). The Plekhm1 target site was conserved throughout the length of the miRNA pairing (Figure 4D), and we could detect cleavage at the same site in the orthologous mRNA in human cells (Supplementary Figure S2). Pfkfb1 showed a weaker Ago2-dependent cleavage site in RACE libraries at a position with complementarity to the let-7 miRNA family. By conventional RACE, cleavage at this site was not detectable in ES cells; however, let-7 expression is quite weak in this cell type. We could readily detect cleavage at the let-7 complementary site in mouse embryonic fibroblasts (Figures 4B, C), where let-7 expression is more robust. Again, the cleavage event occurred in wild-type, but not Ago2−/− MEFs.
Figure 4.
Validation of miRNA-guided cleavage sites. (A) Profile of RACE tag abundances mapping to the Plekhm1 transcript in WT (blue) and Ago2−/− (red) libraries. The identified Ago2-dependent cleavage site is shown by an arrow. (B) Conventional RACE validation of Plekhm1 and Pfkfb1 cleavages in WT, but not Ago2−/− cells. The expected amplicon sizes are indicated by arrows. (C) Complementarity between mir-106b, let-7c and their respective targets, Plekhm1 and Pfkfb1. Triangles indicate the number of RACE clones at the corresponding position. (D) Conservation of the mir-106b target site in the Plekhm1 3′ UTR. See also Supplementary Figure S4.
Drosha-dependent mRNA cleavage
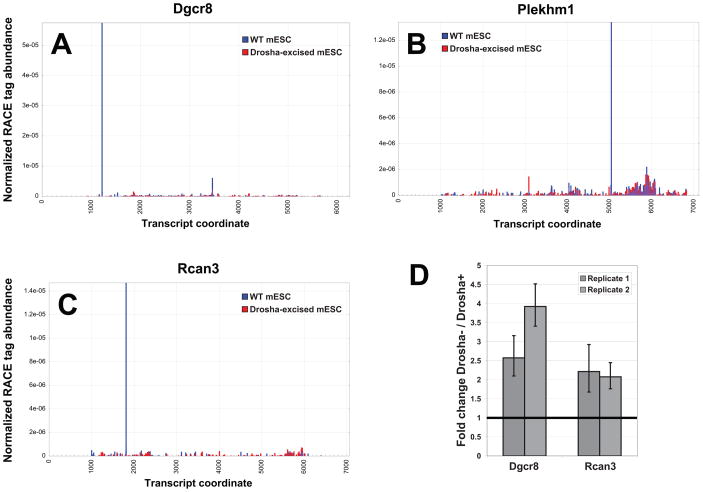
While a number of the sites detected in our global RACE were clearly Ago2-dependent, there were also numerous presumptive endonucleolytic cleavage sites that persisted in the absence of Ago2. Among this class was a strong site near the 5′ end of the Dgcr8 mRNA (Figure 5A). Kim and colleagues previously demonstrated that Drosha, which partners with DGCR8 in the Microprocessor complex, directly regulates DGCR8 production by recognizing a site within its 5′ UTR that mimics a microRNA precursor (Han et al., 2009b). The so-called A2 site, cleaved by Drosha in vitro, falls within one nucleotide of the site detected in our global RACE analysis (Supplementary Figure S3).
Figure 5.
Drosha-dependent cleavage sites in mammalian mRNAs. (A) Profile of RACE tag abundances for the Dgcr8 transcript in WT (blue) and Drosha-excised (red) libraries. (B), (C) Same as in (A), but for Plekhm1 and Rcan3. (D) Transcript levels of Dgcr8 and Rcan3 in Drosha-excised mES cells relative to wild-type controls, as measured by QPCR. See also Supplementary Figure S5 and Table S5.
To determine the extent of the transcript population that might be recognized and cleaved by Drosha, we took advantage of a conditional Drosha-null ES cell line. Following cre-mediated inactivation of Drosha in ES cells, levels of Drosha mRNA decrease by 5–12 fold (data not shown). In accord with these observations, a comparison of global RACE libraries from these cells to those from wild-type (cre-uninduced) ESCs revealed loss of the miR-106b-directed, Ago2-dependent Plekhm1 site (Figure 5B). We also saw loss of the Ago2-independent site in Dgcr8, consistent with its being Drosha mediated (Figure 5A).
Using their behavior in genetic mutants as a guide, we searched for additional sites of Drosha-dependent cleavage within mRNAs. We noted a substantial number of sites (Supplementary Table S3), which varied in the strength of their cleavage signature within wild-type libraries and in the degree to which they responded to Drosha loss. The novel genes in this category are exemplified by Rcan3 (Figure 5C). Furthermore, we observed an increase in total Dgcr8 and Rcan3 levels in Drosha-excised cells by QPCR (Figure 5D), strongly suggesting a regulatory nature for these cleavage events.
The vast majority of these sites are not likely to be responding to Drosha loss indirectly via microRNA depletion because they are not strongly changed in Ago2-null cells nor do they overlap with predicted sites of microRNA-mRNA interaction. Many of the sites that we identified can be folded into local or more long-range secondary structures that could provide the double-stranded substrates preferred by the RNaseIII family. Notably, after exposing in vitro transcripts of candidate targets (e.g., Dvl2) to immunoaffinity purified Drosha, we do detect bands consistent with predicted cleavage sites (not shown). However, pending their further analysis, we cannot definitively state that the sites we proposed are cleaved directly by Drosha. Thus, a direct enzyme-substrate relationship remains the most parsimonious explanation for our observations; however, further experiments will be required to provide definitive support for this proposition.
Additional endonucleolytic mRNA cleavage events
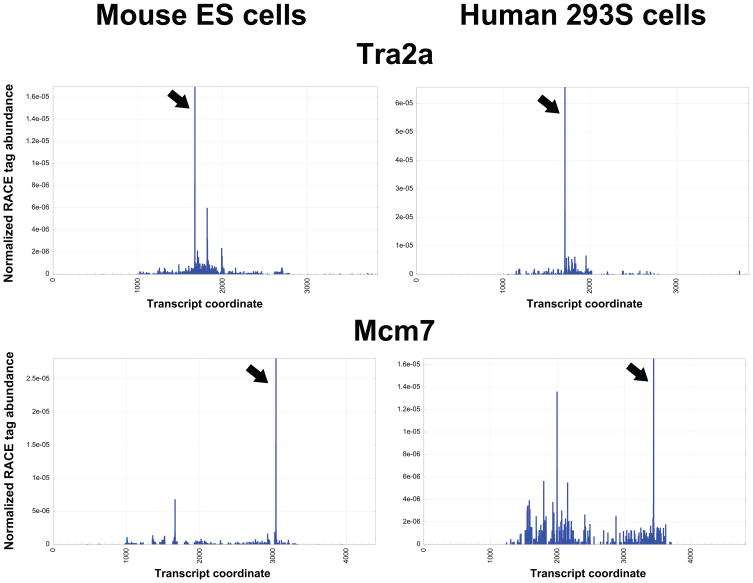
If we eliminate sites conforming to our expectations for Ago2- and Drosha-mediated cleavage, the global RACE data still reveals many robust cleavage events that might be catalyzed by other endonucleases. In many cases, these signals were strikingly strong and arose from relatively abundant transcripts (Figure 6 and Supplementary Table S4).
Figure 6.
Examples of conserved Ago2- and Drosha-independent cleavage sites in mammalian mRNAs. Profiles of RACE tag abundances for TRA2A and MCM7 in mouse ES (left) and human 293S (right) libraries. See also Supplementary Table S6.
Because the nucleases that catalyze such events are unknown, it is difficult to assess the biological significance of these observations. One way to prioritize potentially significant sites might be to examine conservation, both of the cleavage event and of sequence contexts that might ultimately lead to the identification of instructive motifs. We therefore compared global RACE data from human 293S cells to signatures seen in mouse ES cells. Although these cell types are not particularly similar, we were able to find many instances where orthologous genes were expressed in both species and where we observed similar or identical sites of presumptive endonucleolytic processing (Supplementary Table S4). Two examples are shown in Figure 6. We confirmed the veracity of the genome-wide data with individual, gene-specific RACE for TRA2A in 293S cells (Supplementary Figure S4).
Discussion
We began the present work in part to gain insight into the evolutionary pressure to conserve Argonaute catalysis in vertebrates. Catalytic potential is maintained despite overwhelming evidence that microRNAs can operate largely without the need for an intact Ago catalytic site. Toward this end, we sought to determine the repertoire of microRNA-directed Ago cleavage targets through the use of a genome-wide RACE method that captures RNAs bearing a 5′ phosphate termini.
A strong indication that we were identifying relevant sites came from our observed enrichment for a 10 nucleotide overlap between the 5′ end of microRNAs and the 5′ ends of RACE products. This depended on the integrity of Ago2 and on the comparison of cleavage sites being made to bona fide rather than scrambled microRNA sequences. The Ago2 target set was further enriched through bioinformatic predictions, and these combined approaches have now added substantially to the number of known microRNA-directed cleavage sites in mammals. Previously, a site within a coding mRNA, HOXB8, had been validated (Yekta et al., 2004). A non-coding RNA from the Rtl1 locus has also been proposed to be regulated by miRNA-directed cleavage (Davis et al., 2005; Seitz et al., 2003).
Thus far, we have examined only one cell type, ES cells, exhaustively, and these are relatively microRNA poor. This likely causes us to minimally estimate the true extent of Ago cleavage targets that might be present were we to analyze many separate cell types. For example, we could only weakly detect cleavage of Pfkfb1 in ES cells, while this was readily apparent in MEFs. Similarly, we saw no signature in the global data of the HOXB8-miR-196 interaction because neither the microRNA nor its partner are abundant in ES cells (Yekta et al., 2004). Thus, we propose that a wide variety of miRNA cleavage targets may provide one factor which contributes to the evolutionary pressure to maintain Ago2 catalysis.
The majority of 5′ monophosphorylated mRNA fragments that we detected were not dependent upon the presence of Ago2, but a number of Ago2-independent sites did depend upon another RNAi pathway component, Drosha. Previous studies had demonstrated the ability of Drosha to regulate gene expression by cleaving a microRNA-mimetic structure in the 5′ end of the DGCR8 gene. We also uncovered this site as a Drosha-dependent event in a global RACE comparison of wild-type and Drosha-mutant ES cells. However, we also noted a variety of other Drosha cleavage sites, which appeared with varying strengths in our dataset. In contrast, a study by Shenoy and Blelloch concluded that mRNA targeting by the Drosha/Dgcr8 complex is only limited to Dgcr8 (Shenoy and Blelloch, 2009). It is possible that lack of sequencing data on longer RNAs (>32nt) from Microprocessor-depleted cells limited the sensitivity of their approach.
Computational folding predictions often place the cleavage sites in regions of extensive secondary structure; however most do not present canonical pri-miRNA-like hairpins (data not shown). Our current understanding suggests that Drosha complexes recognize and cleave helical regions adjacent to single-stranded segments. This is likely determined in part by the co-factors with which it associates, namely DGCR8/Pasha for the canonical RNAi pathway. It is presently unclear whether the sites that we observe are dependent upon this cofactor or whether alternative cofactors might confer upon Drosha a preference for different structural motifs, which might be common to the cleavage sites that we observe.
Over the past several years, a remarkable variety of new long and small RNA products have come to light. Overall, it seems as if the vast majority of the mammalian genome is transcribed, and many of its transcriptional products become further processed after their production. As one example, we recently reported the identification of new small RNA species that appear to be derived from mature mRNAs by endonucleolytic cleavage and cytoplasmic capping (Fejes-Toth et al., 2009), a hypothesis that has gained support through the identification and isolation of a cytoplasmic capping complex capable of acting on 5′ monophosphorylated substrates (Otsuka et al., 2009). This suggested that there must exist a wide variety of endonucleolytic processing events that goes far beyond what is currently appreciated.
In accord with this notion, we observe a large collection of endonucleolytic cleavage sites that are independent of both Ago2 and Drosha. Both their abundance and their evolutionary conservation are suggestive of important functions. Though we have not yet identified the nucleases responsible for such events, a number of possibilities present themselves. Several endonucleases are known to leave 5′ phosphate termini, including Dicer and RNase P. Several more are likely to leave 5′ phosphates as judged from their similarity to known nuclease families. These include the stress-induced endonuclease IRE1, RNase L, and APE1. The PIN domains of Dis3/Rrp44 and SMG6 nucleases adopt an RNase H-like fold that could generate 5′-phosphorylated products (Glavan et al., 2006). As most of these enzymes lack inherent substrate specificity, their targeted cleavage sites are likely to be selected by partner binding proteins.
Considered as a whole, our findings not only reveal a previously unappreciated breadth in the roles of RNAi family enzymes in mRNA cleavage but also suggest that mRNAs are subject to a surprising diversity of endonucleolytic, post-transcriptional processing events. The path toward understanding the precise biological impact of these processing events will likely require our linking individual cleavage sites with the nucleolytic complexes that generate them.
Experimental Procedures
Please see Supplementary Materials and Methods for additional information
Bioinformatic Predictions of near-perfect mRNA targets
For human and mouse analyses, the reverse complementary sequences of all miRBase Release 14 miRNAs were aligned against Ensembl Release 54 mRNA transcripts, allowing for up to 5 mismatches and no insertions/deletions. The resulting predictions were dominated by a multitude of hits generated from miRNAs with degenerate or simple repetitive sequences. To eliminate such questionable hits, the analysis was limited to miRNAs numbering below 400 as a proxy of their abundance. The predictions were filtered for perfect matches at miRNA positions 9–11, and sorted by the number of non-GU mismatches and total mismatches. As a control, we predicted targets for control miRNAs, which were created by shuffling the real miRNAs while preserving the dinucleotide composition (Workman and Krogh, 1999).
Individual 5′-phosphate dependent 5′ RACE
Individual cleavage products were detected using the Invitrogen GeneRacer kit from 5 μg of total cellular RNA. The GeneRacer 5′ oligo was ligated directly to total RNA samples, requiring the presence of 5′ phosphates on ligated molecules. PCR reactions were carried out in a final 50 μl reaction using KOD Hot Start DNA polymerase (Novagen) containing 5 μl 10X buffer, 3 μl 25mM MgSO4, 5 μl 2mM dNTPs, 1.5 μl 10μM primers, 1 μl template DNA, and 1 μl enzyme. PCR conditions: 94 °C for 2 minutes, followed by 35 cycles of 94 °C for 30 seconds, 58–63 °C (gene-specific) for 30 seconds, and 70 °C for 1 minute, with a final extension at 70 °C for 10 minutes.
Global 5′ RACE library preparation
Poly(A)+ mRNAs were isolated from 200–500 μg of total RNA using the Invitrogen Dynabeads mRNA Direct kit. Briefly, 500 μl of oligo(dT) beads were washed once in binding buffer, resuspended in 500 μl binding buffer plus 500 μl of total RNA, and rotated at room temperature for 5 minutes. Beads were washed twice with 500 μl buffer B and eluted with 200 μl 10mM Tris-HCl, pH 7.5 by heating to 65 °C for 2 minutes and rapidly removing the eluate. The eluate was re-purified over the same beads, eluted in 100 μl, and ethanol-precipitated. The RNA was ligated to a 5′ linker, requiring the presence of a 5′ phosphate on the ligated RNA, in a 20 μl volume (2 μl 10X T4 RNA ligase buffer, 2 μl DMSO, 2 μl 50 μM SBS3 linker, 2 μl T4 RNA ligase (Ambion), 12 μl reconstituted RNA pellet) at 37 °C for 1.5 hours. The reaction was phenol-chloroform-extracted, chloroform extracted, purified over 50 μl of oligo(dT) beads as above, and eluted in 25 μl. Reverse transcription reactions were carried out by handom hexamer priming: 11 μl of ligated RNA, 1 μl of 100 μM SBS8-N6 primer, and 1 μl of 10mM dNTPs were annealed at 65 °C for 5 minutes and mixed with 4 μl 5X 1st strand buffer, 1 μl 100mM DTT, 1 μl RNaseIN, 1 μl SuperScript III (Invitrogen), and incubated at 50 °C for 1 hour, followed by 70 °C for 15 minutes. Template RNA was degraded by addition of 1 μl RNase H (Invitrogen) and incubating at 37 °C for 20 minutes. PCR reactions on the resulting cDNAs were carried out in a final 50 μl reaction using KOD Hot Start DNA polymerase (Novagen) containing 5 μl 10X buffer, 3 μl 25mM MgSO4, 5 μl 2mM dNTPs, 3 μl 10μM PE-P5-SBS3 and PE-P7-SBS8 primers, 1 μl template DNA, and 1 μl enzyme. PCR conditions: 95 °C for 2 minutes, followed by 30 cycles of 95 °C for 15 seconds, 60 °C for 30 seconds, and 72 °C for 1 minute, with a final extension at 72 °C for 7 minutes. Products were run on a 2% low-melt agarose gel, and the 150–500 bp (sometimes 150–1000bp) range was excised, purified, and sequenced by Illumina high-throughput sequencing. For Ago2-dependent studies, a total of 6 independent wild-type samples and 10 Ago2−/− samples were prepared and sequenced on one Illumina lane each. For 293S samples, a total of 7 independent samples were sequenced. For Drosha-dependent studies, two conditional knockout replicates were performed. Drosha flox/flox, CreER mouse embryonic stem cells were treated with 100nM 4-OH tamoxifen for 4–9 days, along with untreated controls. Each replicate sample was sequenced on 4–5 Illumina lanes. Sequencing data has been deposited to GEO/SRA, accession number GSE21975.
Bioinformatic analysis of sequenced libraries
We used the ENSEMBL transcripts as our transcriptomic reference (Birney et al., 2004). Reads were mapped with the RMAP mapping tool (Smith et al., 2009), allowing up to 2 mismatches. All reads were 36nt. Mapping was done to a reference that included a non-redundant set of all exonic sequence from the mm9 and hg18 assemblies from the UCSC Genome Browser (Kent et al., 2002), for mouse and human, respectively. A non-redundant junction reference was constructed from all pairs of exons within a gene; unique pairs of genomic positions for donor and acceptor sites defined junctions. If a read mapped inside an exon or junction corresponding to a given transcript, the read was assigned to that transcript (in this way reads may be assigned to multiple transcripts).
Based on the read counts at each site within each transcript, endonuclease sites were identified for a given biological replicate as follows. First, within each transcript, the read counts were fit (max likelihood) to a negative binomial (modeling over-dispersed counts data) assuming any sites with read counts of 0 were latent data. We assigned p-values to each given site based on the negative binomial fit for each transcript containing that site. Finally, for each sample we applied FDR (< 0.05) to the complete set of site p-values, and kept each site with a p-value lower than the cutoff.
Drosha-dependent sites were required to be identified as significant in both wild-type replicates, but in neither Drosha Cre-out replicate. The set of Drosha-independent sites was obtained using the above procedure, but using pooled reads from sequencing runs for multiple biological replicates.
Plotting of RACE data
Individual sample libraries were aligned to a database of Ensembl transcripts including 1000nt of 5′ and 3′ flanking sequence. Matching reads were filtered for unique, sense, Ensembl mRNA-matching reads. Read counts were normalized to the total of such reads. Normalized libraries from biological replicates were combined and averaged.
QPCR of transcript levels
Changes in transcript levels were detected using the Taqman RNA-to-Ct 1-step kit (Applied Biosystems) with 18S probes as the internal control.
Supplementary Material
Acknowledgments
The authors would like to thank Assaf Gordon and Oliver Tam for bioinformatic support. FVK is supported by a postdoctoral fellowship from the American Cancer Society, PF-07–058-01-GMC. This work was supported by grants from the NIH and by a kind gift from Kathryn W. Davis. GJH is a professor of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010 doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, Gingeras TR. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. Embo J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009a;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009b;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. Rna. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Shenoy A, Blelloch R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS One. 2009;4:e6971. doi: 10.1371/journal.pone.0006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. Embo J. 2000;19:295–305. doi: 10.1093/emboj/19.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.