Abstract
We report initial results from studies using SWeep Imaging with Fourier Transformation (SWIFT) to detect superparamagnetic iron oxide (SPIO) particle-labeled stem cells in the rat heart. In experiments performed on phantoms containing titanium balls or SPIO-labeled cells, frequency-shifted signals surrounding the paramagnetic objects produced a pile-up artifact in SWIFT images, while total signal intensity was retained to a much greater extent on SWIFT images as compared to gradient echo images. SWIFT images of excised and in vivo hearts showed (a) reduced blooming artifact as compared with gradient echo images, which helped reduce ambiguity in the detection of SPIO labeled cells, (b) enhancement of off-resonance signals relative to the background in the imaginary component of images, and (c) detailed myocardial anatomy in magnitude images which provided anatomical reference. These features suggest SWIFT can facilitate the detection of SPIO-labeled cells in the cardiovascular system.
Introduction
Labeling of stem cells with superparamagnetic iron-oxide (SPIO) particles (1) enables MRI tracking of such cells with high sensitivity and can under specific conditions be used for single cell detection (2,3). High sensitivity to detect SPIO-labeled cells results from the large susceptibility-induced gradients and consequently shortened T2* of the water molecules in the vicinity of the SPIOs.
The detection of SPIOs using T2*-weighted gradient-echo (GRE) sequences has a number of shortcomings. First, the size of the signal void in the image (due to short T2*) is much larger than the SPIO-containing cells, and thus, underlying tissue anatomy is obscured. Secondly, the signal voids arising from high concentrations of SPIOs are sometimes difficult to differentiate from the lack of spins or other intrinsic sources of T2* shortening, such as tissue/air boundary, hemorrhage, and calcification. Consequently, T2*-weighted GRE detection leads to “negative” contrast with low signal-to-noise ratio and poor specificity. A number of “positive” contrast methods have been developed based either on mapping T2* using post-processing methods or selectively exciting or refocusing water signals. Liu et al introduced a susceptibility-gradient mapping (SGM) method to obtain positive contrast from SPIO-labeled cells (4,5). Likewise, Mills et al described a post-processing algorithm, called phase map cross-correlation detection and quantification, to identify dipoles induced by SPIOs (6). Cunningham et al proposed to visualize these spins by exciting selectively the off-resonance spins surrounding the labeled cells (7). In the Inversion-Recovery with ON-resonant water suppression (IRON) method proposed by Stuber et al, the on-resonance water is saturated with a spectrally-selective pre-pulse to obtain signals from off-resonance protons in close proximity to the SPIO-labeled cells (8). Seppenwoolde (9) and Mani (10) proposed a “white marker” method in which a slice-selection gradient is utilized to cancel the local field gradient induced by the presence of SPIOs.
When using any of the above mentioned methods that excite or refocus only a fraction of the off-resonance spins, an additional anatomical reference image is usually required, which poses a limitation in applications like cardiovascular imaging since tissue motion is likely to introduce errors in the sequentially acquired images.
Here we examined a different approach to visualize SPIO-labeled cells grafted in the myocardium, by exploiting the positive contrast possible with the relatively new technique called SWeep Imaging with Fourier Transformation (SWIFT) (11). SWIFT is based on a frequency-swept excitation, with virtually simultaneous signal acquisition with the RF excitation; thus, it is particularly well suited to image objects with extremely fast transverse relaxation rates.
Here, the utility of SWIFT to image water signals around paramagnetic objects was studied using phantoms containing titanium balls and SPIO-labeled stem cells. In addition, ex vivo and in vivo images of rat hearts were obtained following injection of SPIO-labeled stem cells. The results show that the “blooming” susceptibility artifacts associated with GRE detection are suppressed by the use of SWIFT. Perhaps more interesting is the finding that SPIO-related signal can be visualized as positive contrast in the imaginary (Im) component of the SWIFT image, while the magnitude (Mag) image contains all the anatomical details of the heart. These results point to SWIFT as an alternative positive-contrast method for detecting paramagnetic objects, with the additional advantage of simultaneously providing a T1-weighted or proton-density-weighted image for anatomical reference.
Methods
Labeling of stem cells with SPIO
Murine embryonic stem cells (ESC) (16) were labeled with SPIO particles (Feridex®, Berlex Laboratories) by incubation in a labeling media containing 50 µg Fe/ml and 0.4 µg/ml of poly-L-Lysine (Sigma, St. Louis, MO) for 12–16 hours (overnight) using gelatin coated flasks, followed by extensive washing with PBS to remove uninternalized particles. Such labeling scheme leads to cell viability of 95 ± 5%, labeling efficiency of 86 ± 5%, and an iron content of 5.2 ± 0.7 (average ± stdev) picogram per cell estimated by inductive plasma mass spectroscopy (12).
Titanium ball phantom studies
To investigate the effects of large frequency shifts on GRE and SWIFT images, a phantom was constructed from seven plastic centrifuge tubes (50 ml), each containing agarose gelatin and an imbedded plastic mesh. Six of the tubes also contained one titanium ball (TiB) (Abbott Ball Company, West Hartford, CT) imbedded in the gelatin. The TiBs had different diameters (2.38, 3.18, 3.97 and 4.76 mm). The phantom was prepared by partially filling each tube with liquefied gelatin. After solidification, the TiBs and plastic mesh were added, and then the tubes were filled to the top with gelatin. The tube containing no TiB was positioned in the center of the outer six tubes. After taping the tubes together, they were placed in a 700 ml plastic jar filled with a 1% saline solution (Fig. 1A). 3D radial SWIFT and 3D Cartesian GRE images were acquired using a 4 T whole-body magnet interfaced to a Varian INOVA console. A TEM head coil was used for RF transmission and reception (13). 3D SWIFT acquisition was performed with a hyperbolic secant pulse (HS) (14) having a time-bandwidth product (R) of 256 and an excitation bandwidth (= acquisition bandwidth (sw)) of 62.5 kHz with oversampling factor of 32 (15). The signal was acquired in 256 gaps in the HS pulse, with each gap having a length of 10 µs. Data in k-space consisted of 98304 spokes (including positive and negative readout gradient directions) with 128 post-correlation complex points in each radial direction over a field-of-view (FOV) = 203 cm3. The flip angle was 4°, the repetition time (TR) was 5 ms, and no signal averaging was performed, to give a total scan time of ~8 min.
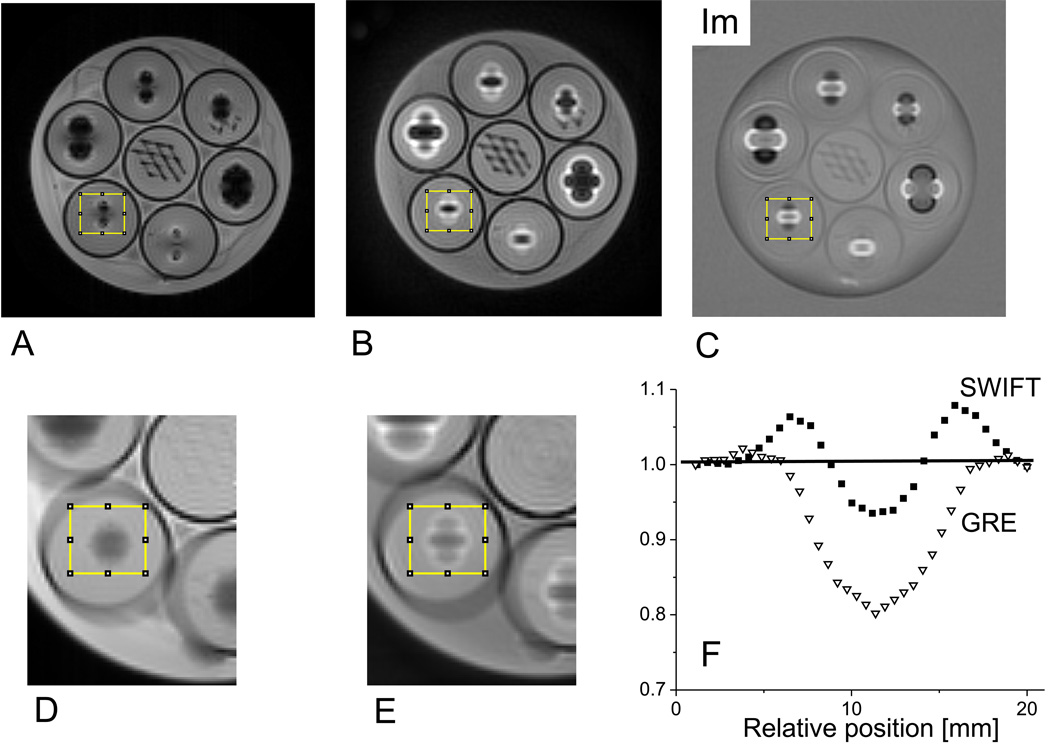
Figure 1.
Imaging studies of a phantom consisting of a plastic jar filled with saline and 7 tubes containing gelatin, titanium balls, and plastic mesh. The images are magnitude mode GRE (A), magnitude mode SWIFT (B), and imaginary mode SWIFT (C). The slice shown contains the titanium balls in the outer 6 tubes and the plastic mesh in the center tube. The balls are situated at slightly different heights; therefore, the slice shown does not cut through the center of every ball. The diameters of the titanium balls in the clockwise direction starting from the top are: 3.97, 2.38, 3.18, 4.76, 2.38 and 3.18 mm. The rims of the tubes are clearly identifiable from the black edge. The displaced and piled-up signals around the balls are clearly visible in the SWIFT images. Expanded views of the GRE and SWIFT images obtained by averaging 20 adjacent slices are also shown with the volume-of-interest (yellow box) from which the summed profiles (F) were obtained.
3D GRE images of the phantom were acquired using the following parameters: flip angle = 11°, TR = 13.5 ms, echo time (TE) = 4 ms, sw = 62.5 kHz, FOV = 203 cm3, and matrix size = 2563, which yielded a total scan time of ~15 min. The 3D data sets were analyzed using ImageJ (v1.38x, NIH, Bethesda, MD).
3D radial SWIFT data were processed using an in-house program developed in LabVIEW (National Instruments) and interpolated with a Kaiser-Bessel function onto a Cartesian grid. After gridding, a complex 3D FT was performed to convert k-space to image space. Image pixel intensity could be displayed as real (Re), imaginary (Im), magnitude (Mag), or phase images. In the case of on-resonance signals, phased k-space data have negligible phase distortion, in which case, Re= Mag and Im= 0. On the other hand, large magnetic susceptibility gradients produce off-resonance signals, in which case, Im ≠ 0.
Cell phantom studies
To investigate whether the concentrations of SPIO-labeled cells can be quantified from SWIFT images, two different phantoms containing SPIO-labeled ESCs were prepared and imaged. To construct the first phantom, which will be called the “uniform cell phantom”, cells were suspended in agarose solutions at concentrations of 0, 1 × 103, 5 × 103, 1 × 104, 2 × 104, and 5 × 104 cells/µl. These solutions were then placed in 5-mm NMR tubes (Optima, Elk Grove Village, IL) and allowed to solidify. A second phantom, referred to as the “cell pellet phantom”, was constructed to investigate the ability to quantify cells on the basis of (displaced) signal intensity in the imaginary image, after phase correcting the dataset. In this case, the NMR tubes were first filled with cell-free agarose solution, and then a well was formed by inserting the tip of a 200-µl pipette 2.5 cm into the solution during solidification. Different numbers of cells (0, 2 × 103, 1 × 104, 5 × 104, 2 × 105, and 1 × 106), each suspended in 20 µl of saline, were placed in each well. The cells were then spun down at 1000 rpm. After removing the supernatant, the NMR tubes were filled completely with agarose solution. This procedure resulted in cell pellets embedded in gelatin, with no needle track.
3D radial SWIFT and 3D Cartesian GRE images of the cell phantoms were acquired using a 9.4 T magnet interfaced to a Varian INOVA console. A single surface coil (diameter= 2 cm) was used for radiofrequency transmission and reception. The spatial resolution was 0.1 × 0.2 × 0.1 mm3 using FOV of 3 × 6 × 3 cm3 and matrix of 2563 in both SWIFT and GRE acquisitions. The flip angle was manually varied from 5– 20° in separate SWIFT acquisitions. A flip angle of 15° was used in GRE acquisitions, with TR = 11 ms and TE = 3 ms.
Cell phantom images were analyzed using ImageJ and a custom program written in MatLab (The Mathworks, Natick, MA). The dependency of image intensity on cell concentration in the uniform cell phantom was determined from region-of-interest (ROI) measurements on the magnitude (Mag) images. The average signal in the ROI was obtained from 30 adjacent axial slices. To minimize confounding effects from occasional cell clusters, ROIs were carefully selected to encompass the most homogeneous appearing regions in each tube. The dependency of image intensity on cell concentration in the cell pellet phantom was determined from the imaginary mode (Im) image after applying a phase correction procedure which involved two steps: 1) determining the average phase from a ROI placed in the gelatin tube which did not contain cells, and 2) subtracting this phase from the phase of the original SWIFT images. As a result, the imaginary image of the phase-corrected SWIFT dataset preserved only the off-resonance signal component. ROIs were contoured around the cell pellet region in each tube, and root of the summation of the square (RSOS) of the Im signal was taken from each ROI and summed over the slab of 15 axial slices (covering the cell pellet).
Injection of stem cells into the heart
All surgical and imaging procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Adult athymic rats (female, 150– 200 g) were purchased from Frederick Cancer Center (Frederick, MD). Intramyocardial injection of stem cells was performed as previously described (16). Briefly, under gaseous anesthesia (1– 2 % isoflurane mixed with oxygen), the rat was intubated and ventilated through a small- animal ventilator (Harvard Apparatus, MA). The tidal volume (1.5– 2.5 ml) was determined for each animal according to its respiratory rate and body weight. The body temperature was maintained by a heating lamp. The electrocardiogram (ECG) was monitored throughout the surgery. After the thoracic cavity and pericardium was opened, 5.0, 2.0, 0.5, or 0.2 million (M) cells suspended n 100 µl (for 5.0– 2.0 M) or in 50 µl (for 0.5- 0.2 M) serum- free culture media were injected in one spot into the mid- anterior wall of the left ventricle. An insulin syringe with a 29.5-gauge needle that is bent 45° at 3 mm from the tip was used for injection; the needle tip penetrated about 2 mm into the epicardium to deliver the cells. A total of five rats were injected. Two animals received 2.0 M cells, while the other three received 5.0, 0.5, or 0.2 M cells.
In vivo MRI detection of SPIO- labeled cells in the heart
A cardiac and respiratory gated GRE sequence (17) was used initially to visualize SPIO-labeled cells. Measurements were performed using a 4.7 T magnet interfaced to a Varian INOVA console. Volume coil transmission and surface coil reception were used as described previously (12). In-plane resolution was 1562 µm2 with slice thickness of 1 mm.
ECG-gated in vivo SWIFT images were acquired using a 9.4 T scanner (Varian Direct Drive). The animal was sedated by 1– 1.5% isoflurane mixed with oxygen and was placed prone on a surface coil (diameter = 4.8 cm). The ECG was monitored and core temperature was maintained at 37°C. The 3D SWIFT sequence used the follow parameters: TR = 2.5 ms, HS pulse flip angle = 10°, matrix = 2563, FOV = 7 × 7 × 14 cm3, sw = 125 kHz, acquisition gaps of 6 µs, and 32000 spokes (including positive and negative readout gradients) in k-space with 128 post-correlation complex points in each radial FID, which resulted in spatial resolution of 0.5 × 0.5 × 1 mm3. Four spokes were acquired after each R-wave trigger to capture the end-of-diastolic phase of the heart cycle, with no signal averaging, which resulted in a total scan time of 27.5 min.
For comparison with SWIFT, multi-slice 2D-GRE images were acquired while the animal remained sedated in the magnet using the following parameters: TR = 6 ms, flip angle = 30°, TE = 3.3 ms, sw = 50 kHz, matrix of 1282 (zero filled to 2562), FOV = 7 × 7 cm2, and slice thickness of 0.5 mm, to match the spatial resolution used in SWIFT. Twenty slices were acquired to cover the entire heart. Twelve signal averages were performed to yield a total scan time of 9 min.
Ex vivo MRI of excised heart
After performing in vivo imaging, the heart was harvested, perfused (retrograde) with 4% formalin, and then suspended in the Fomblin oil (Thorofare, NJ), a perfluorinated polyether, which provides a homogeneous media while yielding no proton signal. For ex vivo imaging, 3D SWIFT (radial) and GRE (Cartesian) images were acquired sequentially on the same heart using the 9.4 T INOVA system. A single-loop (1 cm diameter) surface coil was used for radiofrequency transmission and reception. Both SWIFT and GRE images were acquired with matched resolution of 983 µm3.
In the SWIFT sequence, excitation was performed with the HS pulse producing flip angle = 22°. Data in k-space consisted of 96000 spokes (including positive and negative readout gradient directions) with 128 complex points in each radial direction. Other parameters included: T R= 2.5 ms, sw = 125 kHz, and four signal averages, giving a total scan time of ~15 min.
In the GRE sequence, experimental parameters were: flip angle = 30°, TR = 13.5 ms, TE = 3 ms, sw = 80 kHz, with 256 complex points in each of 65536 (256 × 256) phase encode steps. No signal averaging was used to match the scan time of SWIFT (~15 min.).
Histology and Prussian blue staining for iron oxide
After performing ex vivo imaging, the heart was embedded in paraffin and cut into 5 µm sections. The position of the SPIO-labeled cells was obtained from in vivo or ex vivo images, and sections from the corresponding slab of tissue were incubated with Pearl’s reagent at room temperature for 30 minutes in the dark, and cell nuclei were counterstained with 4',6- diamidino-2-phenylindole (DAPI, Molecular probe, Eugene, OR).
Results
Experiments conducted on the TiB phantom revealed how large magnetic susceptibility variations affect SWIFT and GRE images differently. In both types of images, the TiBs could be visualized indirectly via the large dipole field effect they produced (Fig. 1). With the GRE sequence, extensive signal loss occurred in regions surrounding the TiBs (Fig. 1A), whereas with SWIFT, frequency-shifted resonances near the TiBs yielded measurable signal that was displaced and consequently resulted in a “pileup” artifact in both the Mag image (Fig. 1B) and the Im image (Fig. 1C). It is apparent that the net signal contained in the proximity of a given TiB is much greater in the SWIFT image than in the GRE image. Hence, it appears that signals can be retained with SWIFT despite having extremely short T2* values. The difference in net signal intensity with GRE versus SWIFT was quantified in a volume-of-interest (VOI) encompassing the entire distorted signal region around one of the TiBs. In the analysis, the VOI was placed identically in the GRE and SWIFT images and contained 20 adjacent slices (Figs. 1D, E). Horizontal projections of the VOI (obtained by averaging pixels in the vertical direction, with normalization to the undistorted signal intensity close to the edges of the VOI), are shown for GRE (triangles) and SWIFT (squares) in Fig. 1F. The solid line connecting the intensities at the edges of the VOI represents approximately the level of the undistorted signal. It can be seen that, in the VOI where large local field variations were present, the GRE method yielded a net signal loss when the signals below and above the solid line are summed up. In contrast, SWIFT did not show such loss; instead, it attained a slight gain in integrated signal possibly due to T1 effect of water spins near the ball.
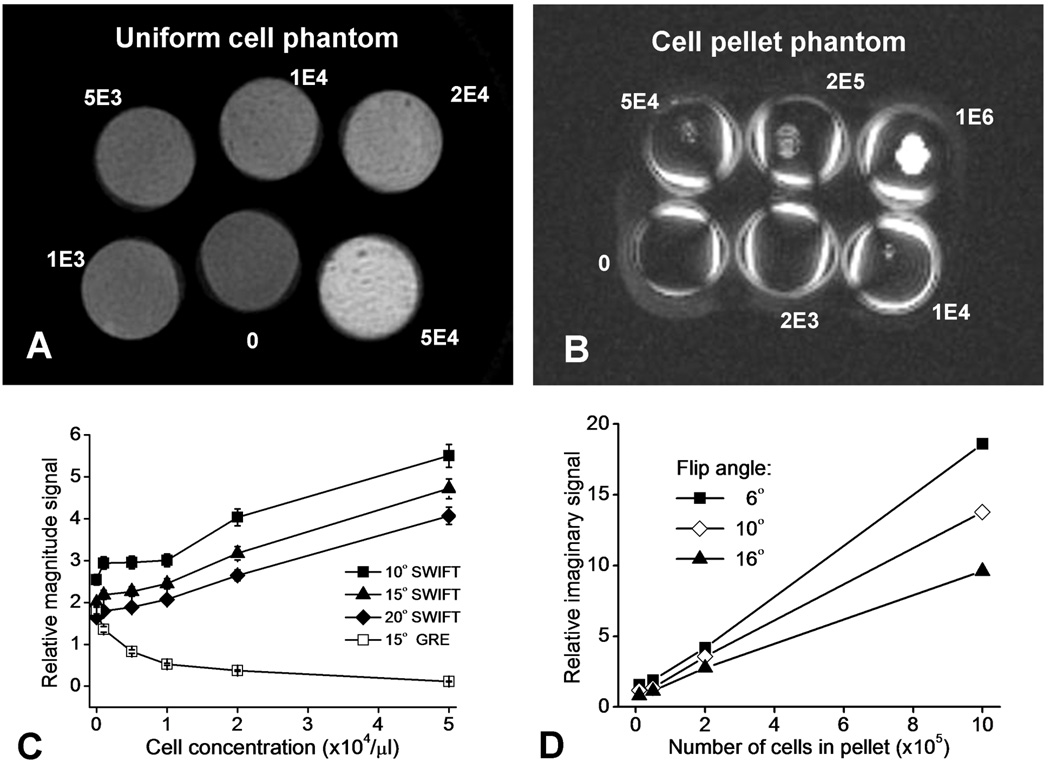
The experiments performed on the SPIO-labeled cells in gelatin provided clear evidence that signal intensity in SWIFT images is proportional to cell concentration, for cells that are clustered (in the pellet) or uniformly distributed. SWIFT Mag images of the uniform cell phantom are shown in Fig. 2A, and the plot in Fig. 2C displays the signal magnitude as a function of cell concentration, for both SWIFT and GRE images. It can be seen that the signal intensity in the GRE Mag images decreased approximately exponentially as a function of cell concentration, whereas the signal intensity in the SWIFT Mag images increased proportionally with cell concentration. As the flip angle increased, a linear relationship between SWIFT Mag intensity and cell concentration became more apparent, most likely due to increased T1 weighting. A direct relationship between signal intensity and cell concentration was also observed with the cell pellet phantom, both in Mag (not shown) and Im images (Fig. 2B). The Im images contain only off-resonance signals resulting from a significant magnetic susceptibility effect. Thus, air interfaces at the tube surfaces, as well as SPIO-labeled cells, are visible in the Im images. As can be seen from the plot (Fig. 2D), the relationship between SWIFT Im signal intensity versus cell number is highly linear, when measured in ROIs surrounding the cell pellets. Interestingly, the slope of the linear function decreases as the flip angle increases. Such an inverse relationship between off-resonance signal intensity and flip angle might be explained by increasing T1 saturation as the flip angle increased. With the choice of acquisition parameters used in these SWIFT experiments, the pellet containing the lowest number of cells (2000) was below the detection threshold.
Figure 2.
Results from the cell phantom studies. The SWIFT magnitude mode image of the uniformly distributed cell phantom (A) and the maximal intensity projection of the phase-corrected SWIFT imaginary mode image of the cell pellet phantom (B) are shown. The signal magnitude as a function of cell concentration in the uniform cell phantom is plotted, for GRE and SWIFT acquisitions using different flip angles (C). The SWIFT imaginary mode signal (root of the summation of the square, RSOS) as a function of number of cells in the cell pellet phantom is plotted, for SWIFT acquisitions using different flip angles (D). The error bars in C represent standard deviation of the measured signal intensity in 30 ROIs.
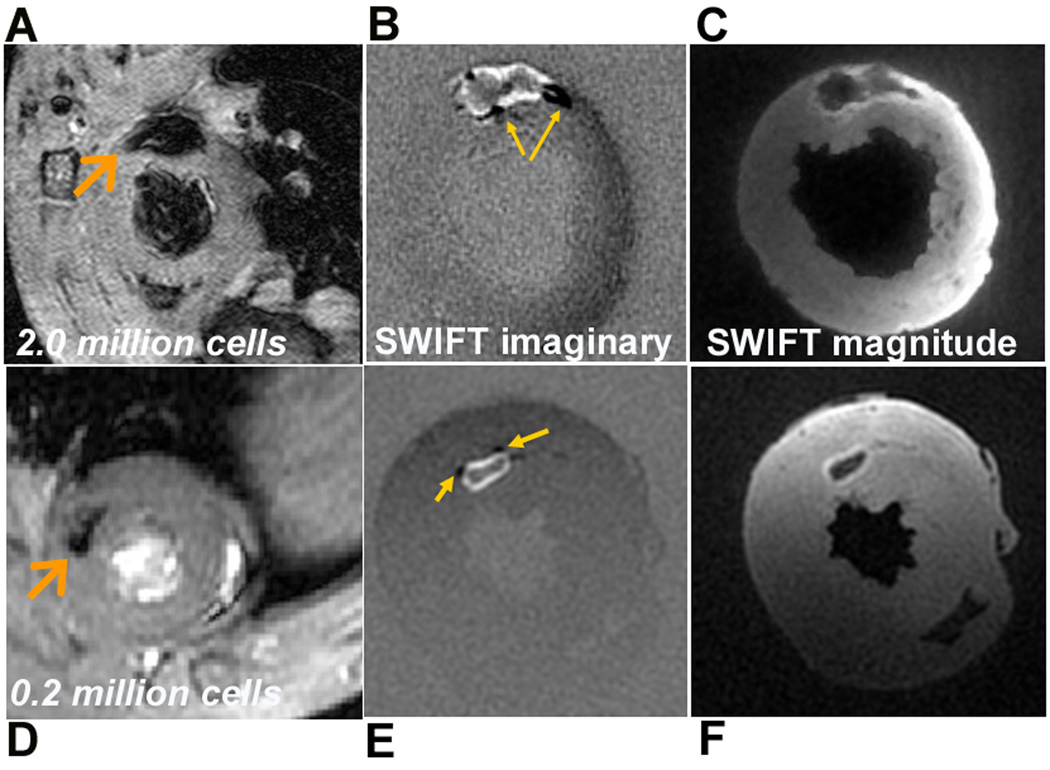
In Fig. 3, ex vivo 3D SWIFT images of the hearts receiving 2.0 and 0.2 million cells are displayed in Mag (Figs. 3B, E) and Im images (Figs. 3C, F), along with the in vivo GRE images (Figs. 3A, D). The in vivo GRE images show the typical “blooming” signal voids associated with magnetic field susceptibility induced by the presence of SPIO-labeled cells. The signal voids are reduced substantially in the SWIFT images displaying different contrasts. In the Im images, positive signals associated with SPIO-labeled cells are intense and shifted from the center to form a bright rim (“positive contrast”), intermingled with areas of negative signal (indicated by arrows on Figs. 3B and E) relative to background. Since both positive and negative SWIFT Im signal originates from off-resonance spins (frequency-shifted due to the presence of SPIO-labeled cells), the negative Im signal does not represent a signal void. Therefore both positive and negative Im signals can be displayed as positive contrast while the on- resonance spins are close to invisible in the Im images. In addition, due to the extremely short acquisition delay, other moderate signal losses due to T2* effects are not visible in the Im images, which facilitates unambiguous detection of SPIO- labeled cells.
Figure 3.
In vivo GRE detection and ex vivo SWIFT detection of SPIO-labeled ESCs in the heart. Short axis GRE images and SWIFT imaginary and magnitude images obtained from the heart receiving 2.0 million (A-C) and 0.2 million (D-F) cells. Arrows in A and D point to locations of the stem cells, while those in B and E point to regions where negative contrast (compared to the background) was generated by some of the off-resonance spins.
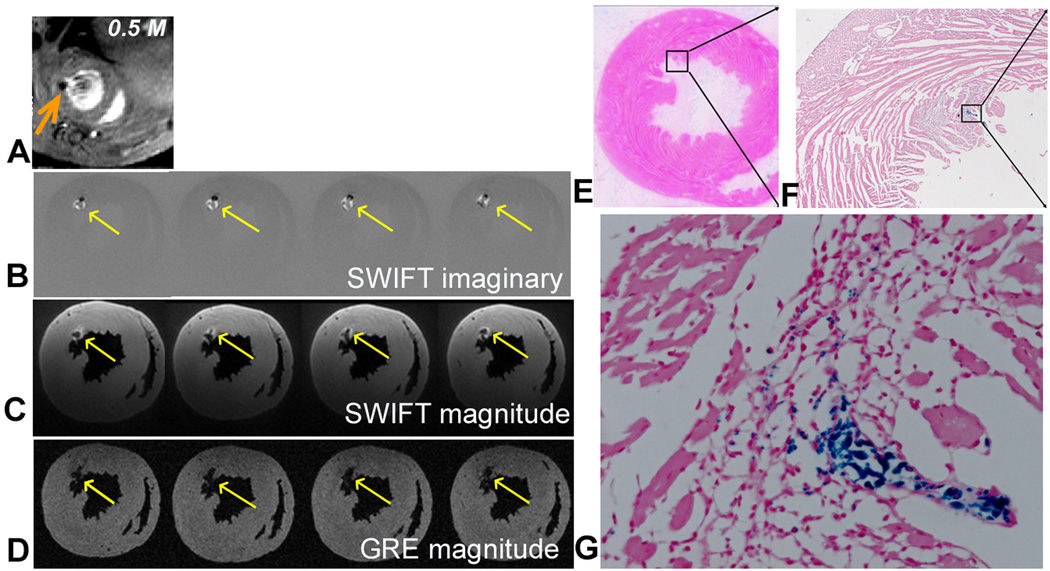
The utility of SWIFT for the detection of SPIOs is further demonstrated in the heart that received 0.5 million stem cells (Fig. 4). As shown in the in vivo GRE image (Fig. 4A), the stem cells are located very close to the endocardial border. Therefore, the signal void produced with the GRE sequence could easily be mistaken for ventricular lumen as demonstrated in the ex vivo images (Fig. 4D). However, this region was visualized conspicuously in the corresponding SWIFT Im images (Fig. 4B, arrows) as a positive rim with dark spots; all the myocardial anatomies were presented in the Mag images (Fig. 4C). Prussian blue staining was performed to confirm that a cluster of SPIO-labeled cells was indeed present close to the endocardial border (Fig. 4E- G). While the thickness of the histological sections (5 µm) was only a fraction of the 3D SWIFT resolution (nominal 98 µm, isotropic resolution), the Prussian blue positive cells are clearly visible and resemble the morphology of embryonic stem cells with small cell size and a large ratio between nucleus and cytoplasm.
Figure 4.
In vivo GRE image (A), ex vivo SWIFT imaginary (B), magnitude (C), and ex vivo GRE magnitude (D) images obtained from a heart receiving 0.5M labeled stem cells. Arrows indicate location of cells. E-G: Prussian blue staining of the heart sections at the corresponding level of MR images. Magnified views of the box in E and F are presented in F and G, respectively.
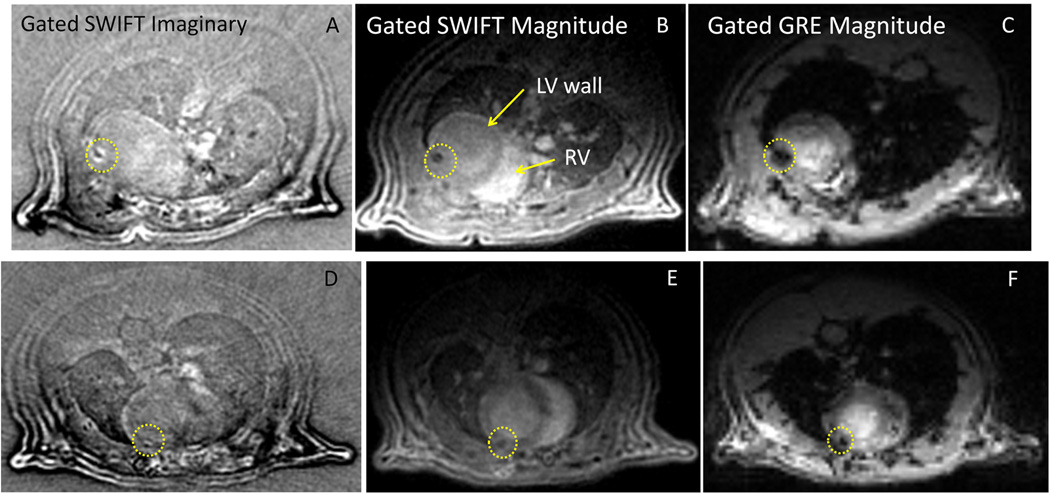
Finally, Fig. 5 shows in vivo ECG-gated SWIFT and GRE images of animals that received 2 and 5 M stem cells. It can be seen that ECG-gated SWIFT acquisition produced end-diastolic images of excellent quality. In SWIFT Im (Fig. 5A, D) and Mag images (Fig. 5B, E), myocardial wall, left and right ventricular lumen, major vessels in the chest, and lung parenchyma are well delineated. SPIO-containing cells were visualized as a hypointense region on the Mag image, which appears more focused compared to the hypointense region on the GRE image (Fig. 5C, F). Correspondingly, in the Im images, the signals related to SPIO-labeled cells are shifted to form a bright rim (positive contrast). Both positive and negative SWIFT Im signal originates from off- resonance spins due to the presence of SPIO-labeled cells. Large frequency shifts at tissue boundaries also gave rise to Im signals that did not significantly interfere with the ability to detect SPIO-labeled cells in the heart. In addition, because the Mag and Im images are perfectly registered (generated from the same data set), SPIO spots can be easily separated from other off-resonance signals by reading both images. Accordingly, the Mag image serves as the reference image, without potential errors introduced by separate acquisition of the positive contrast image and the reference image. In addition, as compared with the GRE image, the myocardium wall appears homogenous in the SWIFT image, and thus, reduces ambiguity in the detection of SPIO-labeled cells. While SWIFT images were acquired only during end-of-diastole (see Methods), GRE images were acquired over the entire cardiac cycle in order to reduce the scanning time. Therefore, Figs. 5C, F do not show diastolic images, although the anatomical location is similar to that shown in the SWIFT images.
Figure 5.
ECG-gated in vivo SWIFT and GRE images of two rats receiving 5 million (A-C) and 2 million (D-F) SPIO-labeled stem cells. SPIO-associated signals are marked by a yellow circle on the SWIFT imaginary (A, D) and magnitude images (B, E) and GRE magnitude images (C, F). While SWIFT images were acquired only during end-of-diastole (see Methods), GRE images were acquired over the entire cardiac cycle in order to reduce the scanning time. Therefore, panels C and F are not diastolic images, but they represent similar anatomical location.
Discussion
Broadband frequency-swept excitation and extremely short acquisition delay are unique features of SWIFT which make it particularly well suited for imaging frequency-shifted resonances and for minimizing signal loss from T2*-induced dephasing. The described experiments on the titanium-ball phantom were designed to demonstrate these advantages and to help explain the contrast observed in ex vivo and in vivo images of stem cells in myocardium. Interestingly and unexpectedly, a small amount of net signal increase was observed when the signal intensity was integrated over a region encompassing the titanium ball. Although this phenomenon remains unexplained at this time, it is conceivable that some of the signal increase occurs as a result of T1-shortening of water spins at the surface of the titanium ball.
The mechanisms by which magnetic susceptibility variations produce signal voids in 2D and 3D Cartesian sampled GRE images have been discussed in detail in the literature (18,19). The appearance of the void detected by radial sequences is less well described, but is similar. The cause of any overall signal loss is the finite delay from excitation to acquistion. In a GRE sequence, this finite delay is TE, the time elapsed between the center of the excitation pulse and acquisition of the zero k-space point. In SWIFT, the finite delay is the time between the center of a pulse element and the acquisition in the gap (= 7 µs in the experiments described herein). By sampling data so quickly after each pulse element (as shown in the pulse sequence diagram found in (11)), SWIFT retains the signal of fast relaxing spins. Furthermore, the short acquisition delay of SWIFT makes it highly insensitive to motion.
SWIFT is sensitive to all spins having transverse relaxation times larger than the reciprocal value of the spectral width, T2 > 2π/sw (11), i.e., T2 > 5 µs using the current sw value of 125 kHz. As compared to the ultra-short TE method (UTE) (20), SWIFT more effectively captures and resolves signals from fast relaxing spins, since frequency encoding commences almost immediately after excitation, with no time used for gradient ramping (11). As compared to GRE-based techniques, in the presence of SPIO particles the signal loss is considerably minimized with SWIFT. Instead, the off-resonance signal is displaced over a few voxels and "piles up" in magnitude and imaginary mode images. In this case, the positive and negative signal in the imaginary component of the SWIFT image facilitates the localization of SPIO particles. It is anticipated that, because almost all signals are preserved, the displacement of signal intensity can be corrected with the appropriate field map leading to a positive contrast visualization of SPIO-related signals. Initial results using an iterative refocusing method suggest that the displaced and piled-up signals can be refocused, resulting in postive contrast in the injection site (21).
The experiments performed on the cell phantoms demonstrated the expected dependence of SWIFT signal intensity on SPIO-labelled cell concentration. The experiments also revealed interesting dependencies on flip angle, both in Mag and Im images. Although the flip angle dependencies might be explained by changes in T1-weighting, further experiments will be necessary to obtain definitive evidence to fully explain these dependencies. Also, as compared with GRE imaging, SWIFT has diminished sensitivity to detect the lowest concentrations of SPIO-labeled cells. A detailed theoretical analysis of how paramagnetic objects affect SWIFT signal intensity, including its dependency on acquistion parameters (e.g., flip angle, TR, and sw), is currently underway and will be presented separately. In the future, it may be possible to increase the sensitivity of SWIFT to detect low concentrations of small paramagnetic objects by using optimal acquisition parameters.
The results from ex vivo studies on rat heart demonstrate that over a wide range of biologically relevant doses of SPIO-labeled stem cells (0.2– 5.0 million), SWIFT Im images can provide superb contrast for detecting SPIO-related signals, whereas the Mag images can provide an anatomical reference. Here, the first ECG-gated SWIFT cardiac images have been presented. These data demonstrate the feasibility of in vivo SWIFT detection of SPIO-labeled cells in the heart. Off-resoance signals associated with SPIO appear in the imaginary component images and can be readily registered on the magnitude images, which clearly reveal myocardial anatomy; thus, acquisition of a separate reference image is not required. Taken together, our data suggest that SWIFT offers an alternative to currently available positive contrast methods (3–6), with potential advantages for cardiovascular applications ranging from stem cell tracking to detection of vulnerable atheroplaques that take up paramagnetic imaging probes. Current effort is focused on implementing ECG and respiratory self-gating strategy (22) to achieve cardiac cine SWIFT.
Acknowledgement
This study has been supported by NIH grants R01- HL81185 and P41- RR008079, and the W.M. Keck Foundation. We thank Dr. Bin Huang for surgical assistance in grafting the stem cells in the hearts, Steady State Imaging, LLC for supplying a SWIFT compatible coil and Ti ball bearings, and Penn Small Animal Imaging Facility (SAIF). In addition we acknowledge and thank Dr. Ryan Chamberlain for development and support of ifort gridding software, and Mr. Brian Hannah for support of high performance Linux servers and remote access.
Footnotes
Part of this study was presented at the 17th Scientific Meeting of the ISMRM (April 18– 24, 2009, Honolulu, Hawaii).
Disclosure: Dr. Garwood has an equity interest in SSI and is entitled to sales royalty through the University of Minnesota for products related the research described in this paper. These relationships have been reviewed and managed by the University of Minnesota in accordance with its conflict of interest policies.
References
- 1.Bulte JW, Duncan ID, Frank JA. In vivo magnetic resonance tracking of magnetically labeled cells after transplantation. J Cereb Blood Flow Metab. 2002;22(8):899–907. doi: 10.1097/00004647-200208000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Heyn C, Ronald JA, Mackenzie LT, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55(1):23–29. doi: 10.1002/mrm.20747. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proc Natl Acad Sci USA. 2004;101(30):10901–10906. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Dahnke H, Jordan EK, Schaeffter T, Frank JA. In vivo MRI using positive-contrast techniques in detection of cells labeled with superparamagnetic iron oxide nanoparticles. NMR Biomed. 2008;21(3):242–250. doi: 10.1002/nbm.1187. [DOI] [PubMed] [Google Scholar]
- 5.Dahnke H, Liu W, Herzka D, Frank JA, Schaeffter T. Susceptibility gradient mapping (SGM): a new postprocessing method for positive contrast generation applied to superparamagnetic iron oxide particle (SPIO)-labeled cells. Magn Reson Med. 2008;60(3):595–603. doi: 10.1002/mrm.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills PH, Wu YJ, Ho C, Ahrens ET. Sensitive and automated detection of iron-oxide-labeled cells using phase image cross-correlation analysis. Magn Reson Imaging. 2008;26(5):618–628. doi: 10.1016/j.mri.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham CH, Arai T, Yang PC, McConnell MV, Pauly JM, Conolly SM. Positive contrast magnetic resonance imaging of cells labeled with magnetic nanoparticles. Magn Reson Med. 2005;53(5):999–1005. doi: 10.1002/mrm.20477. [DOI] [PubMed] [Google Scholar]
- 8.Stuber M, Gilson WD, Schar M, Kedziorek DA, Hofmann LV, Shah S, Vonken EJ, Bulte JW, Kraitchman DL. Positive contrast visualization of iron oxide-labeled stem cells using inversion-recovery with ON-resonant water suppression (IRON) Magn Reson Med. 2007;58(5):1072–1077. doi: 10.1002/mrm.21399. [DOI] [PubMed] [Google Scholar]
- 9.Seppenwoolde JH, Viergever MA, Bakker CJ. Passive tracking exploiting local signal conservation: the white marker phenomenon. Magn Reson Med. 2003;50(4):784–790. doi: 10.1002/mrm.10574. [DOI] [PubMed] [Google Scholar]
- 10.Mani V, Adler E, Briley-Saebo KC, Bystrup A, Fuster V, Keller G, Fayad ZA. Serial in vivo positive contrast MRI of iron oxide-labeled embryonic stem cell-derived cardiac precursor cells in a mouse model of myocardial infarction. Magn Reson Med. 2008;60(1):73–81. doi: 10.1002/mrm.21642. [DOI] [PubMed] [Google Scholar]
- 11.Idiyatullin D, Corum C, Park JY, Garwood M. Fast and quiet MRI using a swept radiofrequency. J Magn Reson. 2006;181(2):342–349. doi: 10.1016/j.jmr.2006.05.014. Epub 2006 Jun 2019. [DOI] [PubMed] [Google Scholar]
- 12.Qiao H, Zhang H, Zheng Y, Ponde DE, Shen D, Gao F, Bakken AB, Schmitz A, Kung HF, Ferrari VA, Zhou R. Embryonic Stem Cell Grafting in Normal and Infarcted Myocardium: Serial Assessment with MR Imaging and PET Dual Detection. Radiology. 2009;250(3):821–829. doi: 10.1148/radiol.2503080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan JT, Hetherington HP, Otu JO, Pan JW, Pohost GM. High frequency volume coils for clinical NMR imaging and spectroscopy. Magn Reson Med. 1994;32(2):206–218. doi: 10.1002/mrm.1910320209. [DOI] [PubMed] [Google Scholar]
- 14.Silver M, Joseph R, Hoult D. Highly selective π/2 and π pulse generation. J Magn Reson. 1984;59(2):347–351. [Google Scholar]
- 15.Idiyatullin D, Corum C, Moeller S, Garwood M. Gapped pulses for frequency-swept MRI. J Magn Reson. 2008;193(2):267–273. doi: 10.1016/j.jmr.2008.05.009. Epub 2008 May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiao H, Surti S, Choi SR, Raju K, Zhang H, Ponde DE, Kung HF, Karp JS, Zhou R. Death and proliferation time course of stem cells transplanted in the myocardium. Molecular Imaging and Biology. 2009;11(6):408–414. doi: 10.1007/s11307-009-0222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou R, Pickup S, Glickson JD, Scott C, Ferrari VA. Assessment of Global and Regional Myocardial Function in the Mouse Using Cine- and Tagged MRI. Magn Resone Med. 2003;49:760–764. doi: 10.1002/mrm.10423. [DOI] [PubMed] [Google Scholar]
- 18.Posse S, Aue WP. Susceptibility artifacts in spin-echo and gradient-echo imaging. J Magn Reson. 1990;88(3):473–492. [Google Scholar]
- 19.Kim J, Kucharczyk W, Henkelman R. Cavernous hemangiomas: dipolar susceptibility artifacts at MR imaging. Radiology. 1993;187(3):735–741. doi: 10.1148/radiology.187.3.8497623. [DOI] [PubMed] [Google Scholar]
- 20.Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. J Comput Assist Tomogr. 2003;27(6):825–846. doi: 10.1097/00004728-200311000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Moeller S, Idiyatullin D, Corum C, Zhou R, Garwood M. Iterative reconstruction for off-resonance effects in SWIFT imaging. Proceedings of ISMRM. 2009;17:2864. [Google Scholar]
- 22.Hiba B, Richard N, Janier M, Croisille P. Cardiac and respiratory double self-gated cine MRI in the mouse at 7 T. Magn Reson Med. 2006;55(3):506–513. doi: 10.1002/mrm.20815. [DOI] [PubMed] [Google Scholar]